Abstract
Bulb rot is one of the most destructive diseases of tulip (Tulipa gesneriana L.). In November 2022, rotten tulip bulbs and terminal buds were found in Songjiang District, Shanghai, China. Fungal isolates were isolated from the rotten bulbs and identified as Fusarium based on colony morphology and ITS sequences. Further analyses of tef1, rpb1, and rpb2 barcoding sequences and conidial micromorphology identified the Fusarium isolates as F. annulatum. The pathogenicity of the F. annulatum isolates was verified with Koch’s postulates. This is the first report of F. annulatum causing bulb rot disease of tulip.
1. Introduction
Tulip (Tulipa gesneriana L.), belonging to the family Liliaceae, has a wide array of flower colors and shapes. Used as cut flowers, pot plants, and landscape plants, tulip is one of the most important bulbous crops worldwide due to its ornamental value and economic value. Many diseases threaten tulip production. Bulb rot is one of the most destructive diseases of tulip, accounting for 20–30% annual loss of tulip bulbs in storage and field worldwide [1]. Fungal pathogens belonging to genera Fusarium, Botrytis, Penicillium, Pythium, Rhizoctonia, Aspergillus, and Rhizopus [1,2] and bacterial pathogen Pectobacterium [3] have been reported to be associated with the bulb rot of tulip.
Fusarium oxysporum f. sp. tulipae is the most harmful and studied fungal pathogen of the basal rot disease of tulip, causing direct bulb loss during cultivation and storage and a much larger problem by producing a large amount of ethylene. This leads to flower abortion, uneven and stunted growth, reduced rooting, and gummosis, in addition to the negative effects on non-infected bulbs during storage and shipment [4]. Formae speciales or special form is an informal subspecific rank in F. oxysporum classification based on plant pathogenicity and host specificity [5,6]. During the last three decades, multigene phylogeny based on multiple DNA barcoding genes played a critical role in Fusarium taxonomy subject to the International Code of Nomenclature for algae, fungi, and plants (https://www.iapt-taxon.org/nomen/main.php) (accessed on 1 May 2025) [6,7,8,9]. Lombard et al. designated the epitype for F. oxysporum to resolve the taxonomy of the Fusarium oxysporum species complex and proposed multiple cryptic species as species [6]. Yet F. oxysporum f. sp. tulipae has not been classified to a species subject to the International Code of Nomenclature for algae, fungi, and plants.
In China, F. oxysporum f. sp. tulipae was first isolated from tulip bulbs in Xi’an botanical garden in Xi’an City, Shaanxi Province, and identified by macro- and micro-morphological analyses in 1997 [10]. Meanwhile, F. moniliforme was also isolated from tulip bulbs in Xi’an botanical garden and identified by morphology [10]. The name F. moniliforme was later replaced by F. verticillioides [11,12]. In 2010, Chang et al. reported that F. proliferatum causing bulb wilt of tulip was isolated from tulip bulbs in Xi’an botanical garden and identified by morphological and ITS sequence analyses [13]. In 2021, Yilmaz et al. designated the epitypes of F. verticillioides and F. proliferatum within the Fusarium fujikuroi species complex and resolved the taxonomy of F. verticillioides and F. proliferatum [9]. Yet the Fusarium pathogens associated with the bulb rot disease of tulip in China have not been clarified to species subject to the International Code of Nomenclature for algae, fungi, and plants and under a modern taxonomic framework using multigene phylogeny [6,9,14].
In November 2022, rotten tulip bulbs and terminal buds were found in a flower and plant nursery base in Shanghai. To ascertain the causal agent of the suspected bulb rot of tulip, we isolated fungal pathogens and identified the pathogens by macroscopic and microscopic observations, multiple DNA barcoding sequence analyses, and pathogenicity tests.
2. Materials and Methods
2.1. Isolation of Fungi
On 23 November in 2022, rotten tulip bulbs and terminal buds were found in the Wuku base of a flower and plant nursery (121.128042 E, 30.941657 N) in the Songjiang District of Shanghai. Ten rotten tulip bulbs and terminal buds (Figure 1) were used to isolate the pathogens. After removing the tunics, the bulbs were surface sterilized with 75% ethanol. Pieces of bulb scales (5 mm × 5 mm) containing healthy and diseased tissues were cut off and transferred onto potato dextrose agar (PDA) containing ampicillin (100 mg/L) and streptomycin (100 mg/L) and then incubated at 25 °C in the dark. After fungal mycelia grew and covered about 40 mm of the PDA, the leading edges of the fungal mycelia were picked with the tip of a sterilized needle, transferred onto fresh PDA plates, and incubated at 25 °C in the dark. This process was repeated three times to obtain purified fungal colonies.
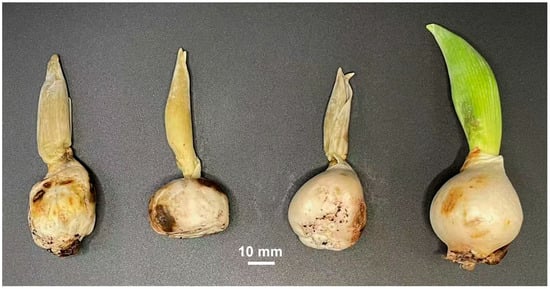
Figure 1.
Rotten tulip bulbs and terminal buds found in the Songjiang District of Shanghai.
2.2. Amplification and Analysis of Fungal DNA Barcoding Sequences
Genomic DNA was extracted from mycelia of fungal isolates grown on PDA using a FastPure Plant DNA Isolation Mini Kit (Vazyme, Nanjing, China). DNA barcoding sequences of the internal transcribed spacer (ITS), translation elongation factor 1-α gene (tef1), and the largest subunit and second largest subunit of the RNA polymerase genes (rpb1 and rpb2) were amplified using the primers (Table 1) and protocols described by Wang et al. [8]. The amplicons were sequenced using the Sanger method by Beijing Tsingke Biotech Co., Ltd. (Hangzhou, China). The ITS and tef1 amplicon sequences were analyzed after removing the primer regions.

Table 1.
Primers for amplification of fungal ITS, tef1, rpb1, and rpb2 barcoding sequences.
The sequences of tef1, rpb1, or rpb2 from the isolates YJX-0509 and YJX-0511, the representative strains including type, epitype, or neotype strains of the closely related species within the Fusarium fujikuroi species complex, and the outgroup F. solani strain NRRL 43468 (Table S1) were aligned using the MUSCLE program integrated in the MEGA software version 5.2.2 [19]. After eliminating positions containing gaps and missing nucleotides at both ends of the aligned sequences, the partial tef1, rpb1, and rpb2 sequences were concatenated; the final 3704 aligned nucleotides were constructed to a phylogenetic maximum-likelihood tree using IQ-TREE multicore version 1.6.12 [20] with the TIM2e+I+G4 model.
2.3. Microscopy
Fungal conidia for microscopy were generated on PDA and water agar. Conidia and other microstructures of fungal isolates were examined using an Eclipse 80i microscope and recorded by a Nikon digital camera with the NIS-Elements F3.0 image system (Nikon, Tokyo, Japan).
2.4. Pathogenicity Test of Fungal Isolates
Pathogenicity of fungal isolates were tested on tulip bulbs by inoculating fresh mycelia of isolates YJX-0509 and YJX-0511. After removing tunics, tulip bulbs were surface-sterilized and wounded with a cross scratch (5 mm) by a sterilized scalpel; fresh marginal mycelia on PDA were punched and transferred onto the cross scratches on the bulbs; sterile round PDA blocks without fungi were used as mock inoculation. The bulbs were then incubated in plastic pots under a transparent plastic cover in a gnotobiotic condition in a growth chamber with a setting of 25 °C, 75% relative humidity, and 12 h light and 12 h dark photoperiod. Disease severity index after inoculation was scored using the following symptom severity scale of 0–4: 0 = no symptom, smooth surface and uniform color of the bulb, healthy internal tissue; 1 = rot restricted to the site of inoculation, slight surface depression or discoloration, local browning of internal tissue; 2 = rot extending from the site of inoculation, obvious brown or rot area ≤ 25%, partial necrosis of internal tissues; 3 = rot area 26–50% accompanied by mold covering, half necrosis of internal tissues; 4 = complete rot, complete necrosis of internal tissues.
3. Results
3.1. Colony Morphology of Fungal Isolates Resembling Fusarium
Six fungal isolates (YJX-0509, YJX-0510, YJX-0511, YJX-0513, YJX-0514, and YJX-0515) were isolated from the tulip bulbs showing rot symptoms. Their colonies on PDA showed cottony aerial mycelia with white filamentous margin on the surface; three isolates (YJX-0509, YJX-0510, and YJX-0514) produced violet pigments from the surface view (Figure 2). The reverse sides of about half of the colonies (YJX-0510, YJX-0511, and YJX-0514) on PDA showed annual-ring-like patterns (Figure 2). The pigment production and annual-ring-like pattern were variable for the subcultures of the same isolate under the same conditions.
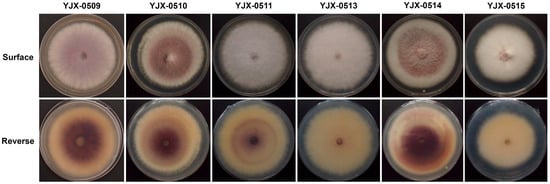
Figure 2.
Colony morphology of fungal isolates from tulip bulbs on potato dextrose agar (diameter 90 mm) at 25 °C in the dark for 7 d.
3.2. Identification of Fungal Isolates to Fusarium annulatum by DNA Barcoding Sequence Analyses
The ITS sequences (519 bp) from the six fungal isolates are identical; their tef1 sequences (670 bp) are also identical. Their ITS sequences show the closest sequence identities (>99%) to those of strains belonging to the Fusarium fujikuroi species complex. Their tef1 sequences show the highest sequence identities (98.6–99.7%) and the closest phylogeny to F. annulatum (Figure S1). The six Fusarium isolates probably belong to the same species, F. annulatum.
The isolate YJX-0509, representing the violet pigment-producing isolates (YJX-0509, YJX-0510, and YJX0514), and the isolate YJX-0511, representing the isolates with pale-surfaced colonies (YJX-0511, YJX-0513, and YJX-0515) (Figure 2), were used to amplify rpb1 and rpb2 sequences for species identification. PCR amplicons about 1900 bp were amplified from isolates YJX-0509 and YJX-0511 using the rpb1 primer pairs RPB1-Fa/RPB1-G2R. The amplicon sequencing obtained rpb1 sequences of 1780 bp without sequences of both primers and downstream near the primers; the resulting rpb1 sequences of isolates YJX-0509 and YJX-0511 (GenBank accession no. PQ787493 and PQ787494) are identical and show sequence identity of 99.9% (1778/1780 bp) to the rpb1 sequence of the F. annulatum-type strain CBS 258.54T. PCR amplicons about 2000 bp were amplified from isolates YJX-0509 and YJX-0511 using the rpb2 primer pairs RPB2-5f2/RPB2-11ar. The amplicon sequencing obtained rpb2 sequences of 1889 bp without sequences of both primers and downstream near the primers; the resulted rpb2 sequences of isolates YJX-0509 and YJX-0511 (GenBank accession no. PQ791804 and PQ791805) are also identical and show sequence identity of 99.5% (1880/1889 bp) to the rpb2 sequence of the F. annulatum type strain CBS 258.54T.
On the phylogenetic tree based on the concatenated tef1, rpb1, and rpb2 sequences, the isolates YJX-0509 and YJX-0511 are clustered together with F. annulatum strains (Figure 3). The DNA barcoding sequence analyses classified the Fusarium isolates from tulip bulbs as F. annulatum.

Figure 3.
Phylogenetic maximum-likelihood tree of Fusarium fujikuroi species complex based on concatenated tef1, rpb1, and rpb2 sequences. The maximum-likelihood tree on 3704 sites was generated using IQ-TREE multicore version 1.6.12 with the TIM2e+I+G4 model. The support percentages of ultrafast bootstrap (1000 tests) are shown at the nodes. The scale bar indicates 0.01 substitutions per site. The GenBank accession numbers of the sequences from the isolates YJX-0509 and YJX-0511, the representative strains including type (T), epitype (ET), or neotype (NT) strains of the closely related species within the Fusarium fujikuroi species complex, and the outgroup F. solani strain NRRL 43468 are listed in Table S1.
3.3. Micromorphology of Fusarium annulatum
The isolates YJX-0509 and YJX-0511 formed abundant microconidia after they grew on water agar for 7 d while they formed microconidia and macroconidia on PDA over 14 d. Microconidia are single-cells without septum, in fusiform, obovoid, or rod shape, 4.2–13.9 × 0.9–3.8 μm, arranged in false heads or chains on monophialides (Figure 4a–c). Macroconidia are straight or slightly curved, 12.5–65.6 × 0.9–4.9 μm, three to five septa with blunt apical cells and foot-shaped basal cells (Figure 4d–g). Chlamydospores have not been observed.
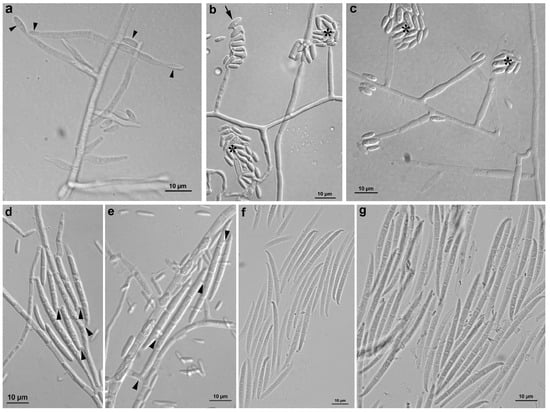
Figure 4.
Micromorphology of Fusarium annulatum isolates YJX-0509 and YJX-0511 from tulip bulbs. (a–c) Various morphologies of microconidia and monophialidic conidiogenous cells. (a) Multiple monophialidic conidiogenous cells generating single microcondia (arrowheads). (b) Microconidia forming chains (arrow) and false heads (asterisks). (c) Microconidia forming false heads (asterisks). (d,e) Multiple conidiogenous loci (arrowheads) generating macroconidia. (f,g) Macroconidia with three to five septa. Scale bar: 10 μm.
3.4. Pathogenicity of Fusarium annulatum
After inoculation, mycelia of the isolates YJX-0509 and YJX-0511 grew on the surface of the bulb scales, forming white cottony colonies and causing shrinkage and rot of the bulbs and terminal buds while the mock-inoculated bulbs remained without symptoms (Figure 5). At 10 d after inoculation, the severity of the bulb rot reached level 3 (rot area 50% with mold coverage), and the disease index was 75%. The fungal isolates reisolated from the inoculated tulip plants had similar morphology and identical tef1 sequences to those of the original isolates YJX-0509 and YJX-0511 (GenBank accession no. PQ791635 and PQ791636), which have been deposited in the China Center for Type Culture Collection with the culture numbers CCTCC AF 2024021 and CCTCC AF 2024022.
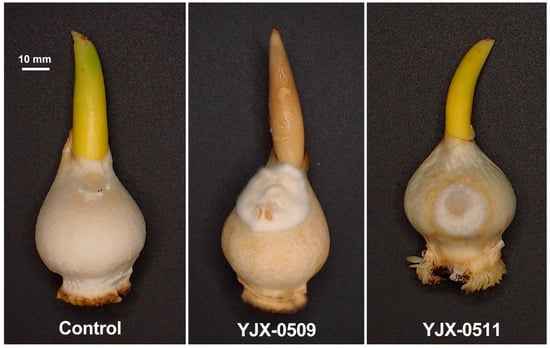
Figure 5.
Tulip bulb rot appeared at 10 d after inoculation of Fusarium annulatum isolate YJX-0509 and YJX-0511.
4. Discussion
We isolated six fungal isolates from the rotten tulip bulbs, which showed colony morphology resembling Fusarium. Although the colony morphologies of the six isolates were diverse and variable under the same growth condition, they have identical ITS and tef1 sequences. The sequence identity and phylogenetic analyses of the tef1 sequences, the primary species identification marker [14], located the six Fusarium isolates to the same species F. annulatum (Figure S1). The isolate YJX-0509, representing the violet pigment-producing isolates, and the isolate YJX-0511, representing the isolates with pale-surfaced colonies, were selected for further analyses. Their identical rpb1 sequences, identical rpb2 sequences, and high sequence identities (rpb1 99.9% and rpb2 99.5%) to those of the F. annulatum-type strain CBS 258.54T, and the closest phylogeny to F. annulatum based on multigene tef1, rpb1, and rpb2 determine that the Fusarium isolates from the tulip bulbs belong to F. annulatum. Moreover, the micromorphology of the representative isolates, such as predominantly straight macroconidia, is typical of F. annulatum [9].
The pathogenicity of the F. annulatum isolates to tulip bulbs was verified with Koch’s postulates. To promote Fusarium pathogenicity, we inoculated fresh mycelia of isolates YJX-0509 and YJX-0511 onto wounded bulbs’ surfaces after removing the tunics and kept the bulbs under 25 °C, a general optimum temperature for mycelial growth of Fusarium, and high relative humidity (>75% under the plastic cover) in a gnotobiotic condition. The resulting white cottony colonies on the bulb surface and dry rot symptoms (Figure 5) are not the same as the original diseased tulips (Figure 1), probably because the wounding inoculation, temperature, relative humidity, the gnotobiotic condition, and incubation time were different from the original conditions with relatively low temperature and humidity and relatively complex microbiome.
Fusarium annulatum is a morphologically diverse species associated with over 200 plant host species worldwide [9]. F. annulatum had been extensively studied in the past under the name F. proliferatum, a segregate of F. moniliforme [9,11]. Yilmaz et al. distinguished F. annulatum from F. proliferatum by their distinct phylogeny and micromorphology [9]. The F. moniliforme from tulip bulbs described by Duan et al. produced chlamydospores but no macroconidia [10], whereas the F. proliferatum causing bulb wilt of tulip described by Chang et al. [13] and the F. annulatum described here produce macroconidia but no chlamydospores. The F. proliferatum isolate causing bulb wilt of tulip produced rare macroconidia [13]; its ITS sequence (GenBank accession no. PV291676) shows an identity of 99.2% (503/507) to the ITS sequences (GenBank accession no. PQ775112 and PQ775113) of the F. annulatum isolates, higher than that of 98.6% (500/507) to the ITS sequence (GenBank accession no. AF158302) of the F. proliferatum epitype strain CBS 480.96ET. The F. proliferatum isolate causing the bulb wilt of tulip is close to the F. annulatum isolates causing the bulb rot of tulip but cannot be reclassified as F. annulatum based on the morphological and molecular information provided by Chang et al. [13]. Together, previous studies on Fusarium pathogens in tulips have not presented clear species taxonomy subject to the International Code of Nomenclature for algae, fungi, and plants. Here, our study clarifies the species taxonomy of F. annulatum causing bulb rot of tulip and shows tulip as a new host of F. annulatum.
5. Conclusions
In conclusion, the sequence identity and phylogenetic status of the DNA barcoding sequences, conidial morphology, and pathogenicity of the fungal isolates from the rotten tulip bulbs support the idea that the isolates belonging to F. annulatum are the causal agents of the bulb rot of the tulips. This is the first report of F. annulatum causing bulb rot disease of tulip.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae11050518/s1, Figure S1: Phylogenetic maximum-likelihood tree of Fusarium fujikuroi species complex based on tef1 sequences. Table S1: DNA barcoding sequences used in constructing the three-locus tef1-rpb1-rpb2 phylogenetic tree for the Fusarium fujikuroi species complex.
Author Contributions
Conceptualization, J.L. and Q.A.; methodology, Q.L., S.M., and T.L.; validation, Q.A.; formal analysis, Q.L., S.M., and T.L.; investigation, Q.L., S.M., T.L., and C.L.; resources, J.L., L.C., and Y.S.; data curation, Q.A.; writing—original draft preparation, Q.L., S.M., and T.L.; writing—review and editing, J.L. and Q.A.; supervision, B.L. and Q.A.; project administration, L.C., Y.S., and B.L.; funding acquisition, J.L., Y.S., and B.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Shanghai Agricultural Science and Technology Innovation Project (T2023101), the Zhejiang Major Agricultural Technology Collaborative Promotion Plan Project, and Zhejiang Government Scholarship.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.
Conflicts of Interest
The authors declare no conflicts of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Nisa, Q.; Gulzar, G.; Dar, M.S.; Shahnaz, E.; Banday, S.; Bhat, Z.A.; El-Sheikh, M.A.; Nabi, S.U.; Arya, V.M.; Anwar, A.; et al. New reports of pathogen spectrum associated with bulb rot and their interactions during the development of rot in tulip. BMC Genom. Data 2024, 25, 40. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, N.T.; Reyad, N.-H.A.; Halawa, A. Management of root and bulb rot affecting some flower bulb production in Egypt. Egypt. J. Biol. Pest Control. 2017, 27, 271–278. [Google Scholar]
- Boyraz, N.; Bastas, K.; Maden, S.; Yasar, A. Bacterial leaf and peduncle soft rot caused by Pectobacterium carotovorum on tulips in Konya, Turkey. Phytoparasitica 2006, 34, 272–280. [Google Scholar] [CrossRef]
- Miller, W.B. Fusarium in tulips: Ethylene, gum, and aborted flowers. Greenh. Prod. News 2002, 12, 36–39. [Google Scholar]
- Michielse, C.B.; Rep, M. Pathogen profile update: Fusarium oxysporum. Mol. Plant. Pathol. 2009, 10, 311–324. [Google Scholar] [CrossRef] [PubMed]
- Lombard, L.; Sandoval-Denis, M.; Lamprecht, S.C.; Crous, P.W. Epitypification of Fusarium oxysporum–clearing the taxonomic chaos. Persoonia 2019, 43, 1–47. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.; Sutton, D.A.; Rinaldi, M.G.; Sarver, B.A.; Balajee, S.A.; Schroers, H.J.; Summerbell, R.C.; Robert, V.A.; Crous, P.W.; Zhang, N.; et al. Internet-accessible DNA sequence database for identifying fusaria from human and animal infections. J. Clin. Microbiol. 2010, 48, 3708–3718. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.M.; Crous, P.W.; Sandoval-Denis, M.; Han, S.L.; Liu, F.; Liang, J.M.; Duan, W.J.; Cai, L. Fusarium and allied genera from China: Species diversity and distribution. Persoonia 2022, 48, 1–53. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, N.; Sandoval-Denis, M.; Lombard, L.; Visagie, C.M.; Wingfield, B.D.; Crous, P.W. Redefining species limits in the Fusarium fujikuroi species complex. Persoonia 2021, 46, 129–162. [Google Scholar] [CrossRef] [PubMed]
- Duan, M.; Yuan, Y.; Zhao, J. The occurrence and control of the basal rot disease of tulip. Plant Prot. 1997, 23, 35–36. (In Chinese) [Google Scholar]
- Seifert, K.A.; Aoki, T.; Baayen, R.P.; Brayford, D.; Burgess, L.W.; Chulze, S.; Gams, W.; Geiser, D.; de Gruyter, J.; Leslie, J.F.; et al. The name Fusarium moniliforme should no longer be used. Mycol. Res. 2003, 107, 643–644. [Google Scholar] [CrossRef]
- Summerell, B.A. Resolving Fusarium: Current status of the genus. Annu. Rev. Phytopathol. 2019, 57, 323–339. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Tao, G.; Li, Y. Identification of bulb wilt pathogen in Tulipa. Acta Agric. Boreali Sin. 2010, 19, 120–123, (In Chinese with English abstract). [Google Scholar]
- Crous, P.W.; Lombard, L.; Sandoval-Denis, M.; Seifert, K.A.; Schroers, H.J.; Chaverri, P.; Gené, J.; Guarro, J.; Hirooka, Y.; Bensch, K.; et al. Fusarium: More than a node or a foot-shaped basal cell. Stud. Mycol. 2021, 98, 100116. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to the Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: New York, NY, USA, 1990; pp. 315–322. [Google Scholar]
- O’Donnell, K.; Kistler, H.C.; Cigelnik, E.; Ploetz, R.C. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Nat. Acad. Sci. USA 1998, 95, 2044–2049. [Google Scholar] [CrossRef] [PubMed]
- Reeb, V.; Lutzoni, F.; Roux, C. Contribution of RPB2 to multilocus phylogenetic studies of the euascomycetes (Pezizomycotina, Fungi) with special emphasis on the lichen-forming Acarosporaceae and evolution of polyspory. Mol. Phylogenet. Evol. 2004, 32, 1036–1060. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Whelen, S.; Hall, B.D. Phylogenetic relationships among ascomycetes: Evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 1999, 16, 1799–1808. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef] [PubMed]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).