Genome-Wide Analysis of Mlo Genes and Functional Characterization of Cm-mlo38 and Cm-mlo44 in Regulating Powdery Mildew Resistance in Melon
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of Mlo Genes in Cucurbitaceae
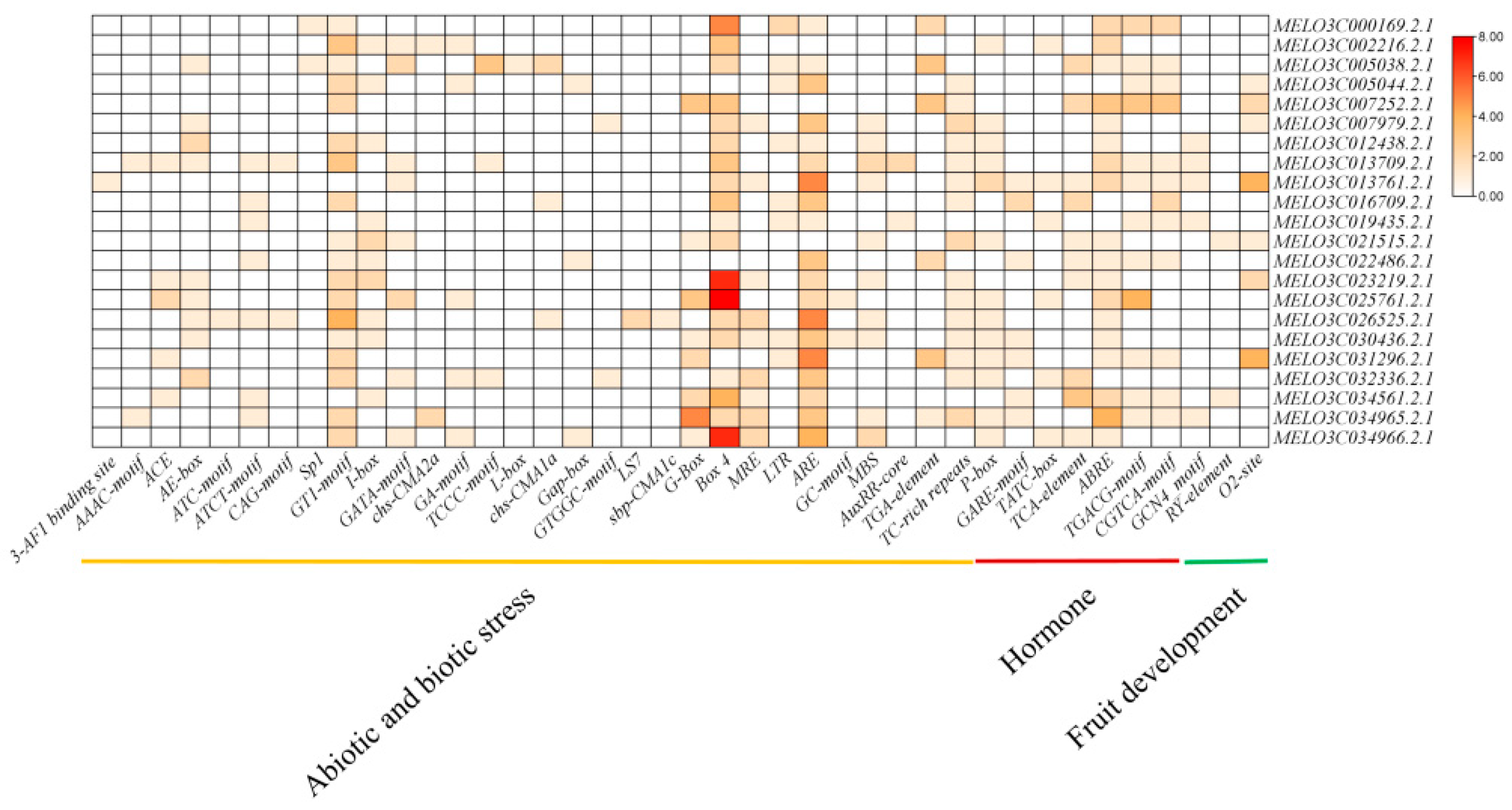
2.2. Sequence Analysis of Mlo Genes
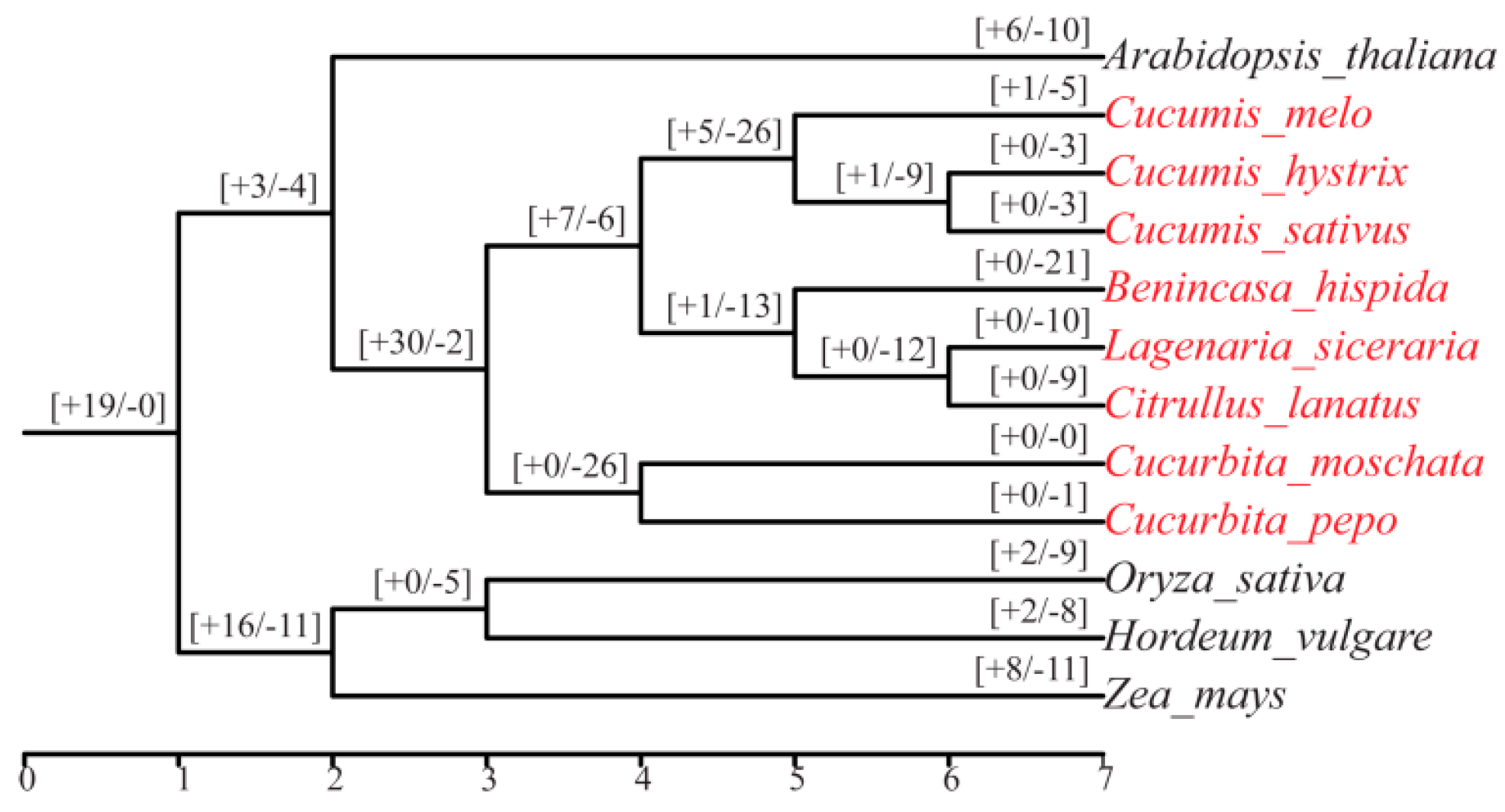
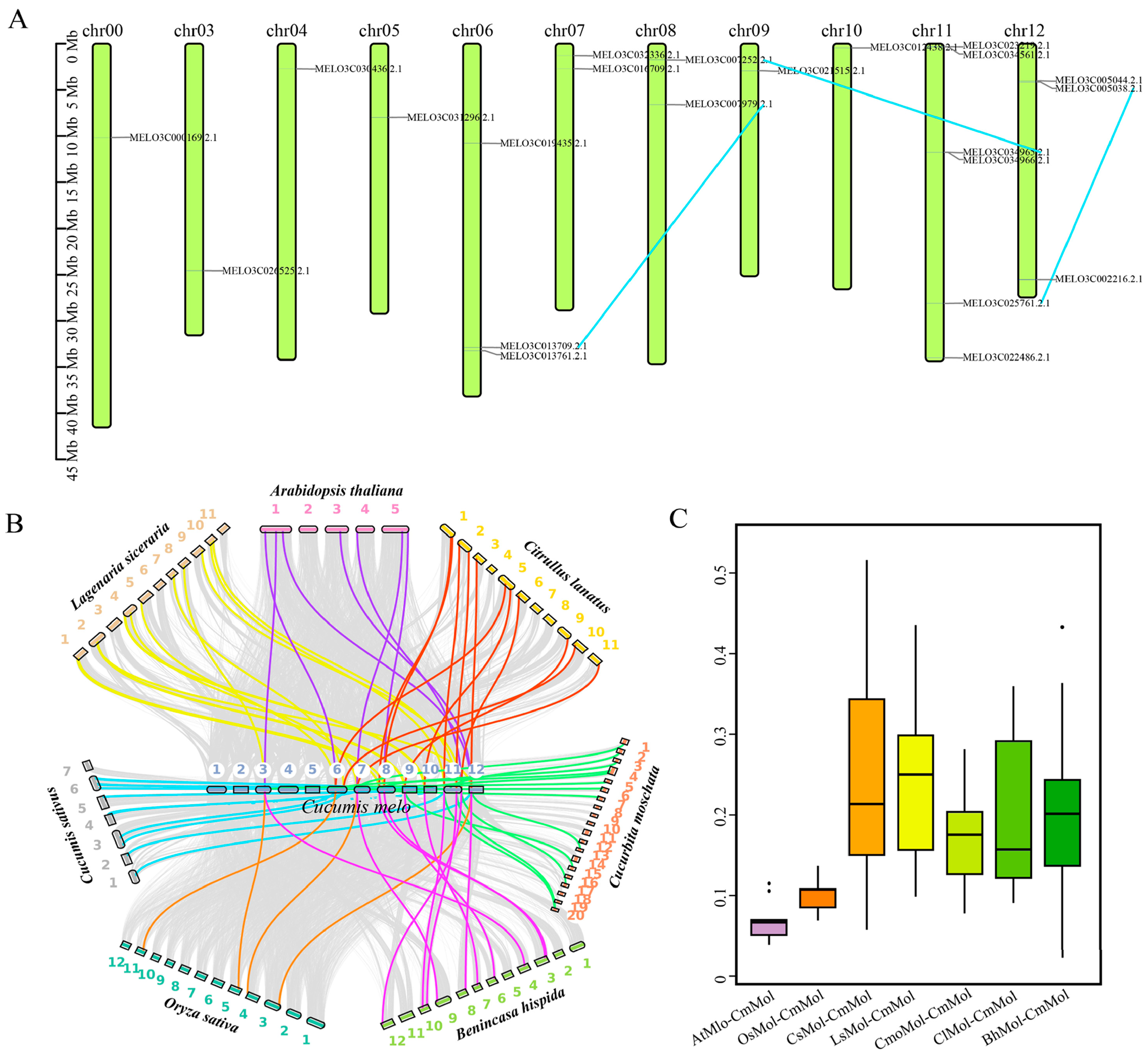
2.3. Phylogenetic and Collinearity Analyses of Mlo Genes
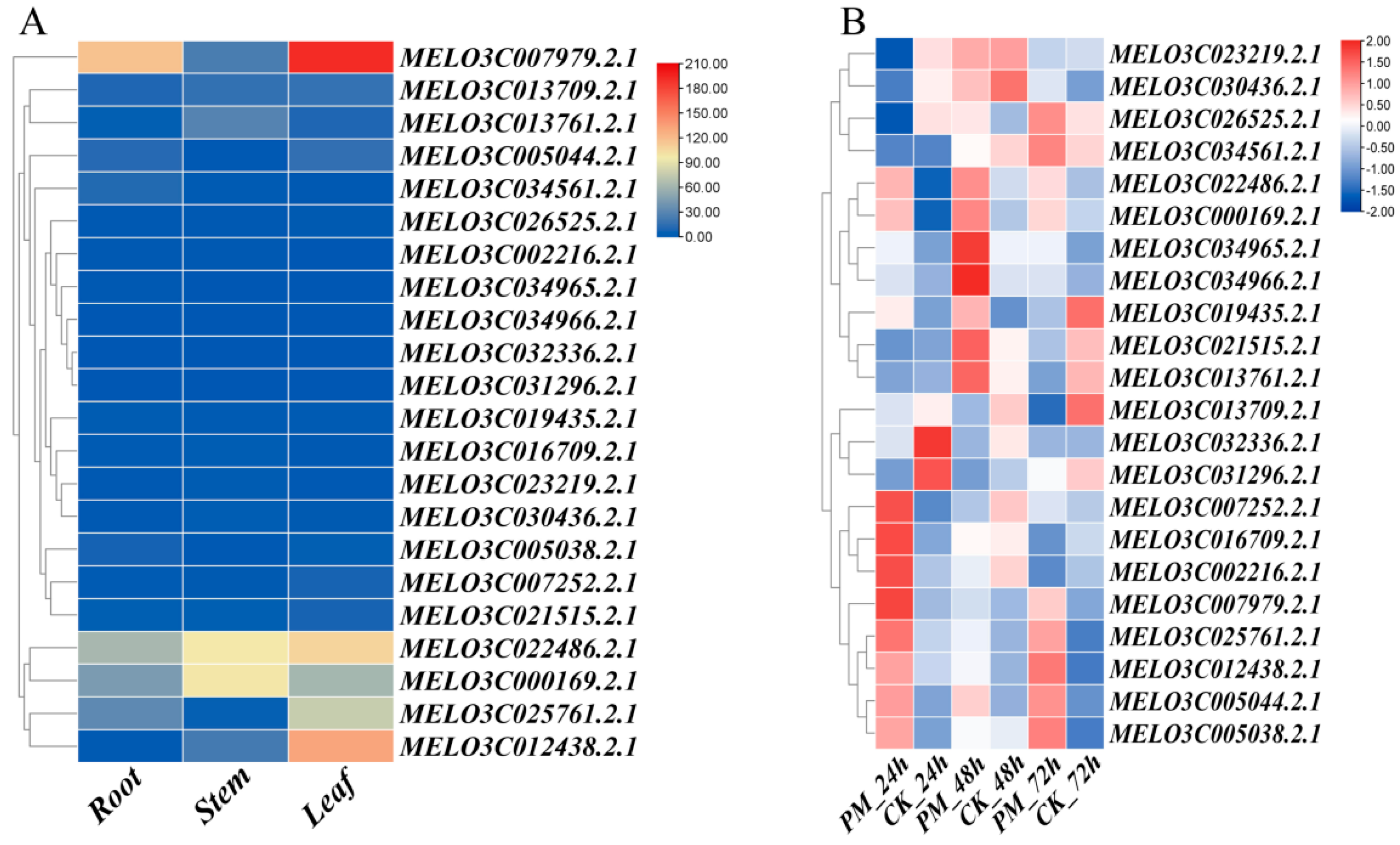
2.4. Expression Analysis of Mlo Genes
2.5. RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
2.6. Virus-Induced Gene Silencing (VIGS)
2.7. Subcellular Localization
2.8. Protein Interaction Network Analysis
2.9. Statistical Analysis
3. Results
3.1. Identification and Sequence Analysis of Mlo Family Members
3.2. The Phylogenetic and Subcellular Localization Analysis of Mlo Genes
3.3. Intragroup and Intergroup Collinearity Analysis of Mlo Genes
3.4. Promoter Analysis of Cm-mlo Genes
3.5. Expression Patterns in Different Tissues and Expression Response Analysis to PM
3.6. The Functional Verification of Cm-mlo38 and Cm-mlo44
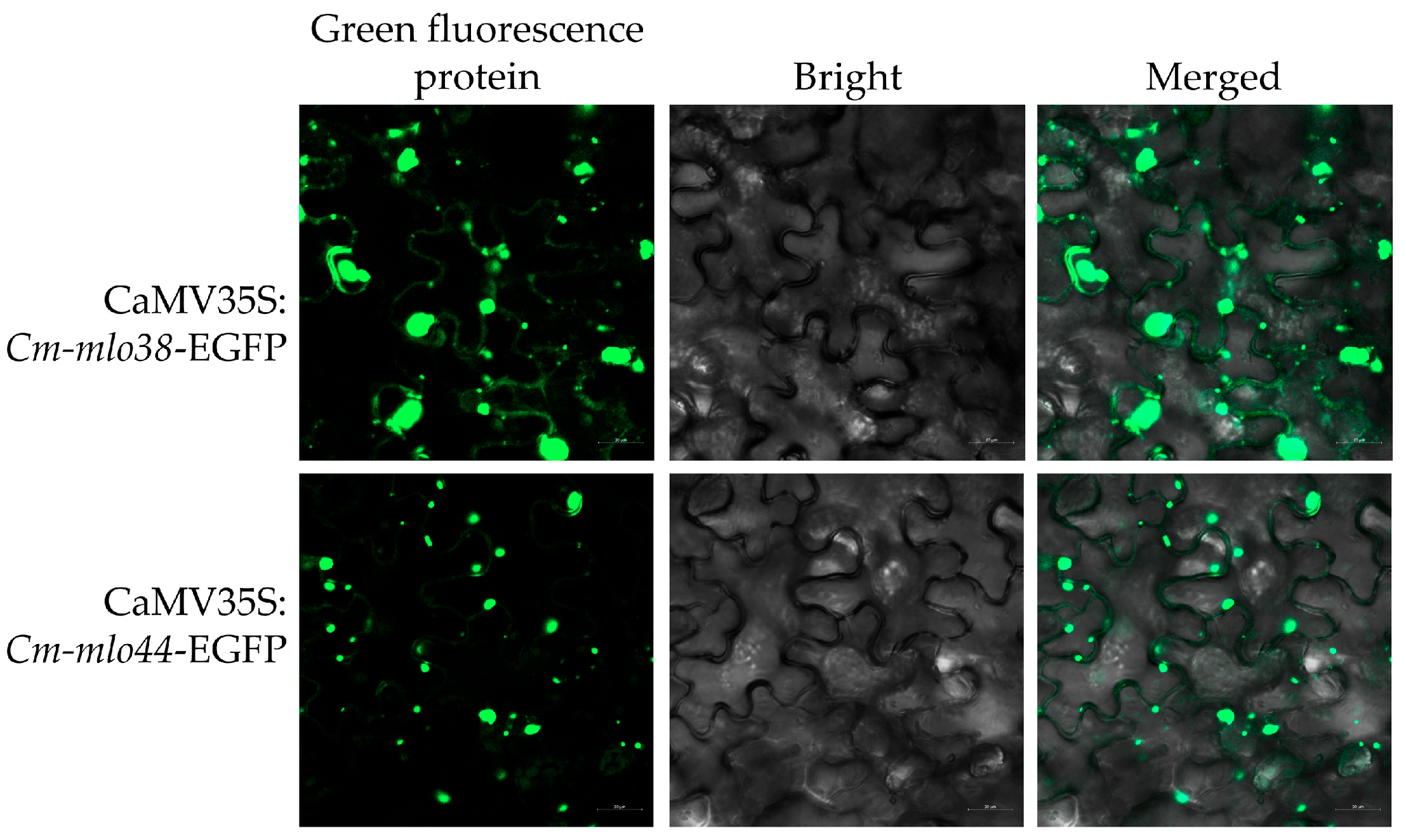
3.7. Subcellular Localization Analysis of Cm-mlo38 and Cm-mlo44 Proteins
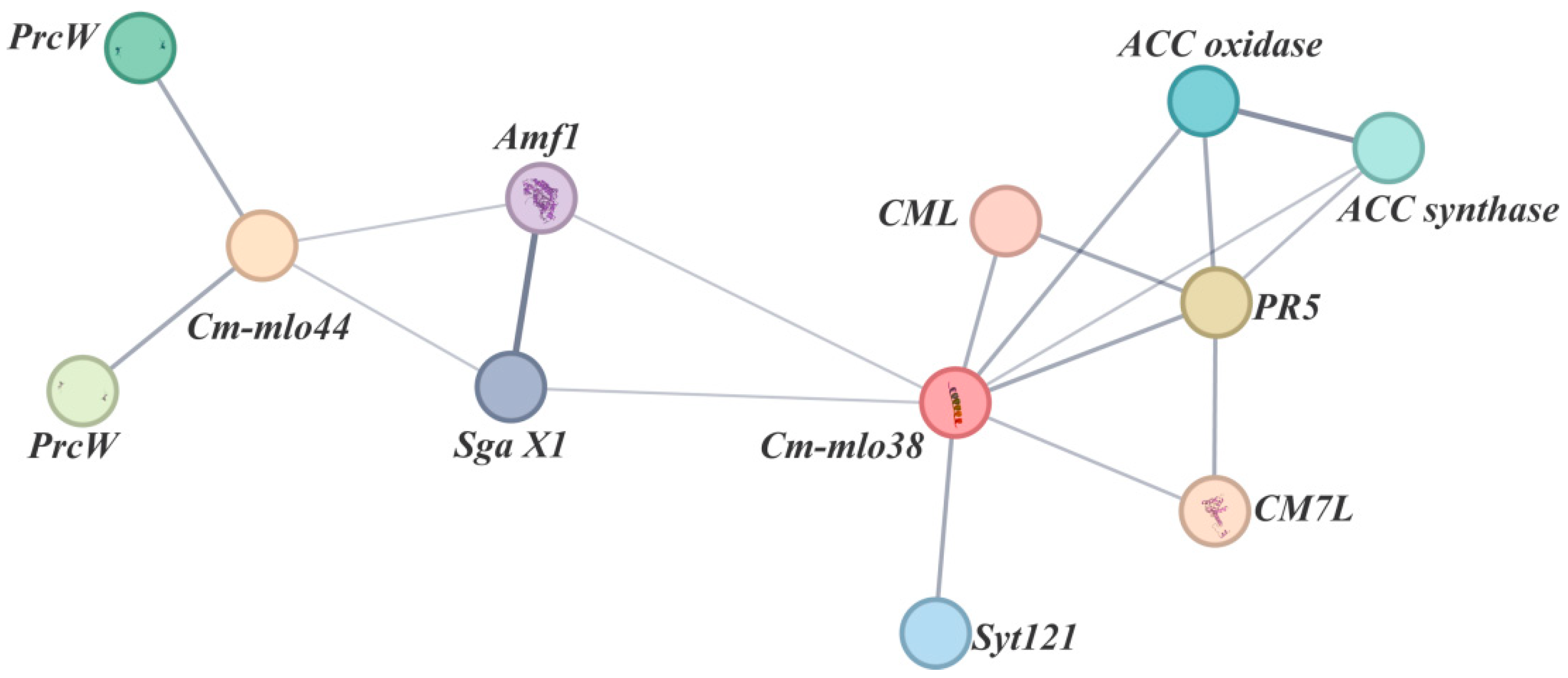
3.8. Protein Interaction Network Analysis of Cm-mlo38 and Cm-mlo44 Proteins
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| Mlo | Mildew resistance locus |
| PM | Powdery mildew |
| Px | Podosphaera xanthii |
| GC | Golovinomyes cichoracearum |
| HMM | Hidden Markov Model |
| NCBI | National Center for Biotechnology Information |
| CDD | Conserved Domain Database |
| PI | Theoretical isoelectric point |
| Mw | Molecular weight |
| NJ | Neighbor joining |
| VIGS | Virus-induced gene silencing |
| CDS | Coding sequence |
References
- Tian, S.; Zhang, Z.; Qin, G.; Xu, Y. Parthenocarpy in Cucurbitaceae, advances for economic and environmental sustainability. Plants 2023, 12, 3462. [Google Scholar] [CrossRef] [PubMed]
- Chomicki, G.; Schaefer, H.; Renner, S.S. Origin and domestication of Cucurbitaceae crops, insights from phylogenies, genomics and archaeology. New Phytol. 2020, 226, 1240–1255. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Siskos, L.; Wang, C.; Schouten, H.J.; Visser, R.G.; Bai, Y. Breeding melon (Cucumis melo) with resistance to powdery mildew and downy mildew. Hortic. Plant J. 2022, 8, 545–561. [Google Scholar] [CrossRef]
- Lebeda, A.; Křístková, E.; Mieslerová, B.; Dhillon, N.P.S.; McCreight, J.D. Status, gaps and perspectives of powdery mildew resistance research and breeding in cucurbits. Crit. Rev. Plant Sci. 2024, 43, 211–290. [Google Scholar] [CrossRef]
- Glawe, D.A. The powdery mildews, a review of the world’s most familiar (yet poorly known) plant pathogens. Annu. Rev. Phytopathol. 2008, 46, 27–51. [Google Scholar] [CrossRef]
- Křístková, E.; Lebeda, A.; Sedláková, B. Species spectra, distribution and host range of cucurbit powdery mildews in the Czech Republic, and in some other European and Middle Eastern countries. Phytoparasitica 2009, 37, 337–350. [Google Scholar] [CrossRef]
- Jørgensen, J.H. Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica 1992, 63, 141–152. [Google Scholar] [CrossRef]
- Panstruga, R. Serpentine plant MLO proteins as entry portals for powdery mildew fungi. Biochem. Soc. Trans. 2005, 33, 389–392. [Google Scholar] [CrossRef]
- Büschges, R.; Hollricher, K.; Panstruga, R.; Simons, G.; Wolter, M.; Frijters, A.; Schulze-Lefert, P. The barley Mlo gene, a novel control element of plant pathogen resistance. Cell 1997, 88, 695–705. [Google Scholar] [CrossRef]
- Davis, T.C.; Jones, D.S.; Dino, A.J.; Cejda, N.I.; Yuan, J.; Willoughby, A.C.; Kessler, S.A. Arabidopsis thaliana MLO genes are expressed in discrete domains during reproductive development. Plant Reprod. 2017, 30, 185–195. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, Y.; Zhang, H. Genome-wide analysis of the mildew resistance locus o (‘MLO’) gene family in tomato (‘Solanum lycopersicum’ L.). Plant Omics 2014, 7, 87–93. [Google Scholar]
- Konishi, S.; Sasakuma, T.; Sasanuma, T. Identification of novel Mlo family members in wheat and their genetic characterization. Genes Genet. Syst. 2010, 85, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, R.; Singh, V.K.; Singh, B.D. Comparative phylogenetic analysis of genome-wide Mlo gene family members from Glycine max and Arabidopsis thaliana. Mol. Genet. Genom. 2014, 289, 345–359. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zhu, H. Molecular evolution of the MLO gene family in Oryza sativa and their functional divergence. Gene 2008, 409, 1–10. [Google Scholar] [CrossRef]
- Zhou, S.J.; Jing, Z.; Shi, J.L. Genome-wide identification, characterization, and expression analysis of the MLO gene family in Cucumis sativus. Genet Mol. Res. 2013, 12, 6565–6578. [Google Scholar] [CrossRef]
- Zhang, T.; Xu, N.; Amanullah, S.; Gao, P. Genome-wide identification, evolution, and expression analysis of MLO gene family in melon (Cucumis melo L.). Front. Plant Sci. 2023, 14, 1144317. [Google Scholar] [CrossRef]
- Ge, X.; Deng, W.; Lee, Z.Z.; Lopez-Ruiz, F.J.; Schweizer, P.; Ellwood, S.R. Tempered mlo broad-spectrum resistance to barley powdery mildew in an Ethiopian landrace. Sci. Rep. 2016, 6, 29558. [Google Scholar] [CrossRef]
- Piffanelli, P.; Ramsay, L.; Waugh, R.; Benabdelmouna, A.; D’Hont, A.; Hollricher, K.; Panstruga, R. A barley cultivation-associated polymorphism conveys resistance to powdery mildew. Nature 2004, 430, 887–891. [Google Scholar] [CrossRef]
- Acevedo-Garcia, J.; Spencer, D.; Thieron, H.; Reinstädler, A.; Hammond-Kosack, K.; Phillips, A.L.; Panstruga, R. mlo-based powdery mildew resistance in hexaploid bread wheat generated by a non-transgenic TILLING approach. Plant Biotechnol. J. 2017, 15, 367–378. [Google Scholar] [CrossRef]
- Acevedo-Garcia, J.; Gruner, K.; Reinstädler, A.; Kemen, A.; Kemen, E.; Cao, L.; Takken, F.L.W.; Reitz, M.U.; Schäfer, P.O.; Connell, R.J.; et al. The powdery mildew-resistant Arabidopsis mlo2 mlo6 mlo12 triple mutant displays altered infection phenotypes with diverse types of phytopathogens. Sci. Rep. 2017, 7, 9319. [Google Scholar] [CrossRef]
- Yan, Z.; Appiano, M.; van Tuinen, A.; Meijer-Dekens, F.; Schipper, D.; Gao, D.; Wolters, A.M.A. Discovery and characterization of a novel tomato mlo mutant from an ems mutagenized micro-tom population. Genes 2021, 12, 719. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Appiano, M.; Pavan, S.; Bracuto, V.; Ricciardi, L.; Visser, R.G.; Bai, Y. Genome-wide study of the tomato SlMLO gene family and its functional characterization in response to the powdery mildew fungus Oidium neolycopersici. Front. Plant Sci. 2016, 7, 380. [Google Scholar] [CrossRef] [PubMed]
- Berg, J.A.; Appiano, M.; Bijsterbosch, G.; Visser, R.G.; Schouten, H.J.; Bai, Y. Functional characterization of cucumber (Cucumis sativus L.) Clade V MLO genes. BMC Plant Biol. 2017, 17, 80. [Google Scholar] [CrossRef]
- Hong, C.; Wei, K.; Junfen, L.; Ji, L. Analysis of powdery mildew resistance in wild melon MLO mutants. Hortic. Plant J. 2015, 1, 165–171. [Google Scholar] [CrossRef]
- Yang, M.; Derbyshire, M.K.; Yamashita, R.A.; Marchler-Bauer, A. NCBI’s conserved domain database and tools for protein domain analysis. Curr. Protoc. Bioinform. 2020, 69, e90. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.H.; Xu, J.; Liu, Y.L.; Feng, J.T.; Chen, H.; He, Y.H.; et al. TB tools-II: A “one for all, all for one” bioinformatics platform for biological big-data minin. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Liu, M.; Liang, Z.; Aranda, M.A.; Hong, N.; Liu, L.; Kang, B.; Gu, Q. A cucumber green mottle mosaic virus vector for virus-induced gene silencing in cucurbit plants. Plant Methods 2020, 16, 9. [Google Scholar] [CrossRef]
- Vogeli-Lange, R.; Wagner, G.J. Subcellular localization of cadmium and cadmium-binding peptides in tobacco leaves implication of a transport function for cadmium-binding peptides. Plant Physiol. 1990, 92, 1086–1093. [Google Scholar] [CrossRef]
- Tek, M.I.; Calis, O.; Fidan, H.; Shah, M.D.; Celik, S.; Wani, S.H. CRISPR/Cas9 based mlo-mediated resistance against Podosphaera xanthii in cucumber (Cucumis sativus L.). Front. Plant Sci. 2022, 13, 1081506. [Google Scholar] [CrossRef]
- Kim, M.C.; Panstruga, R.; Elliott, C.; Müller, J.; Devoto, A.; Yoon, H.W.; Schulze-Lefert, P. Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature 2002, 416, 447–451. [Google Scholar] [CrossRef] [PubMed]
- Elliott, C.; Zhou, F.; Spielmeyer, W.; Panstruga, R.; Schulze-Lefert, P. Functional conservation of wheat and rice Mlo orthologs in defense modulation to the powdery mildew fungus. Mol. Plant-Microbe Interact. 2002, 15, 1069–1077. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Xiao, X. Research Progress on Powdery Mildew in Cucurbitaceae Plants, A Systematic Review. Mol. Pathog. 2024, 15, 155–169. [Google Scholar] [CrossRef]
- Nevo, E. Evolution of genome-phenome diversity under environmental stress. Proc. Natl. Acad. Sci. USA 2001, 98, 6233–6240. [Google Scholar] [CrossRef]
- Wang, C.; Gao, G.; Cao, S.; Xie, Q.; Qi, H. Isolation and functional validation of the CmLOX08 promoter associated with signalling molecule and abiotic stress responses in oriental melon, Cucumis melo var. makuwa Makino. BMC Plant Biol. 2019, 19, 75. [Google Scholar] [CrossRef]
- Wang, L.; Wu, X.; Xing, Q.; Zhao, Y.; Yu, B.; Ma, Y.; Qi, H. PIF8-WRKY42-mediated salicylic acid synthesis modulates red light induced powdery mildew resistance in oriental melon. Plant Cell Environ. 2023, 46, 1726–1742. [Google Scholar] [CrossRef]
- Zeng, H.; Xu, L.; Singh, A.; Wang, H.; Du, L.; Poovaiah, B.W. Involvement of calmodulin and calmodulin-like proteins in plant responses to abiotic stresses. Front. Plant Sci. 2015, 6, 600. [Google Scholar] [CrossRef]
- Zeng, H.; Zhu, Q.; Yuan, P.; Yan, Y.; Yi, K.; Du, L. Calmodulin and calmodulin-like protein-mediated plant responses to biotic stresses. Plant Cell Environ. 2023, 46, 3680–3703. [Google Scholar] [CrossRef]
- Breen, S.; Williams, S.J.; Outram, M.; Kobe, B.; Solomon, P.S. Emerging insights into the functions of pathogenesis-related protein 1. Trends Plant Sci. 2017, 22, 871–879. [Google Scholar] [CrossRef]


| Species | Common Name | Number of Mlo Genes | Clade | |||||
|---|---|---|---|---|---|---|---|---|
| I | II | III | IV | V | VI | |||
| Cucurbita moschata | Pumpkin | 21 | / | 4 | 7 | 2 | 5 | 3 |
| Lagenaria siceraria | Gourd | 14 | / | 3 | 4 | 1 | 4 | 2 |
| Benincasa hispida | Wax gourd | 15 | / | 3 | 6 | 1 | 3 | 2 |
| Citrullus lanatus | Watermelon | 15 | / | 3 | 5 | 1 | 4 | 2 |
| Cucumis hystrix | Wild cucumber | 16 | / | 3 | 6 | 1 | 4 | 2 |
| Cucumis sativus | Cucumber | 16 | / | 4 | 5 | 1 | 4 | 2 |
| Cucumis melo | Melon | 22 | / | 4 | 6 | 3 | 5 | 4 |
| Cucurbita pepo | Squash | 20 | / | 4 | 7 | 2 | 4 | 3 |
| Arabidopsis thaliana | Arabidopsis | 15 | / | 5 | 3 | 1 | 3 | 3 |
| Hordeum vulgare | Barley | 14 | 2 | 2 | / | / | 2 | 8 |
| Oryza sativa | Rice | 13 | 2 | 2 | / | / | 2 | 7 |
| Zea mays | Maize | 21 | 2 | 4 | 1 | / | 6 | 8 |
| Total | 202 | 6 | 41 | 50 | 13 | 46 | 46 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gong, F.; Lan, Y.; Zhang, T.; Li, C.; Li, Y.; Xia, F.; Liu, X.; Liu, D.; Liang, G.; Cai, P.; et al. Genome-Wide Analysis of Mlo Genes and Functional Characterization of Cm-mlo38 and Cm-mlo44 in Regulating Powdery Mildew Resistance in Melon. Horticulturae 2025, 11, 509. https://doi.org/10.3390/horticulturae11050509
Gong F, Lan Y, Zhang T, Li C, Li Y, Xia F, Liu X, Liu D, Liang G, Cai P, et al. Genome-Wide Analysis of Mlo Genes and Functional Characterization of Cm-mlo38 and Cm-mlo44 in Regulating Powdery Mildew Resistance in Melon. Horticulturae. 2025; 11(5):509. https://doi.org/10.3390/horticulturae11050509
Chicago/Turabian StyleGong, Fangyi, Yanhong Lan, Tian Zhang, Chun Li, Yifan Li, Feng Xia, Xiaojun Liu, Duchen Liu, Genyun Liang, Peng Cai, and et al. 2025. "Genome-Wide Analysis of Mlo Genes and Functional Characterization of Cm-mlo38 and Cm-mlo44 in Regulating Powdery Mildew Resistance in Melon" Horticulturae 11, no. 5: 509. https://doi.org/10.3390/horticulturae11050509
APA StyleGong, F., Lan, Y., Zhang, T., Li, C., Li, Y., Xia, F., Liu, X., Liu, D., Liang, G., Cai, P., & Fang, C. (2025). Genome-Wide Analysis of Mlo Genes and Functional Characterization of Cm-mlo38 and Cm-mlo44 in Regulating Powdery Mildew Resistance in Melon. Horticulturae, 11(5), 509. https://doi.org/10.3390/horticulturae11050509





