Abstract
Loropetalum chinense is a significant small tree and ornamental shrub known for its colorful foliage and is widely used in landscaping in tropical and subtropical regions. This study aimed to establish an efficient, tissue culture-independent genetic transformation system for L. chinense. Cuttings from two varieties, ‘Xiangnong Xiangyun’ and ‘Hei Zhenzhu’, were infected with different strains of Agrobacterium rhizogenes. The results showed that the K599 strain significantly induced hairy roots in both varieties, with ‘Xiangnong Xiangyun’ demonstrating a higher survival rate (60%), rooting rate (51.66%), and hairy root induction efficiency (45%) compared to ‘Hei Zhenzhu’. Based on these findings, ‘Xiangnong Xiangyun’ and the K599 strain were selected for further optimization through an orthogonal L9 (33) experiment, which focused on optimizing the infection solution composition, bacterial concentration, and infection duration, Finally, the genetic transformation system established at the beginning of the experiment was validated on ‘Xiangnong Xiangyun’ plants using the pre-screening LcDREB-43 gene of our group. Among these factors, infection duration was identified as the most influential for improving transformation efficiency. The optimal conditions were determined as an infection solution containing MES solution, a bacterial concentration of OD600 = 0.8, and a 15 min infection duration. Under these optimized conditions, the survival rate, rooting rate, induction efficiency, and transformation efficiency reached 86.67%, 70%, 61.67%, and 43.33%, respectively. Furthermore, the transgenic plants with LcDREB-43 overexpression and pCAMBIA1305-GFP were obtained through the established transformation system, the authenticity of the system was proved, and the production application was carried out through phenotypic observation, molecular identification, and auxiliary verification of physiological indicators.
1. Introduction
Loropetalum chinense, belonging to the genus Loropetalum in the family Hamamelidaceae, is an evergreen woody plant and an important species for landscape greening in tropical and subtropical regions. Its variety, L. chinense var. rubrum, known for its striking red or purple foliage, further enhances its ornamental value. The species as a whole is widely cultivated due to its strong ecological adaptability, high ornamental value, and broad range of applications [1]. The flowers and leaves of L. chinense are not only visually appealing but also possess medicinal properties, including heat-clearing, detoxification, antibacterial, anti-inflammatory effects, and the regulation of fat metabolism [2]. Additionally, the leaves of L. chinense are rich in anthocyanin, which can be used as non-toxic edible pigments with antimicrobial properties, offering significant potential in both the medical and industrial fields [3].
However, changing environmental conditions and human activities have led to challenges in the cultivation of L. chinense. High-temperature environments frequently result in a phenomenon known as “foliage rejuvenation”, which diminishes the plant’s ornamental value. Concurrently, there is a pressing demand for superior L. chinense varieties, yet the long breeding cycle of woody plants hinders its industrial development. Fortunately, the completion of L. chinense’s whole genome sequencing has marked a turning point, enabling research to advance to the molecular biology level and paving the way for more efficient breeding strategies and scientific exploration [4].
Despite this progress, the regeneration and genetic transformation system of L. chinense remains underdeveloped due to the species’ specific characteristics. Previous studies have primarily focused on tissue culture, particularly the regeneration of leaves, stem segments, and adventitious bud differentiation. For instance, Fu Hongyan et al. [5] reported significant variation in the regeneration ability of leaves across different varieties, with ‘Rose Red Fine Leaf Green’ showing the highest regeneration capacity and ‘Flowering Leaf No. 2’ the weakest. Similarly, Chen Rong et al. [6] optimized conditions for callus induction, adventitious shoot proliferation, and rooting, thereby establishing a preliminary regeneration system. Liu Zhenhua et al. [7] explored in vitro rapid propagation techniques using stem segments with nodes, achieving regrowth roots. Despite these advancements, the current regeneration system faces challenges, such as difficulty obtaining sterile seedlings, prolonged rooting periods, and limited root production, which hinder the establishment of a stable and efficient genetic transformation system. Therefore, a rapid, tissue culture-independent genetic transformation method is urgently needed to advance gene function analysis and genetic improvement in L. chinense.
Agrobacterium rhizogenes, a Gram-negative bacterium in the Rhizobiaceae family, is widely recognized for its broad host range and its ability to induce the formation of hairy roots. These roots exhibit rapid growth and genetic stability, making them ideal materials for gene function verification and secondary metabolism studies [8,9,10,11]. Unlike traditional Agrobacterium tumefaciens-mediated transformations, Agrobacterium rhizogenes-mediated hairy root transformation systems bypass tissue culture requirements and antibiotic screening, offering higher efficiency and practicality [12,13,14,15]. While most current systems rely on sterile seedling leaves or stem segments as explants [16,17,18], the acquisition of sterile materials is labor-intensive and complicated. Woody plants like L. chinense often encounter browning during in vitro culture, further complicating the process. Additionally, in vitro culture methods involve high costs, long cycles, and technical complexity, limiting their scalability and application.
In a non-tissue culture environment, the genetic transformation system mediated by A. rhizogenes offers a promising solution to several challenges associated with woody plant transformation. This approach significantly enhances transformation efficiency and provides a practical pathway for industrial applications [19,20]. Recent advancements have demonstrated the feasibility of A. rhizogenes-mediated hairy root genetic transformation systems in non-tissue culture environments for woody plants. For instance, Xie Xiaoting et al. [21] successfully developed a non-tissue culture-based genetic transformation system for Carya illinoinensis; Ma Haijie et al. [22] used A. rhizogenes strain K599 for root genetic transformation and genome editing of citrus; Wu Mengjie et al. [23] optimized the non-tissue culture-based transformation system of Phoebe bournei through an orthogonal L9 (33) test; and Jiu Linlu [24] explored non-tissue culture-based transformation conditions for peach (Prunus persica). These findings provide valuable insights for establishing a non-tissue culture-based transformation system for L. chinense.
As one of the largest transcription factor families, the AP2/ERF transcription factor family is ubiquitous in plants and is widely involved in important biological processes such as plant growth and development, secondary metabolism regulation, and biotic and abiotic stress response [25]. Anthocyanin belongs to flavonoids, which are one of the secondary metabolites of the flavonoid pathway, and AP2/ERF transcription factors are widely involved in the secondary metabolism regulation of flavonoids. According to the relevant reports, it is shown that AP2/ERF transcription factors are involved in the synthesis of anthocyanosides in plants such as pears, Arabidopsis thaliana, citrus, etc. [26,27,28]. The previous studies showed that the ethylene response factors Pp4ERF24 and Pp12ERF96 could regulate blue light-induced anthocyanoside synthesis in ‘Red Early Crisp’ pear fruits by interacting with MYB114 [26]. In Arabidopsis thaliana, a study has investigated the regulatory signaling pathway of anthocyanin production due to light response using two ERF mutant Arabidopsis thaliana mutants, AtERF4 and AtERF8, which showed that light inhibits anthocyanin production, and that the ERF mutants reduced the rate and extent of anthocyanin production in association with changes in the transcript levels of anthocyanin biosynthesis genes [27]. While in citrus, three AP2/ERF transcription factor members, CitERF32, CitERF33, and CitRAV1, promote flavanone and flavonoid accumulation by regulating the chalcone isomerase gene CitCHIL1 [28]. Anthocyanidins are widely found in the roots, flowers, leaves, and other organs of L. chinense and have important ornamental, medicinal, and industrial values, but the genetic transformation system of L. chinense has not yet been established to efficiently and orderly validate the gene homology and the development of secondary metabolites.
Despite advancements in genetic transformation techniques, research on transformation systems for L. chinense remains limited, with no highly efficient system currently available. This gap restricts its application in molecular breeding, stress tolerance studies, secondary metabolite biosynthesis, and investigations into growth and developmental mechanisms. To address this limitation, we have developed a simple, rapid, and efficient genetic transformation system for L. chinense mediated by A. rhizogenes. This tissue culture-independent system is adaptable to various species and can be completed within 5–8 weeks. Meanwhile, this study optimizes the transformation process by refining the infiltration solution composition, bacterial suspension concentration, and infiltration duration, significantly enhancing transformation efficiency. In addition, based on the transcription factor LcDREB-43 (augustus36771.t1) screened in our previous transcriptome data [29], we ligated the pCAMBIA1305-GFP vector and overexpressed the LcDREB-43 gene, thus applying the system to practical work.
2. Materials and Methods
2.1. Materials and Equipment
Cuttings of L. chinense ‘Xiangnong Xiangyun’ (20220205) and ‘Hei Zhenzhu’ were collected from healthy mature plants at the Garden and Flower Base of Hunan Agricultural University. Three Agrobacterium rhizogenes strains (K599, C58C1, and MSU440) were commercially obtained from Weidi Biotechnology Co., Ltd. (Shanghai, China). The pCAMBIA1305-GFP vector was stored in an ultra-low temperature freezer in the Laboratory of Ornamental Horticulture at Hunan Agricultural University. In this experiment, the infected cuttings of L. chinense were cultivated at the Garden and Flower Base of Hunan Agricultural University. The growth medium was composed of a volumetric mixture containing 25% perlite and 75% vermiculite.
2.2. Preparation of L. chinense Spike Materials
Disease- and pest-free semi-woody branches (approximately 5 cm in length) were cut from the middle and upper sections of ‘Xiangnong Xiangyun’ and ‘Hei Zhenzhu’. Excess leaves were removed, leaving two leaves per cutting. The basal end of each cutting was cut at an angle to increase contact surface with the rooting substrate, and the cuttings were prepared for further use.
2.3. Preparation and Cultivation of A. rhizogenes Strains
The A. rhizogenes strains K599, MSU440, and C58C1 stored at −80 °C were thawed on ice. For each 100 μL of bacterial suspension, 1 μg of pCAMBIA1305-GFP plasmid was added, and the mixture was pipetted thoroughly. The bacteria were then subjected to the following steps: 5 min on ice, 5 min in liquid nitrogen, 5 min in a 37 °C water bath, and another 5 min on ice. After the final ice bath, 700 μL of antibiotic-free TY liquid medium (5 g∙L−1 tryptone, 3 g∙L−1 yeast extract, 10 mmol∙L−1 CaCl2) was added, and the suspension was incubated at 28 °C with shaking for 2 h. The bacterial cells were collected by centrifugation at 6000 r∙min−1 for 1 min, and the supernatant was discarded. The bacterial pellet was gently resuspended in 100 μL of TY medium. K599 and MSU440 were streaked onto TY agar plates (TY liquid medium supplemented with 15 g∙L−1 agar) containing 50 mg∙L−1 kanamycin sulfate and 50 mg∙L−1 streptomycin. C58C1 was streaked onto TY agar plates (TY liquid medium supplemented with 15 g∙L−1 agar) containing 50 mg∙L−1 kanamycin sulfate, 50 mg∙L−1 streptomycin, and 50 mg∙L−1 rifampicin. The plates were inverted and incubated at 28 °C for 2–3 days [21].
2.4. Preparation of A. rhizogenes Inoculation Solution
Single colonies of A. rhizogenes strains K599, MSU440, and C58C1 harboring the pCAMBIA1305-GFP plasmid were inoculated into 1 mL of TY medium. K599 and MSU440 cultures contained 50 mg∙L−1 kanamycin sulfate and 50 mg∙L−1 streptomycin, while C58C1 also contained 50 mg∙L−1 rifampicin. After incubation at 28 °C for 12–16 h, the cultures were expanded at a 1:100 ratio until the OD600 reached 0.4–1.2 [21]. The bacterial cells were harvested by centrifugation at 6000 r∙min−1 for 10 min and then resuspended in an equal volume of Both MES solution (10 mmol∙L−1 MES-KOH, pH 5.6, 10 mmol∙L−1 MgCl2, 200 μmol∙L−1 acetosyringone) and MS liquid medium (4.5 g/L MS, pH 5.6). The infection solution, prepared without resuspension, could be directly used for subsequent experiments. All prepared solutions were stored for further use.
2.5. Establishment of Transformation System and Hairy Root Induction
Cuttings of ‘Xiangnong Xiangyun’ and ‘Hei Zhenzhu’ were submerged in A. rhizogenes inoculation solutions (OD600 = 0.8 for K599, MSU440, and C58C1) for 10 min. The cuttings were then placed in a vermiculite substrate, with 20 cuttings per group and three replicates, and maintained in a greenhouse at 28 °C. The substrate moisture content was monitored daily and maintained at approximately 80%. Watering was conducted on average once every two days during the summer and once every five days in the winter, with adjustments made according to specific conditions as necessary. Hairy root formation was observed after one month.
2.6. Optimization of Transformation System
Based on the A. rhizogenes-mediated transformation system established in preliminary trials, to optimize the conditions for hairy root induction, a three-factor, three-level orthogonal experiment was conducted, focusing on the composition of the infiltration solution, bacterial concentration, and infiltration duration (Table 1). An L9 (33) orthogonal design (Table 2) was implemented, with each treatment group containing 20 cuttings, and each experiment was replicated three times. The infiltration method involved fully submerging the cuttings in the bacterial solution. After one month of cultivation, the rooting conditions of the cuttings were observed and statistically analyzed. The aim is to optimize the existing transformation system and to screen for the critical factors and optimal conditions that influence the induction of hairy roots in L. chinense cuttings.

Table 1.
Three factors and three levels table.

Table 2.
Three factors and three levels of orthogonal experimental design table.
2.7. Identification and Statistical Analysis of Hairy Roots and Transgenic Roots
Hairy roots were identified using a LUYOR-3415 portable GFP excitation light source (LUYOR Instruments Co., Ltd., Joliet, IL, USA) and photographed. Genomic DNA was extracted from both GFP-expressing roots and non-transformed control roots (WT), and primers were designed for the identification of the rolB gene located in the T-DNA region of A. rhizogenes strain K599. Based on the GFP tag sequence in the vector pCAMBIA1305-GFP, GFP-specific identification primers were designed. PCR amplification was performed to verify the presence of hairy roots and transgenes, using primers specific for the rolB and GFP genes. Genomic DNA was extracted from fresh root tissue using the AFTS-Pin Universal Plant DNA Extraction Kit (ABclonal Biotechnology Co., Ltd., Wuhan, China). Primer sequences were synthesized by Qingke Biotechnology Co., Ltd. (Changsha, China); PCR primer sequences are shown in Supplemental Table S1.
The PCR reaction system included 25 μL of 2× Rapid Taq Master Mix, 1 μL of each primer, and 1 μL of DNA template, supplemented with water (ddH2O) to 25 μL. The PCR conditions were as follows: pre-denaturation at 95 °C for 3 min, followed by 35 cycles of denaturation at 95 °C for 15 sec, annealing at 60 °C for 15 sec, and extension at 72 °C for 15 sec, with a final extension at 72 °C for 15 sec. PCR products were analyzed by agarose gel electrophoresis (1%). A fragment of approximately 703 bp was obtained for the rolB gene and 526 bp for the GFP gene. Transformation efficiency was assessed using the following formulas:
Survival rate (%) = (number of viable plants/total number of treated plants) × 100
Rooting rate (%) = (number of rooted plants/total number of treated plants) × 100
Induction efficiency (%) = (number of hairy rooted plants/total number of treated plants) × 100
Transformation efficiency (%) = (number of transgenic plants/total number of treated plants) × 100
2.8. Application of Non-Tissue Culture Genetic Transformation Systems
2.8.1. Cloning of Genes and Vector Construction
Total RNA was isolated from L. chinense leaves using a Plant RNA Extraction Kit (Vazyme Biotechnology Co., Ltd., Nanjing, China) and reverse-transcribed into cDNA using a Reverse Transcription Kit (Accurate Biotechnology Co., Ltd., Changsha, China). The LcDREB-43 gene was amplified by PCR using primers synthesized by Qingke Biotechnology Co., Ltd. (Changsha, China), and the product was cloned into the pEASY-T1 vector (TransGen Biotechnology Co., Ltd., Beijing, China). Positive clones were sequenced (Sangon Bioengineering Co., Ltd., Changsha, China) and analyzed. The confirmed gene fragment was digested with BamHI (Takara Biotechnology Incorporated, Yokkaichi, Japan) and inserted into the pCAMBIA1305-GFP vector using homologous recombination (Genesand Biotechnology Co., Ltd., Beijing, China). Primer and gene sequences are provided in Supplemental Table S2.
2.8.2. Sequence Alignment and Phylogenetic Analysis
Sequence alignment and phylogenetic analyses were performed using clustalw (www.genome.jp/tools-bin/clustalw, accessed on 6 April 2025), and multiple sequence alignment maps were drawn using SnapGene 6 software. The conserved structural domains of LcDREB-43 protein and AP2/ERF transcription factor proteins of other plant species were identified using MEME (https://meme-suite.org/meme/tools/meme, accessed on 6 April 2025). Phylogenetic analyses were constructed using the neighbor-joining method with 1000 bootstrap replicates in MEGA 7 software.
2.8.3. Identification of Root Systems in LcDREB-43 Transgenic Plants
To further validate the feasibility of the genetic transformation system of L. chinense in a non-tissue culture transformation environment, we used a recombinant plasmid of LcDREB-43 transcription factor (pCAMBIA1305-LcDREB-43-GFP) to infest ‘Xiangnong Xiangyun’ plants with A. rhizogenes strain K599, following the optimized protocol established previously. The plants were infested with K599 Agrobacterium root-forming bacteria using the optimal combination of MES solution, OD600 = 0.8, and 15 min of infestation time as determined in the previous stage.
Firstly, the phenotype of the plants was observed, and the green fluorescence was observed under the fluorescence microscope, and then the DNA and RNA of the transgenic root lines successfully infested with pCAMBIA1305-GFP and pCAMBIA1305-LcDREB-43-GFP were extracted using WT as the control for identification, and the DNA and RNA of the transgenic root lines successfully infested with pCAMBIA1305-LcDREB-43-GFP were extracted using WT as the control for identification. The DNA and RNA extraction kits employed were identical to those utilized in Section 2.7 and Section 2.8.1. The primers for rolB and GFP are consistent with the previous section; qRT-PCR was performed according to the previously described method, and all experiments were repeated three times. Relative gene expression was calculated using the 2−ΔΔCT method. The primers used for qRT-PCR are shown in Supplemental Table S2. The fluorescence quantitative PCR reaction system was as follows: qPCR SYBR: 5 μL, template DNA: 0.5 μL, forward primer: 0.2 μL, reverse primer: 0.2 μL, and make up H2O to 10 μL. The fluorescence quantitative PCR reaction procedure was as follows: pre-denaturation at 95 °C for 30 s, (denaturation at 95 °C for 5 s, annealing at 60 °C for 30 s) for 40 cycles; 65 °C for 5 s, 95 °C for 5 min.
Anthocyanin extraction was performed using a Plant Anthocyanin Content Assay Kit (Sino Best Biological Technology Co., Ltd., Shanghai, China), with all experimental treatments replicated three times. Extraction protocols and quantification algorithms strictly adhered to the manufacturer’s guidelines to ensure methodological standardization and analytical reproducibility of the anthocyanin profiling data.
2.9. Data Processing
Experimental data were analyzed using SPSS 26 software. Analysis of variance (ANOVA) was used to assess the effects of the different factors, with a significance level set at p ≤ 0.05. Excel 2019 was used to create a three-line table, Graph Pad Prism 10 was used for graphing, and Adobe Photoshop 2022 and Adobe Illustrator 2010 were used for modifying and stitching the images.
3. Results
3.1. Establishment of Rooting Agrobacterium-Mediated L. chinense Transformation System
To establish an A. rhizogenes-mediated hairy root transformation system for L. chinense, various strains were tested on cuttings of two cultivars, ‘Xiangnong Xiangyun’ and ‘Hei Zhenzhu’. Hairy root formation was observed in most infected cuttings (Figure 1), with strain K599 demonstrating the highest induction efficiency. Under identical conditions, cuttings of ‘Xiangnong Xiangyun’ showed significantly higher survival rates (60%), rooting rates (51.66%), and hairy root induction efficiency (45%) compared to ‘Hei Zhenzhu’, which exhibited survival rates of 36.66%, rooting rates of 25%, and hairy root induction efficiency of 10% (Table 3). These results highlight the superior compatibility of strain K599 with both varieties, particularly ‘Xiangnong Xiangyun’, which consistently achieved the best outcomes across all evaluated parameters. The enhanced performance of ‘Xiangnong Xiangyun’ suggests it is a more suitable candidate for further transformation studies. Consequently, strain K599 and ‘Xiangnong Xiangyun’ were selected for subsequent optimization and genetic transformation experiments.

Figure 1.
Growth conditions of adventitious roots in L. chinense cultivars ‘Xiangnong Xiang yun’ and ‘Hei Zhenzhu’ After Agrobacterium Infection. (a) L. chinense ‘Xiangnong Xiangyun’ cultivar, WT are wild-type plants without Agrobacterium rooting treatment, and TR are grossly treated transgenic plants. (b) L. chinense ‘Hei Zhenzhu’ cultivar, WT are wild-type plants without Agrobacterium rooting treatment, and TR are grossly treated transgenic plants. (c) Close-up view of hairy roots induced in ‘Xiangnong Xiangyun’ following infection treatment. (d) Close-up view of hairy roots induced in ‘Hei Zhenzhu’ following infection treatment.

Table 3.
Induction of hairy roots in rhizomes of L. chinense by different cultivars and strains.
3.2. Optimization of the Agrobacterium-Mediated Rooting Transformation System of L. chinense
To optimize the A. rhizogenes-mediated genetic transformation system for L. chinense, orthogonal experiments were conducted to evaluate the effects of key factors, including infiltration solution composition, bacterial concentration, and infiltration duration, on transformation efficiency. The results (Table 4) revealed significant variations in the survival rate, rooting rate, induction efficiency, and transformation efficiency under different treatments. The highest survival rate (91.67%) was achieved using a bacterial infiltration solution with a bacterial concentration of OD600 = 0.4 and an infiltration duration of 15 min. Optimal rooting rates (70%) were observed with MS liquid medium and MES solution as infiltration solutions, paired with a bacterial concentration of OD600 = 0.8 and infiltration durations of 30 min and 15 min, respectively. The highest hairy root induction efficiency (61.67%) and transformation rate (43.33%) were both achieved using MES solution, a bacterial concentration of OD600 = 0.8, and a 15 min infiltration time. These findings demonstrate that the composition of the infiltration solution, bacterial density, and infiltration duration are critical factors influencing the efficiency of L. chinense transformation, providing a robust foundation for further refinement and application of this system.

Table 4.
Results of three-factor, three-level orthogonal experiment.
3.3. Polar Analysis and Optimal Conditions for L. chinense Transformation
Polar analysis (Table 5) revealed that the factors influencing the survival rate of L. chinense were ranked in order of importance as follows: infiltration time (C) > infiltration solution composition (A) > bacterial concentration (B). Among these factors, infiltration time had the most pronounced effect, with a 15 min treatment achieving the highest survival rate of 88%. However, extending the infiltration time to 60 min significantly reduced the survival rate to 69.67%.

Table 5.
Polar analysis of results of orthogonal experiments.
Similarly, polar analysis of the rooting rate, induction efficiency, and transformation efficiency identified infiltration time as the most critical factor, followed by bacterial concentration, while the infiltration solution composition had the least influence. The highest rooting rate of 61.67% was observed under a 15 min infiltration time, whereas the 60 min treatment resulted in the lowest rooting rate of 34%. The induction and transformation efficiency followed a similar trend, with the 15 min infiltration achieving the highest values at 46.33% and 31.67%, respectively. These values dropped markedly to 22.33% and 12.67%, respectively, when the infiltration time was extended to 60 min. These findings highlight the pivotal role of infiltration time in optimizing the transformation efficiency of L. chinense.
The analysis of variance (Table 6) revealed that infiltration time significantly influenced survival rate, rooting rate, induction efficiency, and transformation efficiency. Although the composition of the infiltration solution did not have a statistically significant effect on any of the evaluated indices, bacterial concentration had a notable impact on rooting and induction efficiency. Based on the polar analysis of the orthogonal experimental design, the optimal transformation conditions were identified as MES solution as the infiltration medium, bacterial concentration at OD600 = 0.8, and an infiltration duration of 15 min. These conditions consistently yielded the highest survival rate, rooting rate, induction efficiency, and transformation efficiency, underscoring their robustness and effectiveness in optimizing the transformation process for L. chinense.

Table 6.
Analysis of variance of the results of orthogonal experiment.
3.4. Identification of Transgenic Hairy Roots in L. chinense
Transgenic hairy roots were successfully induced in L. chinense cuttings using A. rhizogenes (Figure 2a). Fluorescence microscopy (Figure 2b) revealed a strong green fluorescent signal in the transgenic roots, confirming the successful expression of the GFP reporter gene. Morphological comparisons showed that transgenic hairy roots (Figure 2d) exhibited significantly enhanced branching and accelerated growth compared to the wild-type (L. chinense) root system (Figure 2c), demonstrating the efficiency of A. rhizogenes-mediated transformation in producing genetically modified roots.
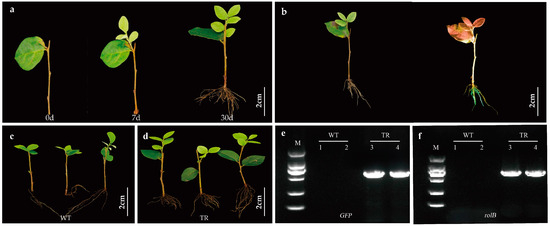
Figure 2.
Detection of transgenic hairy roots of L. chinense. (a) One-month root growth of ‘Xiangnong Xiangyun’ plants. (b) ‘Xiangnong Xiangyun’ plants under natural light (left), ‘Xiangnong Xiangyun’ plants stimulated by portable fluorescent protein detector (right). (c) WT is untransformed roots of ‘Xiangnong Xiangyun’ plants, as negative controls. (d) TR is the hairy root induced by infection. (e) PCR for GFP gene L. chinense hairy roots. (f) PCR for rolB gene in L. chinense hairy roots.
To verify the transgenic nature of the hairy roots, molecular analysis was conducted through PCR amplification. Genomic DNA from transgenic and non-transformed roots was tested for the rolB gene, a marker located in the T-DNA region of A. rhizogenes. The rolB gene was successfully amplified in transgenic hairy roots but not in non-transformed samples (Figure 2e). Additional PCR analysis using GFP-specific primers confirmed the integration of the GFP gene, as indicated by the presence of the expected GFP-specific amplification band in transgenic hairy roots, which was absent in control samples. These results conclusively demonstrate the successful integration of exogenous genes into L. chinense through A. rhizogenes-mediated transformation (Figure 2f).
3.5. Application of a Genetic Transformation System for L. chinense Under Non-Tissue Culture Sequence Alignment and Phylogenetic Analysis
LcDREB-43 has an open reading frame of 870 bp and is predicted to encode 289 amino acids. The theoretical isoelectric point (pI) of LcDREB-43 is 6.80. The molecular weight of LcDREB-43 was calculated to be 32.63 kDa. Sequence comparison analyses showed that LcDREB-43 and proteins from other plants contain a conserved structural domain of homologous structural domains (AP2) (Figure 3a,b). The amino acid sequence of LcDREB-43 was analyzed by multiple sequence comparison using the neighbor-joining method (NJ) to construct a phylogenetic tree. The results showed that LcDREB-43 was highly homologous to the amino acid sequences of AtRAP2-4 and AtRAP2-13 (Figure 3c).
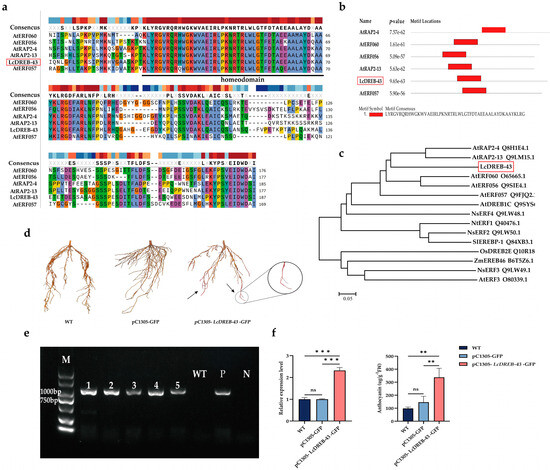
Figure 3.
Validation of LcDREB-43 gene in L. chinense ‘Xiangnong Xiangyun’ cultivar. (a) Multiple sequence alignment. (b) Conserved domain of LcDREB-43 and five orthologous proteins. The orthologous proteins were Arabidopsis thaliana (AtRAP2-4), Arabidopsis thaliana (AtERF060), Arabidopsis thaliana (AtERF056), Arabidopsis thaliana (AtRAP2-13), Arabidopsis thaliana (AtERF057). (c) Phylogenetic tree analysis by the neighbor-joining method was performed on the amino acid sequence comparison of the LcDREB-43 protein with proteins from other plant species. AtRAP2-4 (Arabidopsis thaliana, Q8H1E4.1), AtRAP2-13 (Arabidopsis thaliana, Q9LM15.1), AtERF060 (Arabidopsis thaliana, O65665.1), AtERF056 (Arabidopsis thaliana, Q9SIE4.1), AtERF057 (Arabidopsis thaliana, Q9FJQ2.1), AtDREB1C (Arabidopsis thaliana, Q9SYS6.2), NsERF4 (Nicotiana sylvestris, Q9LW48.1), NtERF1 Q40476.1, NsERF2 Q9LW50.1, SIEREBP-1 (Solanum lycopersicum, Q84XB3.1), OsDREB2E (Oryza sativa Japonica Group, Q10R18.1), ZmEREB46 (Zea mays, B6T5Z6.1), NsERF3 (Nicotiana sylvestris, Q9LW49.1), AtERF3 (Arabidopsis thaliana, O80339.1). (d) Phenotypic observations of the root system of transgenic plants. WT is the untreated root system, which is the control; pC1305-GFP is pCAMBIA1305-GFP; and pC1305-LcDREB-43-GFP is pCAMBIA1305-LcDREB-43-GFP, which is the treatment group. (e) Root identification of transgenic plants. The overexpression plant transformed roots of ‘Xiangnong Xiangyun’ for 1–5; WT is negative control, P is positive control, and N is null control. (f) Differential gene expression levels of LcDREB-43 in explant roots and determination of anthocyanin content. WT is the untreated root system, which is the control; pC1305-GFP is pCAMBIA1305-GFP; and pC1305-43 is pCAMBIA1305-LcDREB-43-GFP. ns indicates p > 0.05, ** indicates significance at p < 0.01, and *** indicates significance at p < 0.001.
3.6. Identification of LcDREB-43 Transgenic Plant
The cloning of the LcDREB-43 gene and the construction of the overexpression vector are shown in the Supplementary Figure (Supplemental Figure S1). The plasmid pCAMBIA1305-LcDREB-43-GFP was introduced into A. rhizogenes strain K599 and used for root transformation. Phenotypic observations revealed distinct rosy red coloration in the root tips of treated plants, in contrast to the yellowish-brown color of wild-type (WT) roots (Figure 3d). This color change was exclusively observed in successfully transformed plants. Microscopic examination of root cross-sections further confirmed significant accumulation of anthocyanin in the root cells of the pC1305-LcDREB-43-GFP treatment group (Supplementary Figure S2).
Subsequently, genomic DNA was extracted from the roots of transformed plants for molecular identification, with wild-type (WT) plants serving as the negative control, the gene plasmid (P) as the positive control, and water (N) as the null control. The results (Figure 3e) demonstrated that the WT samples failed to amplify the overexpression band of the LcDREB-43 (pCAMBIA1305-LcDREB-43-GFP) gene, whereas only the roots of successfully infected positive plants produced the expected target gene band (approximately 1000 bp). Furthermore, only the transformed positive roots were able to amplify the target gene (pCAMBIA1305-LcDREB-43-GFP) band (Figure 3e). RNA was subsequently extracted from the positive roots of transformed plants to analyze the relative expression level of the LcDREB-43 overexpression gene (pCAMBIA1305-LcDREB-43-GFP). The results (Figure 3f) revealed that the expression level of LcDREB-43 was significantly higher in transformed plants compared to WT and pCAMBIA1305-GFP controls.
To further validate the transformation, anthocyanin content was measured in the positive root materials from each treatment. The results (Figure 3f) indicated that the anthocyanin content in roots overexpressing LcDREB-43 (337.34 µg/g) was significantly higher than in WT (97.97 µg/g) and pCAMBIA1305-GFP controls (146.13 µg/g). These findings confirm the successful integration of the LcDREB-43 gene into the root system of L. chinense ‘Xiangnong Xiangyun’ plants.
In summary, the LcDREB-43 overexpression construct (pCAMBIA1305-LcDREB-43-GFP) was utilized to validate the transformation system. Through sequential phenotypic observation, molecular identification, and quantification of anthocyanin content, the feasibility of this system was successfully demonstrated in a non-tissue culture environment. Furthermore, this system has been applied in practical production to enhance the biosynthesis of secondary metabolites, particularly anthocyanin, highlighting its potential for industrial and agricultural applications.
4. Discussion
The traditional genetic transformation of woody plants often relies on in vitro culture techniques. However, previous studies on L. chinense have highlighted significant limitations in the regeneration system, including inefficiencies that fail to meet the needs of genetic transformation [6,30]. In vitro culture techniques require high technical expertise, extended cultivation periods, and significant resource investment, further complicating their application. To date, a comprehensive genetic transformation system for L. chinense has not been established, and no tissue culture-independent transformation methods have been reported for this species.
Recently, non-tissue culture-based transformation techniques have been successfully applied in woody plants such as Carya illinoinensis [21], Citrus sinensis [22], Phoebe bournei [23], Prunus persica [24], and populus euphratica [31]. These advancements provide a valuable reference for developing similar systems in L. chinense. A. rhizogenes-mediated genetic transformation offers notable advantages for woody plants, given its simplicity and efficiency. In this study, L. chinense floral spikes were used as explants, with root induction achieved by directly submerging the spikes into the infiltration solution [32,33]. This straightforward approach bypasses tissue culture, significantly reducing costs, labor, and time requirements. Furthermore, the method is highly efficient, offering a practical and scalable solution for the genetic improvement of L. chinense.
A. rhizogenes-mediated hairy roots are widely employed in plant metabolic pathway studies due to their rapid growth, genetic stability, and ability to produce secondary metabolites [34,35,36]. Traditionally, non-model plants require heterologous validation in model systems. However, the genetic transformation system developed in this study provides an essential tool for homologous gene validation in L. chinense, offering greater accuracy and relevance for functional studies.
The successful establishment of this system depended on multiple factors, including bacterial strain, plant genotype, infiltration solution composition, bacterial concentration, and infiltration duration [37,38,39]. Among the strains tested, K599 exhibited the highest transformation efficiency for both ‘Xiangnong Xiangyun’ and ‘Hei Zhenzhu’ varieties, with a rooting rate and hairy root induction efficiency of 51.7% and 45% for ‘Xiangnong Xiangyun’, respectively. This finding aligns with prior studies on K599’s efficacy in transforming other woody plants, such as P. euphratica [31], P. bournei [23], Caragana intermedia [40], and Vicia faba [41], underscoring its broad applicability. Plant genotype also played a critical role in transformation efficiency, with ‘Xiangnong Xiangyun’ consistently outperforming ‘Hei Zhenzhu’. This variability reflects the influence of genotype on Agrobacterium-mediated transformation, as reported in other studies [42].
Bacterial concentration significantly influenced transformation outcomes. The optimal OD600 value of 0.8 achieved the highest rooting rates and transformation efficiency, consistent with studies in Ziziphus jujuba [43] and peach [24]. However, species-specific differences in response to bacterial concentration were observed, as other studies found an OD600 value of 0.2 optimal for Vernicia fordii [44]. Infiltration duration was identified as the most critical factor affecting survival rate, rooting rate, and transformation efficiency [45]. The 15 min infiltration duration yielded the highest induction and transformation rates of 46.33% and 31.67%, respectively, while extended durations reduction efficiency, consistent with findings in P. persica [24]. Although the composition of the infiltration solution was less significant, the MES solution optimized through orthogonal experiments demonstrated superior results, achieving a rooting rate, induction, and transformation efficiency of 70%, 61.67%, and 43.33%, respectively.
Based on phylogenetic analysis, the LcDREB-43 protein exhibits closer similarity to two members of the DREB subfamily A-6 in Arabidopsis thaliana (AtRAP2-4 and AtRAP2-13). Multiple sequence alignment and conserved motif analysis further confirmed that the LcDREB-43 protein contains a highly conserved DNA-binding domain (AP2). Although DREB transcription factors were initially widely studied for their roles in drought, high salinity, and low-temperature stress, recent research has shown that certain members of the DREB subfamily are also involved in plant responses to high-temperature stress [46]. Additionally, they may enhance plant thermotolerance by regulating secondary metabolic pathways. In existing studies, transcriptomic data of DREB subfamily genes in Brassica napus revealed that the expression level of BrDREB2B increases under high-temperature stress. Transgenic Arabidopsis overexpressing BrDREB2B exhibited significantly higher germination rates, root lengths, and survival rates under polyethylene glycol (PEG), NaCl, and 40 °C heat stress treatments compared to wild-type plants, thereby improving thermotolerance, salt tolerance, and drought tolerance in transgenic Arabidopsis [47]. Similarly, CpRAP2.4 and CpRAP2.4b from Carica papaya L. demonstrated high levels of tolerance to cold and heat stress in transgenic tobacco [48].
Anthocyanin is a natural pigment that serves as a crucial natural antioxidant capable of scavenging free radicals. The diverse compounds they contain are of significant importance in healthcare. Although existing research has extensively explored the ethylene-mediated regulation of anthocyanin biosynthesis in plants through ethylene response factors (ERFs) [49], for LhERF061, a member of the DREB subfamily A-6, this transcription factor has also been shown to respond to ethylene by inhibiting anthocyanin biosynthesis. Specifically, it negatively regulates anthocyanin accumulation by suppressing the expression of key anthocyanin synthesis genes, such as LhMYBSPLATTER and LhDFR [50]. LcDREB-43, identified as a heat stress-responsive factor in our previous research, was also found to promote anthocyanin synthesis through preliminary analyses. In this experiment, independent of heat stress conditions, the overexpression of the LcDREB-43 gene in ‘Xiangnong Xiangyun’ plants resulted in a significant accumulation of anthocyanin in the roots. Furthermore, the extraction and quantification of anthocyanin content in treated roots revealed that LcDREB-43 overexpression lines exhibited markedly higher levels compared to the wild-type (WT) controls. The molecular mechanisms underlying these observations require further investigation. This novel system provides a foundation for future research into gene function, molecular breeding, and secondary metabolite production in L. chinense.
5. Conclusions
This study established a non-tissue culture-based A. rhizogenes-mediated genetic transformation system for L. chinense, providing a rapid, cost-effective, and scalable method for genetic studies. Through optimization of transformation conditions, including the use of MES solution, a bacterial concentration of OD600 = 0.8, and a 15 min infiltration duration, the system achieved high transformation efficiency with rooting, induction, and transformation rates of 70%, 61.67%, and 43.33%, respectively. Validation using the pCAMBIA1305-LcDREB-43-GFP gene confirmed the system’s effectiveness, with transgenic roots displaying significant anthocyanin accumulation and elevated gene expression levels compared to controls. This efficient and tissue culture-independent transformation platform offers a valuable tool for functional genomics, molecular breeding, and germplasm innovation in L. chinense and other woody plants, paving the way for further advancements in plant biotechnology.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/horticulturae11040404/s1. Table S1: PCR primer sequences. Table S2: PCR and qRT-PCR primer sequences. Figure S1: Cloning of genes and vector construction. Figure S2: Cross-sectional view of the root system in L. chinense ‘Xiangnong Xiangyun’ cultivar.
Author Contributions
The authors acknowledge their contributions to the paper as follows: study conception and design: Y.L. (Yanlin Li), X.Y. and X.X.; data collection: T.L., Y.L. (Yang Liu 1) and W.T.; analysis and interpretation of results: Y.L. (Yang Liu 2), D.Z. and C.X.; draft manuscript preparation: T.L. and Y.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Natural Science Foundation of Hunan Province (2024JJ5178); the Natural Science Foundation of Changsha, Hunan Province (kq2402112); the Outstanding Youth Fund of Hunan Forestry Bureau (XLKJ202205); the Key Project of Hunan Provincial Department of Education (22A0155); the Scientific and Technological Tackling Project of Hunan Forestry Bureau (XLK202437); and the Innovative Project of Postgraduate Students of Hunan Province (2023XC108).
Data Availability Statement
Data is contained within the article and Supplementary Material.
Acknowledgments
We acknowledge the editors and all anonymous reviewers for their constructive comments on this manuscript.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- Dong, S.; Ma, J.; Mo, Y.; Li, L. Review of Research on Germplasm Resources and Application of Loropetalum chinense car. rubrum. Guangxi For. Sci. 2022, 51, 290–297. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, Y.; Wu, Z. Study of medical value of Loropetalum chinense. Chin. J. Tradit. Chin. Med. 2014, 29, 2283–2286. [Google Scholar]
- Xu, M.; Ding, Y. Lorpetalum chindense var. rubrum’s Application in Landscape. Chin. Hortic. Abstr. 2010, 26, 92–93+127. [Google Scholar]
- Zhang, X.; Zhang, D.; Zhang, L.; Wang, X.; Xiong, X.; Gan, D.; Yu, X.; Li, Y. The Whole Genome Analysis of Loropetalum chinense var. Rubrum. Mol. Plant Breed. 2020, 18, 7023–7029. [Google Scholar] [CrossRef]
- Fu, H.; Yu, X.; Chen, J.; Shi, A.; Ren, Y. Adventitious Bud Regeneration from Leaves of Loropetalum chinense var. rubrum Cultivars. Chin. Agric. Sci. Bull. 2014, 30, 57–61. [Google Scholar]
- Chen, R.; Zhen, Y.; Zhang, D.; Chen, L.; Gong, J.; Zhou, W.; He, H.; Qin, J. Establishment of high-efficient regeneration system of Loropetalum chinense var. rubrum. J. Cent. South Univ. For. Technol. 2015, 35, 40–45. [Google Scholar] [CrossRef]
- Liu, Z.; Cao, C. Vitro Propagation Techniques of Loropetalum chinense var. rubrum. West. For. Sci. 2018, 47, 80–84. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, D.; Cui, X.; Yu, F. Application research on hairy roots in plants. Food Ferment. Ind. 2023, 1–11. [Google Scholar] [CrossRef]
- Zhang, L.; Luo, Z.; Zhang, X.; Sun, M. Characteristics and Application of Ri Plasmid of Agrobacterium rhizogenes. Appl. Agric. Sci. Anhui Prov. 2010, 38, 8183–8185. [Google Scholar] [CrossRef]
- Xiong, S.; Shu, R.; Zhang, L.; Meng, L.; Yu, Y.; Ni, Z. Research progress and application of transgenic plant Agrobacterium rhizogenes. Agric. Technol. 2017, 37, 72–74. [Google Scholar] [CrossRef]
- Zhu, Y.; Zhu, X.; Wen, Y.; Wang, L.; Wang, Y.; Liao, C.; Zhao, M.; Li, T.; Liu, D.; Li, B.; et al. Plant Hairy Roots: Induction, Applications, Limitations and Prospects. Ind. Crops Prod. 2024, 219, 119104. [Google Scholar] [CrossRef]
- Tsuro, M.; Kubo, T.; Shizukawa, Y.; Takemoto, T.; Inaba, K. Agrobacterium rhizogenes Is a Useful Transporter for Introducing T-DNA of the Binary Plasmid into the Chrysanthemum, Dendranthema Grandiflorum Kitamura, Genome. Plant Cell Tiss. Organ Cult. 2005, 81, 175–181. [Google Scholar] [CrossRef]
- Beigmohammadi, M.; Sharafi, A.; Jafari, S. An Optimized Protocol for Agrobacterium rhizogenes-Mediated Genetic Transformation of Citrullus Colocynthis. J. Apple Biotechnol. Rep. 2019, 6, 113–117. [Google Scholar] [CrossRef]
- Habibi, P.; De Sa, M.F.G.; Da Silva, A.L.L.; Makhzoum, A.; Da Luz Costa, J.; Borghetti, I.A.; Soccol, C.R. Efficient Genetic Transformation and Regeneration System from Hairy Root of Origanum Vulgare. Physiol. Mol. Biol. Plants 2016, 22, 271–277. [Google Scholar] [CrossRef]
- Chen, L.; Cai, Y.; Liu, X.; Guo, C.; Sun, S.; Wu, C.; Jiang, B.; Han, T.; Hou, W. Soybean Hairy Roots Produced in Vitro by Agrobacterium Rhizogenes-Mediated Transformation. Crop J. 2018, 6, 162–171. [Google Scholar] [CrossRef]
- Guo, S. Study on Leaf Regeneration and Genetic Transformation System of Qiuzi Pear and ‘Conference’ Pear. Master’s Thesis, Shandong Agricultural University, Tai’an, China, 2023. [Google Scholar]
- Liu, Y. Rapid Propagation of ‘Luhanliu No.1’ by Tissue Culture and Establishment of Genetic Transformation System Mediated by Agrobacterium rhizogenes. Master’s Thesis, Shandong Agricultural University, Tai’an, China, 2023. [Google Scholar]
- Wang, Y. Establishment of Agrobacterium rhizogenes Mediated Hairy Root Genetic Transformation System in Malus Domestica. Master’s Thesis, Northwest A&F University, Xianyang, China, 2023. [Google Scholar]
- Irigoyen, S.; Ramasamy, M.; Pant, S.; Niraula, P.; Bedre, R.; Gurung, M.; Rossi, D.; Laughlin, C.; Gorman, Z.; Achor, D.; et al. Plant Hairy Roots Enable High Throughput Identification of Antimicrobials against Candidatus Liberibacter Spp. Nat. Commun. 2020, 11, 5802. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Wang, X.; Cao, L.; Ji, J.; Liu, T.; Duan, K. Highly Efficient Agrobacterium Rhizogenes-Mediated Hairy Root Transformation for Gene Functional and Gene Editing Analysis in Soybean. Plant Methods 2021, 17, 73. [Google Scholar] [CrossRef]
- Xie, X.; Huang, Q.; Weng, G.; Yuan, H.; He, W.; Yan, D.; Huang, J.; Wang, X.; Zheng, B. Construction of Agrobacterium rhizogenes- mediated transformation system of Carya illinoinensis without dependence on tissue culture. J. Fruit Sci. 2022, 39, 131–140. [Google Scholar] [CrossRef]
- Ma, H.; Meng, X.; Xu, K.; Li, M.; Gmitter, F.G.; Liu, N.; Gai, Y.; Huang, S.; Wang, M.; Wang, M.; et al. Highly Efficient Hairy Root Genetic Transformation and Applications in Citrus. Front. Plant Sci. 2022, 13, 1039094. [Google Scholar] [CrossRef]
- Wu, M.; Hong, J.; Li, F.; Zhou, S.; Lin, R.; Cheng, L. Construction and optimization of genetic transformation system of Phoebe bournei mediated by Agrobacterium rhizogenes. J. Nucl. Agric. Sci. 2023, 37, 1516–1522. [Google Scholar] [CrossRef]
- Ju, L. Initial Exploration of Peach Non-Graft Transformation System. Master’s Thesis, Huazhong Agricultural University, Wuhan, China, 2023. [Google Scholar]
- Lan, M.; Hong, M.; Xiao, M.; Li, C.; Pan, H.; Zhang, Y.; Lu, L.; Hou, L.; Ge, R.; Wu, W.; et al. Research Progress of AP2/ERF Transcription Factors Participating in Plant Secondary Metabolism and Stress Response. J. Plant Genet. Resour. 2023, 24, 1223–1235. [Google Scholar] [CrossRef]
- Ni, J.; Bai, S.; Zhao, Y.; Qian, M.; Tao, R.; Yin, L.; Gao, L.; Teng, Y. Ethylene Response Factors Pp4ERF24 and Pp12ERF96 Regulate Blue Light-Induced Anthocyanin Biosynthesis in ‘Red Zaosu’ Pear Fruits by Interacting with MYB114. Plant Mol. Biol. 2019, 99, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Koyama, T.; Sato, F. The Function of ETHYLENE RESPONSE FACTOR Genes in the Light-Induced Anthocyanin Production of Arabidopsis Thaliana Leaves. Plant Biotechnol. 2018, 35, 87–91. [Google Scholar] [CrossRef]
- Zhao, C.; Liu, X.; Gong, Q.; Cao, J.; Shen, W.; Yin, X.; Grierson, D.; Zhang, B.; Xu, C.; Li, X.; et al. Three AP2/ERF Family Members Modulate Flavonoid Synthesis by Regulating Type IV Chalcone Isomerase in Citrus. Plant Biotechnol. J. 2021, 19, 671–688. [Google Scholar] [CrossRef] [PubMed]
- Cai, W. Physiological Response and Mechanism of Leaf Color Change of Loropetalum chinense Under Heat Stress. Master’s Thesis, Hunan Agricultural University, Changsha, China, 2023. [Google Scholar]
- Shi, A. Study on Leaf of Tissue Culture and Plantlet Regenration System of Loropetalum chinense var. rubrum. Master’s Thesis, Hunan Agricultural University, Changsha, China, 2015. [Google Scholar]
- Wang, D.; Tang, J.; Shao, M.; Zhang, W.; Wang, H. Poplar tissue culture leaf and spike hairy root occurrence. Chin. Bull. Bot. 2017, 52, 210–217. [Google Scholar]
- Cao, X.; Xie, H.; Song, M.; Lu, J.; Ma, P.; Huang, B.; Wang, M.; Tian, Y.; Chen, F.; Peng, J.; et al. Cut–Dip–Budding Delivery System Enables Genetic Modifications in Plants without Tissue Culture. Innovation 2023, 4, 100345. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Li, S.; Deng, S.; Wang, M.; Wu, Y.; Li, M.; Dong, J.; Lu, S.; Su, C.; Li, G.; et al. A Method of Genetic Transformation and Gene Editing of Succulents without Tissue Culture. Plant Biotechnol. J. 2024, 22, 1981–1988. [Google Scholar] [CrossRef]
- Li, B.; Wang, B.; Li, H.; Peng, L.; Ru, M.; Liang, Z.; Yan, X.; Zhu, Y. Establishment of Salvia Castanea Diels f. Tomentosa Stib. Hairy Root Cultures and the Promotion of Tanshinone Accumulation and Gene Expression with Ag+, Methyl Jasmonate, and Yeast Extract Elicitation. Protoplasma 2016, 253, 87–100. [Google Scholar] [CrossRef]
- Jiao, J.; Gai, Q.; Wang, W.; Zang, Y.; Niu, L.; Fu, Y.; Wang, X. Remarkable enhancement of flavonoid production in a co-cultivation system of Isatis tinctoria L. hairy root cultures and immobilized Aspergillus niger. Ind. Crops Prod. 2018, 112, 252–261. [Google Scholar] [CrossRef]
- Bahramnejad, B.; Naji, M.; Bose, R.; Jha, S. A Critical Review on Use of Agrobacterium Rhizogenes and Their Associated Binary Vectors for Plant Transformation. Biotechnol. Adv. 2019, 37, 107405. [Google Scholar] [CrossRef]
- Han, K.-H.; Meilan, R.; Ma, C.; Strauss, S.H. An Agrobacterium Tumefaciens Transformation Protocol Effective on a Variety of Cottonwood Hybrids (Genus Populus). Plant Cell Rep. 2000, 19, 315–320. [Google Scholar] [CrossRef]
- Luo, P.; Zhang, H.; Xu, J.; Hu, B.; Wang, X.; Li, G.; Fan, C. Establishment of Agrobacterium rhizogenes-Mediated Transgenic System for Eucalyptus tail. Plant Res. 2022, 42, 512–520. [Google Scholar] [CrossRef]
- Feng, Z.; Liu, N.; Bu, Y.; Zhang, G.; Wang, B.; Gong, Y. Establishment of Agrobacterium rhizogenes-Mediated Genetic Transformation System in Vegetable Pea. J. Hortic. 2024, 1–10. [Google Scholar] [CrossRef]
- Yang, C. Establishment of Genetic Transformation System of Hairy Roots of Caragana Intermedia and Its Optimization. Master’s Thesis, Inner Mongolia Agricultural University, Hohhot, China, 2022. [Google Scholar]
- Meng, D.; Yang, Q.; Dong, B.; Song, Z.; Niu, L.; Wang, L.; Cao, H.; Li, H.; Fu, Y. Development of an Efficient Root Transgenic System for Pigeon Pea and Its Application to Other Important Economically Plants. Plant Biotechnol. J. 2019, 17, 1804–1813. [Google Scholar] [CrossRef] [PubMed]
- Tao, N.; Li, M.; Guo, H. Optimization of the sweet potato genetic transformation system mediated by Agrobacterium rhizogenes. Biotechnol. Bull. 2023, 39, 1–9. [Google Scholar] [CrossRef]
- Hao, Z. Agrobacterium-Mediated Transformation of Chinese Jujube. Ph.D. Thesis, Hebei Agricultural University, Baoding, China, 2012. [Google Scholar]
- Jia, H. Root Transgenic System Technology of Tune Tree (Vernicia fordii). Master’s Thesis, Central South University of Forestry and Technology, Changsha, China, 2023. [Google Scholar]
- Gao, X. Establishment and Application of Agrobacterium-Mediated Walnut Genetic Transformation System. Master’s Thesis, Northwest Sci-Tech University of Agriculture and Forestry, Yangling, China, 2021. [Google Scholar]
- Han, F.; Hu, X.; Wang, N.; Xie, Y.; Wang, X.; Zhu, Q. Research Progress in Response of DREBs to Abiotic Stress in Plant. Biotechnol. Bull. 2023, 39, 1–13. [Google Scholar] [CrossRef]
- Wu, Y.; Zhang, L.; Nie, L.; Zheng, Y.; Zhu, S.; Hou, J.; Li, R.; Chen, G.; Tang, X.; Wang, C.; et al. Genome-Wide Analysis of the DREB Family Genes and Functional Identification of the Involvement of BrDREB2B in Abiotic Stress in Wucai (Brassica Campestris L.). BMC Genom. 2022, 23, 598. [Google Scholar] [CrossRef] [PubMed]
- Figueroa-Yañez, L.; Pereira-Santana, A.; Arroyo-Herrera, A.; Rodriguez-Corona, U.; Sanchez-Teyer, F.; Espadas-Alcocer, J.; Espadas-Gil, F.; Barredo-Pool, F.; Castaño, E.; Rodriguez-Zapata, L.C. RAP2.4a Is Transported through the Phloem to Regulate Cold and Heat Tolerance in Papaya Tree (Carica Papaya Cv. Maradol): Implications for Protection Against Abiotic Stress. PLoS ONE 2016, 11, e0165030. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, H.; Zhang, Z.; Zhao, Y.; Zhang, J. The Regulation of Ethylene Responsive Factors(ERFs) in Plant Anthocyanin Synthesis. J. Plant Genet. Resour. 2023, 24, 615–623. [Google Scholar] [CrossRef]
- Yang, Y.; Bi, M.; Luo, K.; Cao, Y.; Wang, J.; Yang, P.; Xu, L.; Ming, J. Lily (Lilium spp.) LhERF061 Suppresses Anthocyanin Biosynthesis by Inhibiting LhMYBSPLATTER and LhDFR Expression and Interacting with LhMYBSPLATTER. Plant Physiol. Biochem. 2025, 219, 109325. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).