Morphological Characteristics and Molecular Marker-Assisted Identification of Ovary Glabrous Phenotype in the Population of Nanchuan Dachashu (Camellia nanchuanica)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Morphological Characterization
2.3. Paraffin Section Observation
2.4. DNA Isolation and KASP Genotyping Assay
2.5. Data Analysis
3. Results
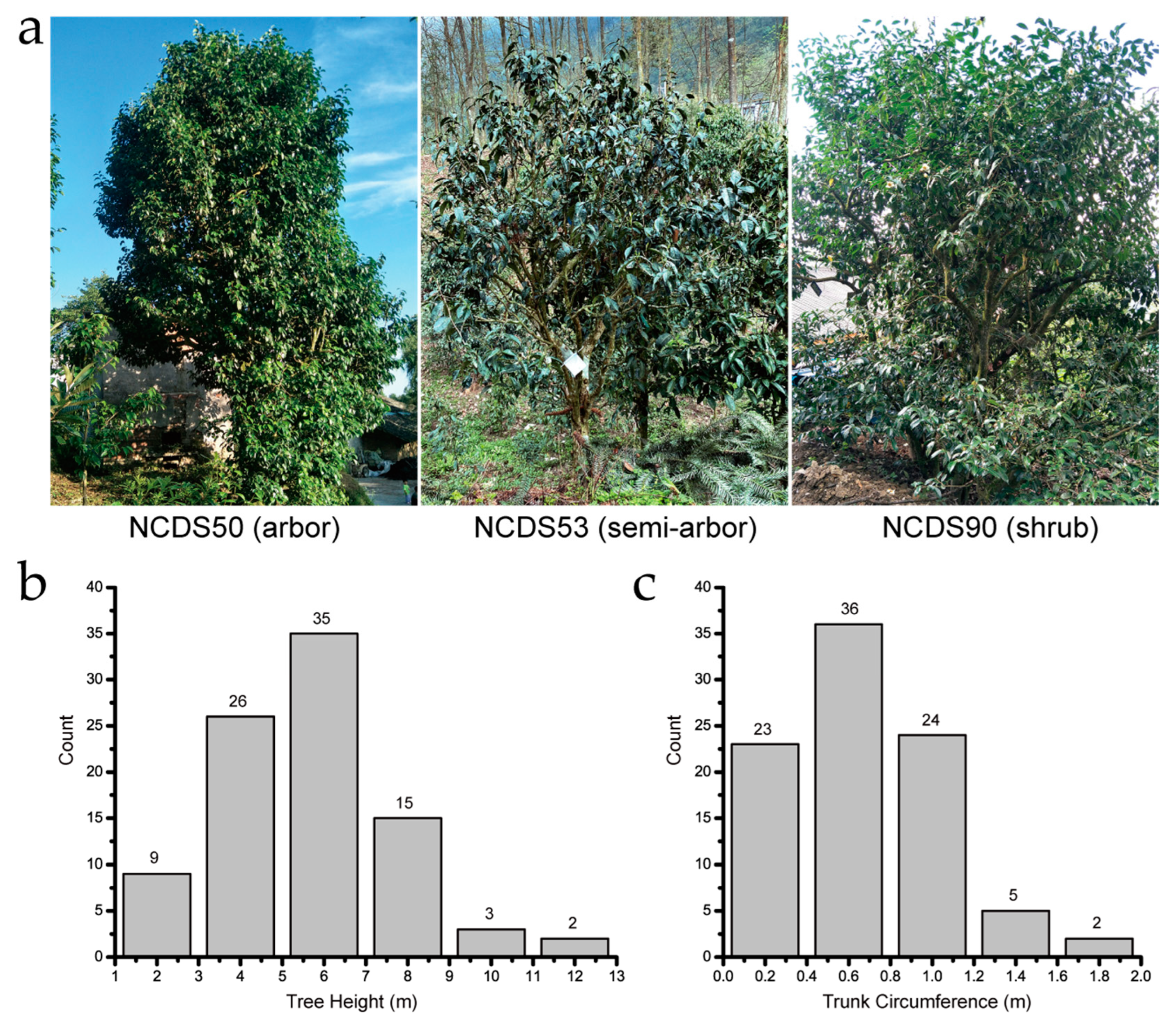
3.1. Natural Distribution and Basic Morphology of the Nanchuan Dachashu Population
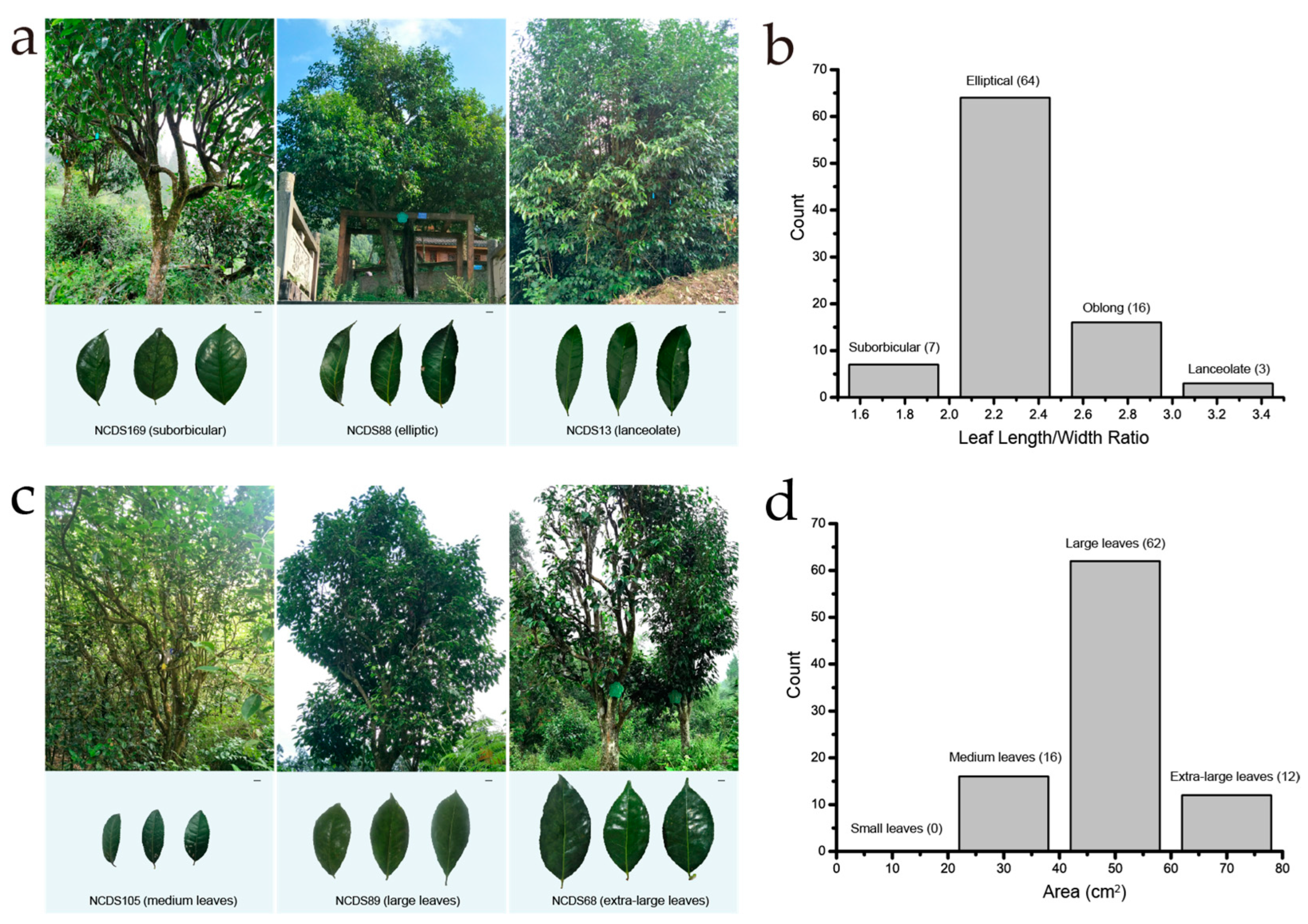
3.2. Leaf Characteristics of the Nanchuan Dachashu Population
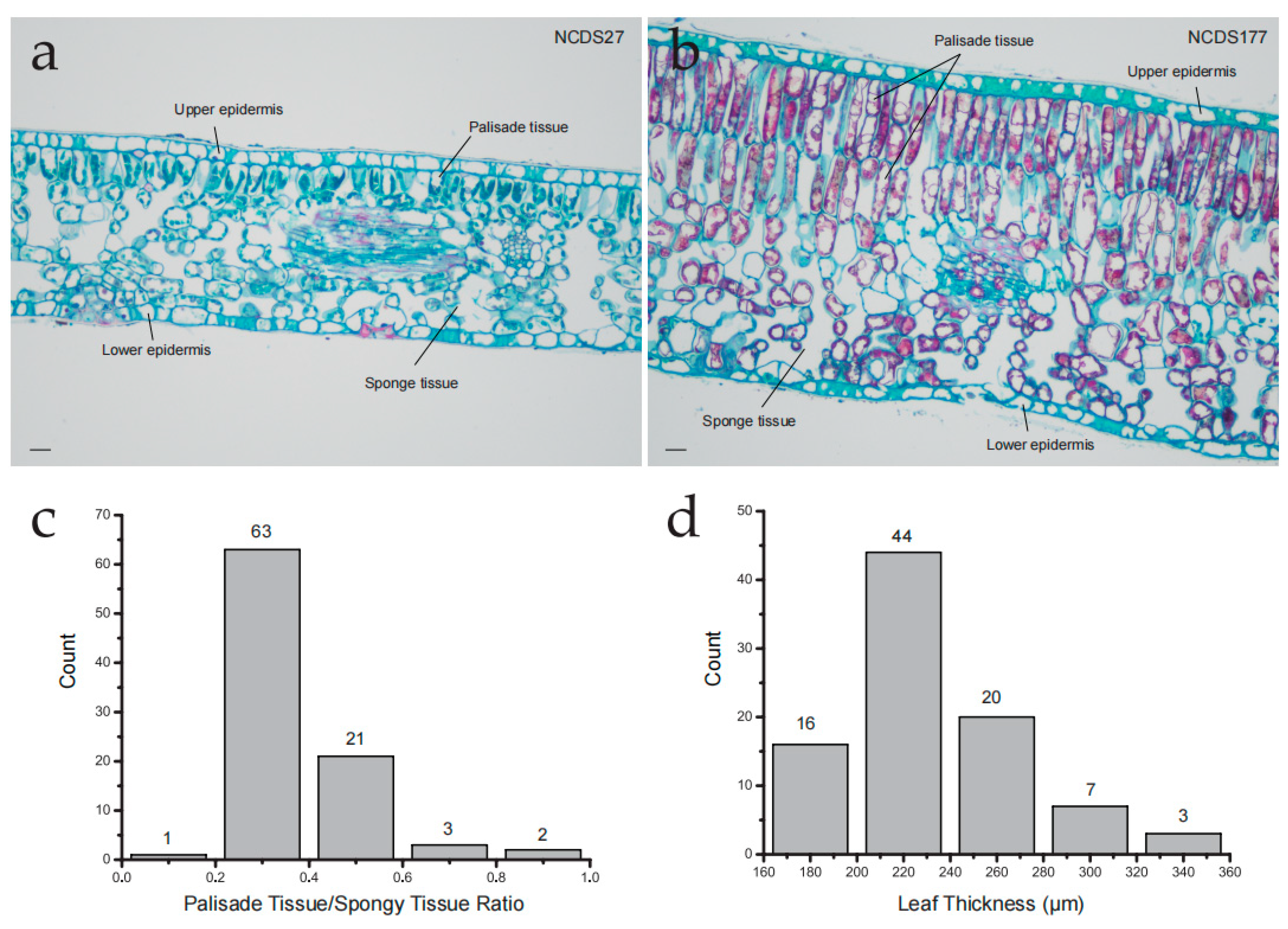
3.3. Cross Sectional Characteristics of Leaves in the Nanchuan Dachashu Population
3.4. Morphology of Ovaries in the Nanchuan Dachashu Population
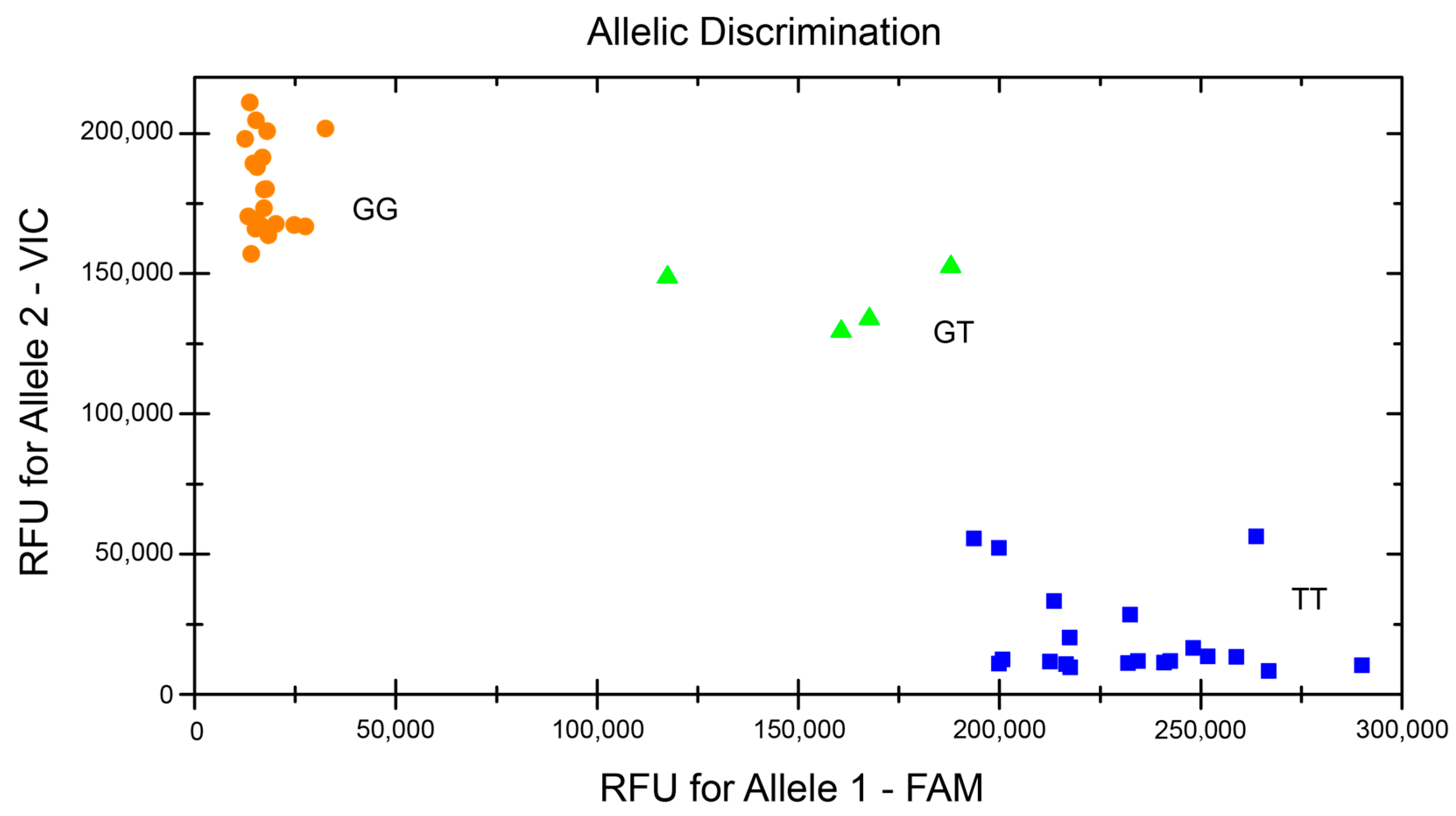
3.5. KASP Genotyping in the Nanchuan Dachashu Population
4. Discussion
4.1. The Nanchuan Dachashu Population Exhibits Rich Morphological Diversity
4.2. Leaf Anatomical Characteristics Provide Clues for Screening Stress-Resistant Germplasm
4.3. Development and Application of KASP Marker for Ovary Glabrous Trait
4.4. Candidate Gene Analysis in the Associated Region of Ovary Wall Trichomes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhao, D.W. Botany and taxonomy of tea (Camellia sinensis, Theaceae) and its relatives. In The Tea Plant Genome; Chen, L., Chen, J.D., Eds.; Springer Nature Singapore: Singapore, 2024; pp. 13–38. [Google Scholar]
- Chang, H.T. New species of Chinese Theaceae. Acta Sci. Natur. Univ. Sunyatseni 1990, 29, 85–87. [Google Scholar]
- Ming, T.L.; Bartholomew, B. Theaceae. In Flora of China; Wu, Z.Y., Raven, P.H., Hong, D.Y., Eds.; Science Press: Beijing, China; Missouri Botanical Garden Press: St. Louis, MO, USA, 2007; pp. 366–478. [Google Scholar]
- Fu, Z.L.; Zhang, S.R.; Li, F.; Chen, J.D.; Chen, L. Tea genetic resources: Diversity and conservation. In The Tea Plant Genome; Chen, L., Chen, J.D., Eds.; Springer Nature Singapore: Singapore, 2024; pp. 59–78. [Google Scholar]
- Wu, Z.; Li, W.; Wang, X.; Zhang, C.; Jiang, X.; Li, S.; Luo, L.; Sun, K.; Zeng, L. Genetic relationship analysis of the population germplasms of section Thea with unstable ovary locule number based on SLAF-seq. Acta Hortic. Sin. 2022, 49, 2455–2470. [Google Scholar]
- Wang, S.; Zhan, F.; Liang, T.; Jiao, C.; Zhang, J.; Liu, Q. Re-investigation of the resources of giant tea trees in Chongqing. Chin. Agric. Sci. Bull. 2003, 19, 87–90. [Google Scholar]
- Yang, S. Thinking on the taxonomy of Camellia sect. Thea. J. Tea Sci. 2021, 41, 439–453. [Google Scholar]
- Luo, Y.; Zhang, X.; Mao, J.; Li, H.; Liu, Q.; Tong, H.; Liang, G.; Wei, X. Analysis of genetic diversity in 35 ancient tea accessions from Chongqing with SSR markers. Mol. Plant Breed. 2019, 17, 6549–6557. [Google Scholar]
- Wang, H.; Suo, H.; Yang, J.; Liu, Q. Analysis of genetic diversity among wild tea plants in Sichuan and Chongqing by ISSR markers. J. Tea Sci. 2009, 29, 168–172. [Google Scholar]
- Wu, Z.; Liu, K.; Zhang, X.; Tang, Q.; Zeng, L. CsNYC1a mediates chlorophyll degradation and albino trait formation in the arbor-type tea plant Camellia nanchuanica. J. Agric. Food Chem. 2024, 72, 14087–14098. [Google Scholar] [CrossRef]
- Wei, F.; Luo, L.; Zeng, L. Characterization of key sweet taste compounds in Camellia nanchuanica black tea. LWT 2023, 182, 114858. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, K.; Tang, Q.; Zeng, L.; Wu, Z. Light intensity regulates low-temperature adaptability of tea plant through ROS stress and developmental programs. Int. J. Mol. Sci. 2023, 24, 9852. [Google Scholar] [CrossRef]
- Li, H.; Wei, X.; Tan, S.; Xu, R.; Liu, Q. Three novel wild tea in Chongqing and their status in botanical classification. J. Tea 2018, 44, 1–5. [Google Scholar]
- Li, J.-W.; Li, H.; Liu, Z.-W.; Wang, Y.-X.; Chen, Y.; Yang, N.; Hu, Z.-H.; Li, T.; Zhuang, J. Molecular markers in tea plant (Camellia sinensis): Applications to evolution, genetic identification, and molecular breeding. Plant Physiol. Biochem. 2023, 198, 107704. [Google Scholar]
- Su, M.H.; Hsieh, C.F.; Tsou, C.H. The confirmation of Camellia formosensis (Theaceae) as an independent species based on DNA sequence analyses. Bot. Stud. 2009, 50, 477–485. [Google Scholar]
- Liu, S.; Liu, H.; Wu, A.; Hou, Y.; An, Y.; Wei, C. Construction of fingerprinting for tea plant (Camellia sinensis) accessions using new genomic SSR markers. Mol. Breed. 2017, 37, 93. [Google Scholar]
- Schmid, K.J.; Sörensen, T.R.; Stracke, R.; Törjék, O.; Altmann, T.; Mitchell-Olds, T.; Weisshaar, B. Large-scale identification and analysis of genome-wide single-nucleotide polymorphisms for mapping in Arabidopsis thaliana. Genome Res. 2003, 13, 1250–1257. [Google Scholar]
- Yu, H.; Xie, W.; Li, J.; Zhou, F.; Zhang, Q. A whole-genome SNP array (RICE6K) for genomic breeding in rice. Plant Biotechnol. J. 2014, 12, 28–37. [Google Scholar]
- Semagn, K.; Babu, R.; Hearne, S.; Olsen, M. Single nucleotide polymorphism genotyping using Kompetitive Allele Specific PCR (KASP): Overview of the technology and its application in crop improvement. Mol. Breed. 2014, 33, 1–14. [Google Scholar]
- Grewal, S.; Hubbart-Edwards, S.; Yang, C.; Devi, U.; Baker, L.; Heath, J.; Ashling, S.; Scholefield, D.; Howells, C.; Yarde, J.; et al. Rapid identification of homozygosity and site of wild relative introgressions in wheat through chromosome-specific KASP genotyping assays. Plant Biotechnol. J. 2020, 18, 743–755. [Google Scholar]
- Jiang, C.-K.; Ma, J.-Q.; Liu, Y.-F.; Chen, J.-D.; Ni, D.-J.; Chen, L. Identification and distribution of a single nucleotide polymorphism responsible for the catechin content in tea plants. Hortic. Res. 2020, 7, 24. [Google Scholar]
- Liu, Y.; Chen, S.; Jiang, C.; Liu, H.; Wang, J.; He, W.; Moon, D.; Chen, J.; Chen, L.; Ma, J. Combined QTL mapping, GWAS and transcriptomic analysis revealed a candidate gene associated with the timing of spring bud flush in tea plant (Camellia sinensis). Hortic. Res. 2023, 10, uhad149. [Google Scholar]
- NY/T 1312-2007; Technical Code for Evaluating Crop Germplasm Tea Plant (Camellia sinensis). China Agricultural Press: Beijing, China, 2007.
- Zhou, L.-H.; Zhao, C.-Y.; Luo, J.-W.; Huang, J.-A.; Tan, H.-P. Speedy method of making template DNA in analysis of tea germplasm based on AFLP fingerprint. Food Sci. 2007, 28, 193–195. [Google Scholar]
- Xia, E.-H.; Li, F.-D.; Tong, W.; Li, P.-H.; Wu, Q.; Zhao, H.-J.; Ge, R.-H.; Li, R.-P.; Li, Y.-Y.; Zhang, Z.-Z.; et al. Tea Plant Information Archive: A comprehensive genomics and bioinformatics platform for tea plant. Plant Biotechnol. J. 2019, 17, 1938–1953. [Google Scholar]
- Chen, L.; Zhou, Z.-X.; Yang, Y.-J. Genetic improvement and breeding of tea plant (Camellia sinensis) in China: From individual selection to hybridization and molecular breeding. Euphytica 2007, 154, 239–248. [Google Scholar]
- Li, W.; Zhang, Q.; Fan, Y.; Cheng, Z.; Lu, X.; Luo, B.; Long, C. Traditional management of ancient Pu’er teagardens in Jingmai Mountains in Yunnan of China, a designated Globally Important Agricultural Heritage Systems site. J. Ethnobiol. Ethnomed. 2023, 19, 26. [Google Scholar]
- Wu, H.; Long, X.; Geng, Y. Companion plants of tea: From ancient to terrace to forest. Plants 2023, 12, 3061. [Google Scholar] [CrossRef]
- Zheng, H.; Fu, T.; Xue, X.; Ye, Y.; Yu, G. Research status and prospect of tea mechanized picking technology. J. Chin. Agric. Mech. 2023, 44, 28–35. [Google Scholar]
- Cao, Q.; Xu, Z.; Xu, B.; Yang, H.; Wang, F.; Chen, L.; Jiang, X.; Zhao, C.; Jiang, P.; Wu, Q.; et al. Leaf phenotypic difference analysis and variety recognition of tea cultivars based on multispectral imaging technology. Ind. Crop. Prod. 2024, 220, 119230. [Google Scholar]
- Zeng, L.; Zhou, X.; Su, X.; Yang, Z. Chinese oolong tea: An aromatic beverage produced under multiple stresses. Trends Food Sci. Technol. 2020, 106, 242–253. [Google Scholar]
- Lv, H.; Zhang, Y.; Lin, Z.; Liang, Y. Processing and chemical constituents of Pu-erh tea: A review. Food Res. Int. 2013, 53, 608–618. [Google Scholar]
- Li, Y.; Luo, Q.; Qin, M.; Xu, W.; Wang, X.; Zhou, J.; He, C.; Chen, Y.; Yu, Z.; Ni, D. Study on color, aroma, and taste formation mechanism of large-leaf yellow tea during an innovative manufacturing process. Food Chem. 2024, 438, 138062. [Google Scholar]
- Qin, X.; Li, F.; He, J.; Liao, S.; Deng, H.; Luo, X.; Peng, Y. The relations between the blade anatomical structure and characters of the new tea varieties in Guangxi. Chin. Agric. Sci. Bull. 2009, 25, 36–39. [Google Scholar]
- Wang, Y.; Hong, Y.; Ding, Z.; Zhang, X.; Wang, Y.; Fan, K. Cold resistant prediction of tea germplasm resources applied by anatomical structure index of leaves. Chin. Agric. Sci. Bull. 2009, 25, 126–130. [Google Scholar]
- Fang, Y.; Meng, Z.; Li, X.; Qiao, Y.; Mei, L.; Liang, Y. Analysis of anatomical structure on tea leaves in Shandong province. J. Tea Sci. 2004, 24, 190–196. [Google Scholar]
- Wang, F.; Wu, S.e.; Feng, H.; Zhao, S.; Zhang, J.; Hong, Y.; Zeng, Z.; Chen, A.; You, X.; Luo, S. The characteristics studies of tea leaves’ anatomical structure on parts of Wuyi Ming Cong Tea. Chin. Agric. Sci. Bull. 2013, 29, 130–135. [Google Scholar]
- Fang, Y.; Li, X.F.; Mu, Z.Z.; Qiao, Y.J.; Yu, L.J.; Sun, L.; Lian, J.G. Development of research on anti-frigidity of tea tree. Non-wood Forest Res. 2004, 22, 69–72. [Google Scholar]
- Huang, F.; Lei, Y.; Duan, J.; Kang, Y.; Luo, Y.; Ding, D.; Chen, Y.; Li, S. Investigation of heat stress responses and adaptation mechanisms by integrative metabolome and transcriptome analysis in tea plants (Camellia sinensis). Sci. Rep. 2024, 14, 10023. [Google Scholar] [CrossRef]
- Xia, E.; Tong, W.; Hou, Y.; An, Y.; Chen, L.; Wu, Q.; Liu, Y.; Yu, J.; Li, F.; Li, R.; et al. The reference genome of tea plant and resequencing of 81 diverse accessions provide insights into its genome evolution and adaptation. Mol. Plant 2020, 13, 1013–1026. [Google Scholar] [CrossRef]
- Wang, X.; Feng, H.; Chang, Y.; Ma, C.; Wang, L.; Hao, X.; Li, A.l.; Cheng, H.; Wang, L.; Cui, P.; et al. Population sequencing enhances understanding of tea plant evolution. Nat. Commun. 2020, 11, 4447. [Google Scholar] [CrossRef]
- Wei, C.; Yang, H.; Wang, S.; Zhao, J.; Liu, C.; Gao, L.; Xia, E.; Lu, Y.; Tai, Y.; She, G.; et al. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc. Natl Acad. Sci. USA 2018, 115, E4151–E4158. [Google Scholar] [CrossRef]
- Wang, Z.; Yu, W.; Wang, P.; Liu, C.; Fan, X.; Gu, M.; Cai, C.; Wang, P.; Ye, N. Wgcna analysis of differentially expressed genes in floral organs of tea germplasms with ovary-glabrous and ovary-trichome. Acta Hortic. Sin. 2023, 50, 620–634. [Google Scholar]
- Sun, B.; Zhu, Z.; Liu, R.; Wang, L.; Dai, F.; Cao, F.; Liu, S. TRANSPARENT TESTA GLABRA1 (TTG1) regulates leaf trichome density in tea Camellia sinensis. Nord. J. Bot. 2020, 38, e02592. [Google Scholar] [CrossRef]
- Umezawa, T.; Sugiyama, N.; Mizoguchi, M.; Hayashi, S.; Myouga, F.; Yamaguchi-Shinozaki, K.; Ishihama, Y.; Hirayama, T.; Shinozaki, K. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl Acad. Sci. USA 2009, 106, 17588–17593. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, J.; Yin, Z.; Pan, Y.; Mao, W.; Peng, L.; Guo, X.; Li, B.; Leng, P. SlPP2C2 interacts with FZY/SAUR and regulates tomato development via signaling crosstalk of ABA and auxin. Plant J. 2024, 119, 1073–1090. [Google Scholar] [PubMed]

| Ovary Wall Trichome Trait | Source | Germplasm Name | Sample ID | Genotyping |
|---|---|---|---|---|
| glabrous ovary | Chongqing | C. nanchuanica | NCDS13 | GG |
| Chongqing | C. nanchuanica | NCDS14 | GG | |
| Chongqing | C. nanchuanica | NCDS15 | GG | |
| Chongqing | C. nanchuanica | NCDS8 | GG | |
| Chongqing | C. nanchuanica | NCDS18 | GG | |
| Chongqing | C. nanchuanica | NCDS19 | GG | |
| Chongqing | C. nanchuanica | NCDS20 | GG | |
| Chongqing | C. nanchuanica | NCDS24 | GG | |
| Chongqing | C. nanchuanica | NCDS26 | GG | |
| Chongqing | C. nanchuanica | NCDS50 | GG | |
| Chongqing | C. nanchuanica | NCDS52 | GG | |
| Chongqing | C. nanchuanica | NCDS88 | GG | |
| Chongqing | C. nanchuanica | NCDS89 | GG | |
| Chongqing | C. nanchuanica | NCDS93 | GG | |
| Chongqing | C. nanchuanica | NCDS94 | GG | |
| Chongqing | C. nanchuanica | NCDS97 | GG | |
| Chongqing | C. nanchuanica | NCDS108 | GG | |
| Chongqing | C. nanchuanica | NCDS109 | GG | |
| Chongqing | C. nanchuanica | NCDS111 | GG | |
| Chongqing | C. nanchuanica | NCDS113 | GG | |
| Hairy ovary | Chongqing | hybrid individual | NCDS17 | TG |
| Chongqing | hybrid individual | NCDS53 | TG | |
| Chongqing | hybrid individual | NCDS90 | TG | |
| Chongqing | hybrid individual | NCDS105 | TG | |
| Chongqing | C. sinensis cv. chuanxiaozhong | BBCX1 | TT | |
| Chongqing | C. sinensis var. assamica | BBYNDY1 | TT | |
| Zhejiang | C. sinensis cv. Baiye1 | BY1 | TT | |
| Fujian | C. sinensis cv. Fuding Dabaicha | FDDB | TT | |
| Fujian | C. sinensis cv. Huangdan | HD | TT | |
| Zhejiang | C. sinensis cv. Huangjinya | HJY | TT | |
| Fujian | C. sinensis cv. Jinguancha | JGC | TT | |
| Fujian | C. sinensis cv. Jinmudan | JMD | TT | |
| Zhejiang | C. sinensis cv. Yuehuang1 | YH1 | TT | |
| Fujian | C. sinensis cv. Mingguan | MG | TT | |
| Chongqing | C. sinensis cv. chuanxiaozhong | NCCX10 | TT | |
| Chongqing | C. sinensis cv. chuanxiaozhong | NCCX12 | TT | |
| Chongqing | C. sinensis cv. chuanxiaozhong | NCCX18 | TT | |
| Chongqing | C. sinensis cv. chuanxiaozhong | NCCX24 | TT | |
| Chongqing | C. sinensis cv. chuanxiaozhong | NCCX8 | TT | |
| Hunan | C. ptilophylla | RCMYC | TT | |
| Zhejiang | C. sinensis cv. Wuniuzao | WNZ | TT | |
| Zhejiang | C. sinensis cv. Yujinxiang | YJX | TT | |
| Zhejiang | C. sinensis cv. Zhonghuang2 | ZH2 | TT | |
| Yunnan | C. sinensis cv. Zijuan | ZJ | TT |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Z.; Tang, W.; Lei, M. Morphological Characteristics and Molecular Marker-Assisted Identification of Ovary Glabrous Phenotype in the Population of Nanchuan Dachashu (Camellia nanchuanica). Horticulturae 2025, 11, 360. https://doi.org/10.3390/horticulturae11040360
Wu Z, Tang W, Lei M. Morphological Characteristics and Molecular Marker-Assisted Identification of Ovary Glabrous Phenotype in the Population of Nanchuan Dachashu (Camellia nanchuanica). Horticulturae. 2025; 11(4):360. https://doi.org/10.3390/horticulturae11040360
Chicago/Turabian StyleWu, Zhijun, Weifeng Tang, and Meng Lei. 2025. "Morphological Characteristics and Molecular Marker-Assisted Identification of Ovary Glabrous Phenotype in the Population of Nanchuan Dachashu (Camellia nanchuanica)" Horticulturae 11, no. 4: 360. https://doi.org/10.3390/horticulturae11040360
APA StyleWu, Z., Tang, W., & Lei, M. (2025). Morphological Characteristics and Molecular Marker-Assisted Identification of Ovary Glabrous Phenotype in the Population of Nanchuan Dachashu (Camellia nanchuanica). Horticulturae, 11(4), 360. https://doi.org/10.3390/horticulturae11040360





