Abstract
Table grape breeding programmes are ongoing in different countries to develop new grape cultivars suitable for rapidly changing consumer demands. Although Türkiye is among the world’s leading countries in table grape production, it does not have quality grape cultivars that can compete in foreign markets. Different breeding programmes are conducted in the country to develop new quality table grape cultivars. These breeding studies’ primary goals are to achieve table grapes that are seedless, with crispy flesh, a large berry size, unique aromas, higher phenolic contents, and improved disease resistance. This study evaluated the basic quality characteristics of 89 hybrid genotypes with different parents obtained from crossbreeding studies and passing the preliminary screening stage. The hybrid genotypes were evaluated comprehensively for many quality criteria. In particular, the hybrid genotypes that received the desired score according to tasting panel results were determined. Among these, genotypes 22BY-097, 14KS-03, 18BK-02, and 22BY-044, which are seedless and have different parents, were the highest-scoring hybrid genotypes, due to their different superior characteristics. High-scoring hybrid genotypes will be registered after these quality characteristics are evaluated in different locations in the next stage. Consequently, the accuracy and consistency of the results obtained will be confirmed, and new cultivars will be transferred to table grape growers in the future. With these high-quality new cultivars, table grape growers can earn more income.
1. Introduction
Globally, 75 million metric tons of grapes are produced worldwide, and approximately 5.6% of this production, 4.1 million tons, is produced in Türkiye. Türkiye is the world’s fifth largest grape producer in terms of area and the sixth largest in terms of production. However, it ranks very low in table grape exports, at around tenth place [1]. One of the main reasons for this situation is that the ‘Sultana seedless’ cultivar is the main cultivar produced [2,3]. As in other countries, new breeding programmes are being carried out in Türkiye to meet rapidly changing market demands. These programmes primarily aim to create cultivars with large berries, seedlessness, a crispy texture, and improved disease resistance or tolerance [4]. Approximately 71% of the world’s grape production is used for wine, 27% for table grapes, and 2% for dried grapes (raisins). With changes in consumption habits in recent years, the demand for newly developed table grapes worldwide has been increasing [5]. New high-quality grape cultivars that can meet the changing consumer demands for table grapes in different countries have begun to be grown commercially [6].
According to data from the International Organization of Vine and Wine (OIV), the most important table and dried grape cultivar in Türkiye and the world is ‘Sultana’ [7]. In Türkiye, 50.4% of grape production is table grapes, 40.4% is dried grapes, and 9.2% is wine grapes/mustard grapes. Although table grape production constitutes half of the total fruit production, its share in exports is only 7.2%. Turkish table grape exports, especially to the European Union countries, are low compared to the current potential. Their export potential can be improved with new cultivars developed as a result of breeding studies [8]. In recent years, breeding studies have been carried out to develop grape cultivars that are seedless, large-berry, resistant/tolerant to critical fungal diseases, rich in phenolic compounds, and of high export quality [9,10,11]. Thanks to the achievements of applied breeding programmes, new seedless cultivars are added to the seedless table grape market every year [6,12]. In surveys that have been conducted, it has been determined that most consumers prefer to eat seedless grapes, which have a unique taste and aroma and positively affect human health, with rich phenolic contents [12,13,14].
There has been a growing demand for hybrid grape cultivars that prioritize environmental and human health, reduce production costs, and exhibit increased tolerance or resistance to diseases. Recently, cultivars rich in phenolic compounds have been preferred, due to their positive effects on human health [15,16]. Different Vitis species have been used in breeding studies for this purpose, and very good results have been obtained. In these studies, cultivars of Vitis labrusca species have been used due to their different superior characteristics [17,18]. They are preferred in breeding studies due to their resistance to fungal diseases (mainly downy and powdery mildew), unique intense foxy aroma, and high yields. In traditional breeding programmes, hybrid crossing combinations are selected based on various breeding objectives. Key goals include seedlessness [19,20], a larger berry size [21], resistance to fungal diseases [22,23,24], berry colour [25], firm fruit flesh [26], early or late harvest [27], and a distinctive aroma [28,29,30].
This study aims to examine the most important quality criteria of hybrid grape genotypes obtained after controlled hybridizations made in 2018, and to determine those with the potential to be cultivar candidates. Eighty-nine hybrid grape genotypes that passed the preliminary selection stage were examined regarding important quality criteria, such as seedlessness, berry size, firm flesh, and unique aroma, related to table quality.
2. Materials and Methods
2.1. Experimental Location
The plant material used in the study was taken from the F1 parcel within the Yalova Atatürk Horticulture Central Research Institute, Türkiye, where 6-year-old hybrid genotypes were established in 2020. Hybrid grape genotypes were obtained after controlled hybridizations made in 2018. The trial area is situated on the borders of Yalova center, between 40°39′40″ north latitude and 29°17′22″ east longitude. The altitude is 3 m above the sea level, and the distance to the sea is 150 m.
2.2. Plant Materials
Details on the parent combinations of the hybrid grape genotypes developed from the breeding programme are given in Table 1 and Figure 1. A total of 10 cultivars of the two Vitis species were used in the crossing programme. Among these cultivars, the ‘K-77’ genotype, used as the female parent, was selected because it belongs to the V. labrusca species, has a foxy aroma, and is quite resistant to diseases. The ‘Beyaz Çavuş’ cultivar was chosen because it does not need emasculation, due to its female flower structure, thin skin, and slightly muscat aroma. The ‘Red Globe’ cultivar was selected because of its large berries, long storage life, and high yield.

Table 1.
Parental forms used in crossing combinations and Vitis species involved in hybrid grape genotypes.

Figure 1.
Parents of grape genotypes used in crossing programme.
‘Superior Seedless’, ‘Ergin Çekirdeksizi’, ‘Autumn Royal’, ‘Crimson Seedless’, and ‘Samancı Çekirdeksizi’ were selected as the male (pollinator) parents, due to their naturally large berries. In addition, ‘Autumn Royal’, ‘Crimson Seedless’, and ‘Samancı Çekirdeksizi’ cultivars were also chosen for their lateness characteristics. ‘Ergin Çekirdeksizi’ was selected for its high yield, while ‘Kishmish Vatkana’ was used as the male parent due to its resistance to powdery mildew disease.
The vines were planted at a distance of 3 × 1 m and trained with the double T training system. The hybrid genotypes used in the study were 6 years old on their own roots, and pruned with a stable cordon system. Summer pruning was performed twice in the summer months. Irrigation was performed by drip irrigation between June and September, by controlling the soil moisture. In general, irrigation was performed once a week with approximately 20 l of water per vine, but no irrigation was performed in weeks with heavy rainfall. The weeds between the rows were mowed regularly once a month with grass mowers attached to the tractor. Additionally, the weeds on the rows were hoed by hand.
In the first stage of the study, the important quality parameters of a total of 89 hybrid genotypes were examined. Then, according to quality analyses and degustation results, the amounts of phenolic compounds were analyzed in 48 hybrid genotypes that showed superior characteristics (especially seedlessness and berry size). Our main goal in this study was to determine potential large-berry seedless grape cultivars that could compete with foreign cultivars. However, if there were hybrid genotypes that had additional characteristics, such as firm flesh, unique aroma, and uniformity, which are sought in some markets, in addition to having large berries and being seedless, they were also selected to be evaluated. In addition, the study aimed to select hybrids that are not seedless, but have other superior characteristics (such as a very large berry size, early or late harvest, and special aroma), as potential cultivar candidates.
2.3. Physical Properties of Berries
Cluster weight (g) (OIV 502; IBPGR 7.1.14): In these analyses, the clusters belonging to each genotype were weighed with a precision scale, and the average weight was calculated. (Table 2).

Table 2.
Average results of cluster weight (g) of hybrid genotypes. (For each trait, sorting was performed from largest to smallest values, and genotypes marked with * are seedless).
Cluster density (OIV 204; UPOV 033; IBPGR 6.2.3): Grape berries were scored according to their density on the bunch. They were scored as 1—very rare, 3—rare, 5—moderate, 7—dense, and 9—very dense (Table 3).

Table 3.
Average results of cluster density of hybrid genotypes. (For each trait, sorting was performed from largest to smallest values, and genotypes marked with * are seedless).
Number of berries in cluster: The number of berries in each bunch was counted, and the average number was calculated (Table 4).

Table 4.
Average results of berry number in cluster of hybrid genotypes. (For each trait, sorting was performed from largest to smallest values, and genotypes marked with * are seedless).
Uniformity of clusters: Clusters were scored as uniform (1) or not uniform (2) (Table 5).

Table 5.
Average results of cluster uniformity of hybrid genotypes. (For each trait, sorting was performed from largest to smallest values, and genotypes marked with * are seedless).
Berry width (mm) OIV (221), IBPGR (6.2.5) (OIV 221; IBPGR 6.2.5): The width was measured with a digital calliper. In these analyses, the average width of 10 berries taken randomly from different parts of each cluster was calculated to represent the hybrids (Table 6).

Table 6.
Average results of berry width (g) of hybrid genotypes. (For each trait, sorting was performed from largest to smallest values, and genotypes marked with * are seedless).
Berry length (mm) (OIV 220; UPOV 035; IBPGR 6.2.5): The length was measured with a digital calliper. In these analyses, the average length of 10 berries taken randomly from different parts of each cluster was calculated to represent the hybrids (Table 7).

Table 7.
Average results of berry length (mm) of hybrid genotypes. (For each trait, sorting was performed from largest to smallest values, and genotypes marked with * are seedless).
Berry weight (g) (OIV 503; IBPGR 7.1.15): The berry weight was measured with a precision scale. In these analyses, the average of 10 berries taken randomly from different parts of each cluster was calculated to represent the hybrids (Table 8).

Table 8.
Average results of berry weight (g) in clusters of hybrid genotypes. (For each trait, sorting was performed from largest to smallest values, and genotypes marked with * are seedless).
Flesh firmness (N) (OIV 235; UPOV 040; IBPGR 6.2.11): A table-type penetrometer was used for berry firmness (N) analysis. Flesh firmness was measured as the penetration depth in grape berries with a 3 mm needle digital penetrometer (FM200, PCE Italia S.R.L. Capannori, Italy) (Table 9).

Table 9.
Average results of flesh firmness (N) of hybrid genotypes. (For each trait, sorting was performed from largest to smallest values, and genotypes marked with * are seedless).
Berry detachment force (N) (OIV 240; UPOV 038; IBPGR 6.2.13): The force required to separate the berries from the stem was measured with a hand-held penetrometer, and expressed in Newtons (Table 10).

Table 10.
Average results of berry detachment force (N) of hybrid genotypes. (For each trait, sorting was performed from largest to smallest values, and genotypes marked with * are seedless).
Seedlessness (visual) (OIV 241; UPOV 043; IBPGR 7.2.7): Hybrids for which no seeds were visually detected were scored as 1, and those for which seeds were detected were scored as 2 (Table 11).

Table 11.
Average results of seedlessness of hybrid genotypes.
Number of seeds: The number of seeds was counted in each berry of the hybrids (Table 12).

Table 12.
Average results of seed number of hybrid genotypes. (For each trait, sorting was performed from largest to smallest values, and genotypes marked with * are seedless).
Dry weight of the seed (mg): (OIV 243; IBPGR 6.2.15): After the seeds or seed traces in each grape were dried, they were weighed to determine their dry weight (mg). Those with a dry weight of 0–18 mg were considered seedless, and given a score of 1, while those weighing more than 18 mg were considered seeded, and received a score of 2 (Table 13).

Table 13.
Average results of seed dry weight (g) of hybrid genotypes. (For each trait, sorting was performed from largest to smallest values, and genotypes marked with * are seedless).
2.4. Chemical Properties of Berries
2.4.1. Total Soluble Solids, Acidity, Maturity Index, Flavour, and Harvesting Time
Total soluble solids (°Brix (%)): The total water-soluble solids (°Brix) value was determined by reading with a digital refractometer (Atago PAL-BX/Acid). All readings were repeated twice in at least 10 samples, and the average value was taken (Table 14).

Table 14.
Average results of TSS (°Brix) of hybrid genotypes. (For each trait, sorting was performed from largest to smallest values, and genotypes marked with * are seedless).
Acidity (g/L): For the acidity value of grape berries, the juices of the berries were diluted 1/50, and then read with a digital refractometer (Atago PAL-1). All readings were repeated twice in at least 10 samples, and the average value was taken (Table 15).

Table 15.
Average results of acidity (g/L) of hybrid genotypes. (For each trait, sorting was performed from largest to smallest values, and genotypes marked with * are seedless).
Maturity index (total soluble solids/acidity): The maturity index was calculated as SSC/Titratable Acid (Table 16).

Table 16.
Average results of maturity index of hybrid genotypes. (For each trait, sorting was performed from largest to smallest values, and genotypes marked with * are seedless).
Particular flavour (OIV 236; UPOV 42; IBPGR 6.2.12): If the hybrid grapes did not have a unique taste, they were scored as 1; if they had a muscat aroma, they were scored as 2; and if they had a foxy or similar aroma, they were scored as 3 (Table 17).

Table 17.
Average results of particular flavour of hybrid genotypes. (For each trait, sorting was performed from largest to smallest values, and genotypes marked with * are seedless).
Harvesting date (OIV 304; IBPGR 7.1.10): The hybrids were harvested when the maturity index value reached approximately 20 (Table 18). Hybrid genotypes were harvested based on a maturity index value of approximately 20, to ensure that they were neither too early, nor too ripe. Also, a taste test was performed to determine the harvest time, and the final decision was made. Only a few hybrid genotypes were harvested with a maturity index value below 20, due to the onset of Botrytis.

Table 18.
Average results of harvesting date of hybrid genotypes. (For each trait, sorting was performed from largest to smallest values, and genotypes marked with * are seedless).
2.4.2. Total Phenolic Content and Antioxidant Activity
In terms of total phenolic content and antioxidant activity (analyzed using 2 different methods), analyses were performed using a total of 48 hybrid genotypes, including 44 seedless hybrid genotypes with sufficient cluster counts, and 4 seeded hybrid genotypes (22BY-106, 21BA-01, 22BY-067, and 21BA-04) with a berry size of 7 g and above. In this part of the study, three bunches were selected from each to represent the 48 hybrid genotypes selected; after being washed and cleaned, the berries were separated from the bunch skeleton. To extract hybrid genotypes, the berries of hybrid grapes were homogenized and crushed. This process was carried out so that all grape berries (skin, flesh, and seeds together) were thoroughly broken. Thus, the total phenol and antioxidant activity values in all the homogenized grapes could be determined. After the peel, pulp, and seeds (seed traces) were homogenized to ensure they were thoroughly broken down, they were divided into three 50 g parts, and total phenol and antioxidant activity analyses were carried out with these extracted samples in 3 replications. Methanol solution was used as a solvent in the extraction of the grapes.
The total phenolic contents of the extracts were determined in three replicates using the Folin–Ciocalteu method [31], with some minor modifications. Briefly, 30 µL of extract and 150 µL of FC reagent were consecutively transferred into tubes containing 2.37 mL of deionized water. After 8 min, 450 µL of saturated Na2CO3 was added to the mixture. The same procedure was run to prepare the blank using 30 mL of deionized water instead of extract. Absorbance values were read at 750 nm, using a spectrophotometer, against the blank, after 30 min incubation at 40 °C. Various concentrations of gallic acid solutions (50, 100, 200, 300, 400, and 500 mg/L) were used to plot a calibration curve. The results are expressed as mg of gallic acid/mL (GAE mg/100 g fresh weight [FW]) (Table 19).

Table 19.
Total phenolic content and antioxidant activity values (analyzed using 2 different methods) of prominent hybrid genotypes*.
The DPPH (2,2-diphenyl-1-picrylhydrazyl) assay procedure described by Thaipong et al. [32] was used, with slight modifications, for the determination of the antioxidant activity. DPPH stock solution was prepared as 24 mg/100 mL methanol (chromatographic grade), and stored at 24 °C before use. The working solution was prepared by diluting the stock solution with methanol to a final absorbance of 1.20 ± 0.02 at 515 nm. For the calibration curve, Trolox® solution, with a final concentration of less than 50 µM in a spectrophotometer cuvette, was used. In the experiments, the extract or standard (150 µL) and DPPH working solution (2850 µL) were mixed in a test tube, and the reaction was continued for 60 min in a dark environment. At the end of this period, the absorbance of the coloured solutions was read at 515 nm, and the results were expressed as mmol TE per 100 g. Extracts were diluted if their absorbance reading was above the linear range of the calibration curve (Table 19).
A cupric reducing antioxidant capacity (CUPRAC) assay was used to determine the antioxidant capacity of the berries of the hybrids. This method is used to measure the ability of polyphenols to reduce copper (II) or cupric ions, vitamin C, and vitamin E [32], and copper (II)-neocuproine (Cu (II)-NC) reagent I is used as a chromogenic oxidizing agent. In this method, an antioxidant solution is mixed with a copper (II) chloride solution, a neocuproine alcoholic solution, and an ammonium aqueous buffer at pH 7, and subsequently, the absorbances of the mixtures are determined at 450 nm after 30 min. The antioxidant activities of hybrid samples were determined by the CUPRAC assay as follows: A 1 mL volume each of copper (II) chloride solution (10 mM), 7.5 mM neocuproine solution (Nc), and ammonium acetate (NH4 Ac) buffer (pH 7) solution were mixed in a test tube, and extract (or standard) solution (x mL) and H2O (1.1 − x) mL) were added to the test tube to make a final volume of 4.1 mL. The test tubes were screw-capped and left for an hour at room temperature, and then the absorbance at 450 nm was recorded against a reagent blank. The standard calibration curve of each antioxidant compound was constructed with Trolox® in this manner, as absorbance versus concentration. The molar absorptivity of the CUPRAC method for each antioxidant was found from the slope of the relevant calibration line, and the antioxidant activity values of the fresh grape berries were expressed as mg Trolox equivalents per 100 g (Table 19).
2.5. Fruit Colour Analyses
The colour of all fruit samples was assessed using the CIELab space with a chromameter (CR–400, Minolta Co., Tokyo, Japan) [33]. The parameters that define the CIELab space are as follows: rectangular coordinates, such as the red/green colour component (a*), yellow/blue colour component (b*), and lightness (L*), and the cylindrical coordinates of chroma and hue angle. The colour differences were calculated in CIELab units, using the parameter (DE*), calculated as DE* = [(DL)2 + (Da)2 + (Db)2 ]1/2 (Table 20).

Table 20.
Average results of berry colours of hybrid genotypes. (For each trait, sorting was performed from largest to smallest values, and genotypes marked with * are seedless).
2.6. Degustation Test (Sensory Analyses)
After harvest, sensory expert analysis of cultivars was performed by a panel consisting of 10 members, using the Atak and Kahraman [34] methods, with some modifications. The panellists who conducted the tasting tests comprised an expert team who had conducted breeding and tasting studies on hybrid grapes for many years. In addition, members of an expert group from the food quality department, who had conducted taste tests on fruits for many years, also participated in these studies as panellists. The list of sensory terms included descriptors of the general acceptance of the bunch (seeded/seedless, skin, flesh, homogeneity of colour), visual appearance (colour, berry shape/size, free disease/pest), odour (fruity/foxy, muscat, special), and taste (sweet, bitter, and sour/acidic). They were rated on a scale ranging from 0 to 5 (0 = minimum; 5 = maximum intensity). The scores were totaled to give a maximum score of 20 points, in terms of 4 criteria. The total scores obtained from the degustation test are given in Table 5, from the highest to the lowest value. It was planned that hybrids with 15 or more points specifically would be taken to the second stage (Table 21).

Table 21.
Average results of degustation score of hybrid genotypes. (For each trait, sorting was performed from largest to smallest values, and genotypes marked with * are seedless).
2.7. Statistical Analysis
Analysis of variance (ANOVA) was performed to determine significant differences among the means of the phenolic compounds. The data were presented as the arithmetic means of three replications ± standard deviations. The least significant difference (LSD) method was used to determine the significance levels for all accessions. Differences were considered significant at p < 0.05. Correlation coefficients (R) from a parametric Pearson’s test were calculated to evaluate covariance relationships among variables. All statistical analyses were conducted using SAS statistical software (JMP 18.1 version) [35].
3. Results
3.1. Physical Properties
As a result of the analyses conducted on 89 different hybrid grape genotypes, an attempt was made to determine the hybrids to be taken to the second stage. In the analyses conducted in terms of essential cluster characteristics, the hybrid genotypes that stood out were as follows (Table 2, Table 3, Table 4, Table 5, Table 6, Table 7, Table 8, Table 9, Table 10, Table 11, Table 12 and Table 13).
The seedless 20BE-15 and 22BY-015 genotypes and the seeded 21BA-01 genotype were the hybrid genotypes with the highest cluster weights (g), and they all had different parental combinations. In crosses where the genotype K77, belonging to the V. labrusca species, was used as the female parent, the cluster sizes and weight remained quite low. Among all the hybrid genotypes, 16 hybrid genotypes with cluster weight of 350 g and above, as well as 29 hybrid genotypes with a cluster weight below 100 g, were determined (Table 2).
For the cluster density evaluation of the hybrid grape genotypes, it was determined that 6 hybrids had a very dense cluster structure, 14 hybrids had a dense cluster structure, 53 hybrids had a medium cluster structure, 16 hybrids had a loose cluster structure, and 1 hybrid had a very loose cluster structure (Table 3). It was determined that the hybrid 20BE-15 (seedless) stood out in terms of both cluster weight and berry number, having the highest number of berries, at 272 berries. A total of 15 hybrid genotypes had more than 100 berries in their clusters (Table 4). Analyzing the uniformity of the structure of the clusters, it was found that approximately half of them were uniform, while the other half were non-uniform (Table 5).
The width (mm), length (mm), weight (g), flesh firmness (N), and berry detachment force (N) values of the hybrid grapes are given in Table 6, Table 7 and Table 8. The eight hybrid genotypes with the greatest berry widths were derived from combinations where the ‘Beyaz Çavuş’ cultivar was used as the female parent. Among these, the 21BA-04 hybrid genotype, with a berry width of 37.62 mm, was determined to have significantly wider berries than the other hybrid genotypes.
In terms of berry length, the 22BY-106 (43.18 mm) and 21BA-04 (39.54 mm) hybrid genotypes were determined to be the genotypes with the longest berries. The female parent of these two hybrid genotypes is the ‘Beyaz Çavuş’ cultivar. The results of berry weight showed that the first ten genotypes with the largest and heaviest berries among the tested hybrid genotypes were obtained from seeded cultivars, in the range of 8.80–5.20 g. However, it was determined that nine hybrid genotypes had a berry weight ranging from 4.00 g to 5.20 g, which could be considered a fairly large berry size for seedless cultivars. It was found that the hybrid genotype 22BY-106 ranked first in terms of both berry length and berry weight.
For evaluations of flesh firmness values, 23 hybrid genotypes had measured values from 6.00 N up to 9.20 N. Since flesh hardness has become a desired trait by consumers in recent years, hybrids with soft flesh are not included in the second stages of breeding programmes unless they have other superior traits (Table 9).
In the assessment of berry detachment force (N), the two hybrid genotypes 23RK-012 and 22BY-096 achieved the highest values of 9.5 N and 9.0 N, respectively. These values are very important regarding the post-harvest performance of the fruit of these two hybrids. Only 14 hybrid genotypes had a detachment force value above 3 N, and the remaining hybrids were revealed to have a value below this (Table 10). It was shown that, of the 89 hybrid genotypes obtained from the crossing combinations of the breeding programme and constituting the material of this study, 32 grape genotypes were seeded, and 57 were seedless (Table 11).
Regarding the number of fruit seeds contained in the seeded genotypes, only one hybrid (14KS-19) had 4.00 seeds, while the hybrids 18BK-03 and 18BK-12 had 3.00 and 3.60 seeds, respectively. The remaining tested seeded hybrids had only one or two seeds in the fruit (Table 12).
For the dry weight of the seeds, grape hybrids with a dry weight of up to 18 mg were accepted as seedless. It was found that the hybrid genotypes with the highest dry seed weights were 12KS-15 (65.00 mg) and 23RK-012 (60.64 mg). Detailed analysis of the results showed that two hybrids were included in the 50–60 mg range, eight were in the 40–50 mg range, 12 were in the 30–40 mg range, eight were in the 18–30 mg range, and the remaining 32 hybrid genotypes were seedless, and had a dry seed weight under 18 mg (Table 13).
3.2. Chemical Composition
The results for some chemical composition and essential fruit quality traits (soluble solids, acidity, maturity index, flavour, and harvesting date) of the hybrid genotypes are presented in Table 14, Table 15, Table 16, Table 17 and Table 18. It was determined that 14 hybrid genotypes had soluble solids (°Brix) values ranging from 20.0% up to 24.9%, while the remaining analyzed genotypes had values ranging from 12.7% to 19.8% (Table 14). In terms of fruit acidity, it was determined that 10 genotypes had acidity values between 1.0 g/L and 1.20 g/L, while the values of 88 tested hybrid genotypes ranged between 0.56 and 0.98 g/L (Table 15). The fruit maturity index of all the assessed grape genotypes ranged between 16.48 and 25.36. The following hybrid genotypes, 21BA-03 (25.36), 22BY-052 (24.78/seedless), and 22BY-075 (24.05/seedless), were determined to have the highest maturity index values (Table 16). The fruits of four hybrid genotypes, 12KS-23 (seedless), 14KS-02 (seedless), 16KY-11 (seedless), and 21BA-03, were determined to have a muscat aroma, while the hybrids 12KS-15, 14KS-04 (seedless), 14KS-13 (seedless), 18BK-14 (seedless), 22BY-032 (seedless), and 22BY-097 (seedless) were found to have a foxy or similar fruit aroma. No distinct fruit aroma was detected in the remaining 80 hybrid genotypes (Table 17). While most of the hybrids were harvested in late July and August, both early and late hybrid genotypes were determined (Table 18).
The results of the total phenolic contents and antioxidant activity values of the fruits, obtained using two methods (DPPH and CUPRAC), are presented in Table 7. It was determined that the seedless hybrid genotypes 22BY-052 and 18BK-02 had the highest contents of total phenolics, totaling 13.23 and 12.92 mg GAE/100 g, respectively. The lowest value (0.62 mg GAE/100 g) of the bioactive compound was obtained in fruit of the seedless hybrid genotype 20BE-29. Analyses of the total antioxidant activity (µM TE/100 g) of fruit samples of the grape genotypes confirmed similar results for both of the abovementioned methods. It was found that fruit of the seedless hybrid genotype 22BY-052 had the highest total antioxidant activity based on the DPPH and CUPRAC methods (1146.82 and/2591.63). In addition, three seedless hybrids, 18BK-02, 18BK-11, and 12KS-09, were also in the group with the highest values based on the two methods. The fruit of the seedless hybrid genotype 22BY-032 had the lowest value (115.37) based on the DPPH method, and the seedless hybrid 12KS-16 had the lowest value (132.12) based on the CUPRAC method (Table 19).
3.3. Fruit Colour Analyses
The results of the fruit colour values (L*, a*, b*, hue, and chroma) of the tested grape hybrid genotypes are shown in Table 20. The hybrid genotype 2BY-078 (seedless) had the highest value in terms of L (perceptual lightness), at 51.18, while the seeded hybrid 11KC-05 achieved the lowest value of 26.47. In terms of a value (−a: green; +a: red), the hybrid genotype 16KY-22 obtained the highest value (+14.91), and 14KS-07 obtained the lowest value (−5.28). For the colour value, b (+b: yellow; −b: blue), the hybrid genotype 11KC-08 obtained the highest value (+11.54), and 11KC-25 had the lowest value (−4.89). Regarding the colour hue value, the highest value was obtained for the hybrid genotype 16KY-01 (+84.97), while the lowest value was obtained for the hybrid 16KY-04 (−76.81). The evaluation of fruit colour by the chroma (saturation or colourfulness) values showed that 11KC-15 and 11KC-07 were the hybrid genotypes with the highest and lowest values, +12.54 and +1.38, respectively (Table 20).
3.4. Degustation Test (Sensory Analyses)
According to the results obtained from the degustation scoring of hybrid grapes, 14 genotypes with at least 15 points out of 20 were determined as promising cultivar candidates. Among the seedless genotypes, 22BY-097, 14KS-03, 18BK-02, and 22BY-044 achieved the highest scores, ranging from 15.0 to 17.5. The fact that the 12 grape genotypes with the highest scores were classified as seedless showed that the sensory panel also preferred seedless grapes to seeded ones (Table 21). In particular, the large size and low acidity of the seedless cultivars were the main reasons for their high scores. The main factors contributing to the highest tasting score of the genotype 22 BY-097 were its seedless berries and muscat aroma. The next highest-scoring genotype, 14KS-03, received high scores from the panellists in the tasting due to its seedlessness and low acid content. The 18BK-02 and 22BY-044 genotypes were also appreciated by the panellists, due to their seedlessness, large fruit size, and high brix content.
3.5. Seedlessness Rates
Table 22 shows the seedlessness rates based on combinations. The combinations with the highest seedlessness rates were the following:

Table 22.
Seedlessness numbers and percentages based on crossing combinations of grape genotypes.
- 24RC (‘Red Globe’ X ‘Crimson Seedless’), with 100% seedlessness
- 12KS (‘K-77’ X ‘Superior Seedless’), with 85.7% seedlessness
- 20BE (‘Beyaz Çavuş’ X ‘Ergin Çekirdeksizi’), with 83.3% seedlessness
On the other hand, the lowest seedlessness rates were observed in the following combinations:
- 21BA (‘Beyaz Çavuş’ X ‘Autumn Royal’), with 0% seedlessness
- 11KC (‘K-77’ X ‘Crimson Seedless’), with 43.9% seedlessness
The overall seedlessness rate across all combinations was determined to be 64%.
3.6. Similarity Dendrogram, Correlation Graph, and PCA
The similarity dendrogram formed as a result of evaluating the different traits of the grape hybrid genotypes is given in Figure 2. The 89 grape hybrid genotypes are divided into nine distinct subgroups in the dendrogram. Some of the hybrids originating from the same parents are classified in the same group, but some of them are classified in different groups. This situation shows that in contrast to a genetic similarity dendrogram based on molecular markers, a similarity dendrogram based on berry traits related to the reflection of the genotype on the phenotype might give quite different results.
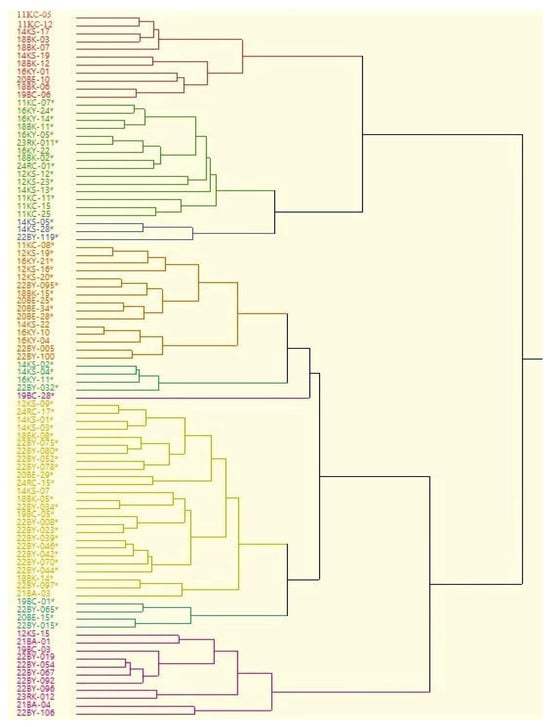
Figure 2.
Similarity dendrogram formed as a result of evaluating fruit traits of hybrid genotypes. (* Seedless hybrids).
Analysis of the results of correlations on a graph of the criteria used in evaluating the different fruit traits of the grape hybrid genotypes indicated that some criteria had a very highly positive relationship with each other. In contrast, other criteria showed a highly negative relationship (Figure 3). It was found that the criteria evaluating cluster-related traits showed a highly positive relationship each other. Similar connections were noticed in the criteria related to berry size/dimensions and seed number. A negative relationship between seed status and sensory test score was determined. It was also confirmed that colour values had both highly positive and negative correlations, due to their inverse relationship.
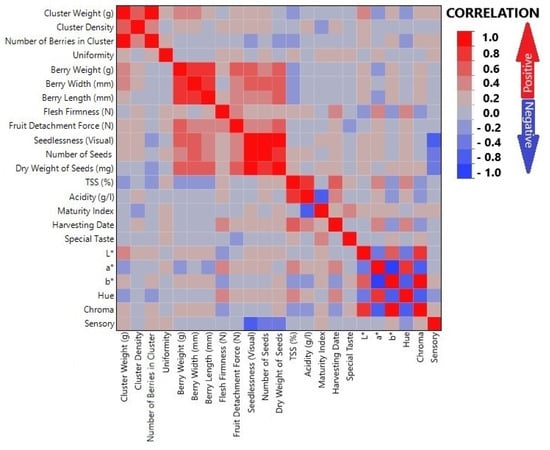
Figure 3.
Correlation graph of criteria used in evaluating fruit traits of hybrid genotypes.
Finally, results obtained from principal component analysis (PCA) data (Figure 4) indicated that some of the evaluated fruit trait criteria acted together. Among these, seedlessness, number of seeds, and dry weight of seeds formed a group. The second group included berry-related traits, and another group contained fruit colour values such as L, b, and chroma, while in the next group there were some criteria related to fruit quality. It was also determined that the results of the sensory evaluation were in a different position to other criteria.
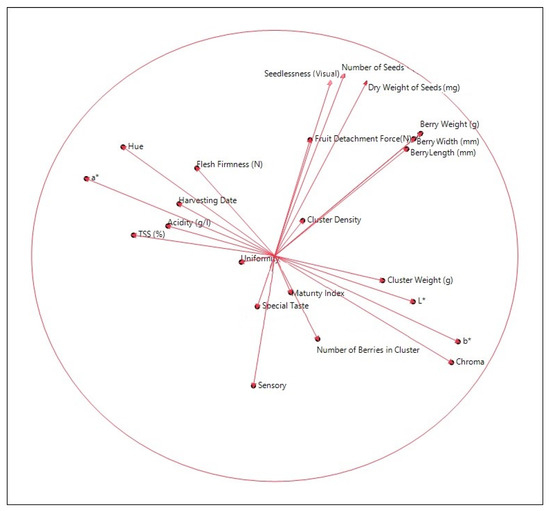
Figure 4.
Principal components analysis (PCA) based on criteria used to define hybrid genotypes.
As a result of the evaluated parameters, and especially the sensory test, the hybrid genotypes and the most important quality characteristics that stood out were as follows: The 22 BY-097 genotype received the highest score in the sensory test because of its seedless berries and muscat aroma. Although it contained seeds, the 12KS-15 hybrid genotype also achieved a high score in the sensory test because of its muscat aroma. One of the seeded genotypes, 23RK-012, was highly scored in terms of fruit taste, and the seedless hybrid 22 BY-039 stood out with its firmness of flesh. In addition, 23RK-012 turned out to be the hybrid genotype with the highest berry–stalk connection, which constitutes an essential advantage for fruit storage and transportation. The seedless hybrid genotype 22 BY-039 was also determined to have larger berries than the others. Two seedless hybrid genotypes, 18BK-02 and 18BK-11, which had dark-coloured berry skin, were included in the group with the highest values. In addition, the seedless grape hybrids 22BY-052 and 12KS-09, which had lighter berry skin colours, were included in the group with the highest total phenolic content and antioxidant activity values.
4. Discussion
Table grape breeding programmes are being carried out in many countries worldwide to meet changing consumer demands [6]. Most of these programmes aim to develop new grape cultivars that are seedless, and have large berries, crunchy flesh, and a unique aroma [36]. This study comprehensively evaluated essential quality criteria of hybrid genotypes obtained from crossbreeding two Vitis species for similar targets. In recent years, interspecific hybridization has been carried out to develop new low-cost hybrid cultivars that are more resistant to fungal diseases and produce quality grapes with less spraying [37,38,39]. Due to the susceptibility of the cultivars of the widely grown V. vinifera species to biotic and abiotic stress conditions, V. labrusca cultivars are particularly preferred in breeding due to their resistance to these adverse conditions. However, many hybrid genotypes may have undesirable traits in terms of fruit quality [40].
As in this study, one of the main focuses of table grape breeding programmes is to develop seedless, large-grape cultivars. Stenospermocarpic genotypes are widely used as pollinator (male) parents to obtain new elite seedless cultivars in classical breeding [41]. Early or late fruit ripening and harvest is among the most critical criteria in table grape breeding work [42]. Cultivars that ripen out of season find buyers willing to pay high prices, because the grape supply in the market is low [5]. Therefore, in many grape breeding programmes, efforts are made to develop early- and late-ripening hybrid genotypes. Volynkin et al. [43] tried to develop early-ripening grape cultivars with berries larger than 6 g, using genotypes as male parents, which produced fruit that could be harvested in the very early or early season. They stated that up to 63.9% earliness of fruit ripening could be achieved in hybrid genotypes, depending on the crossing combination. Similarly, in our study, the male parent of most grape hybrids for which the fruit were harvested in the early period (before August) was the early-ripening cv. ‘Ergin Çekirdeksizi’. In addition, the fact that several late hybrids harvested in September in our study will be offered to the market when the number of grape varieties available is decreasing strengthens the possibility of finding buyers willing to pay a high price.
Xu et al. [44] conducted a long-term table grape breeding programme in China. Hybrid genotypes were obtained by crossing V. vinifera cultivars among themselves, and also with V. labrusca. As a result, twelve table grape cultivars were released and registered in China. Ten cultivars were obtained by the hybridisation among V. vinifera cultivars, and two cultivars were selected from the interspecific hybridisation between V. vinifera and V. labrusca. It was reported that hybrids with a strong muscat aroma were chosen from these genotypes. It was also stated that seedlessness, earliness, and berry size were considered as essential criteria in cultivar selection. In line with this study, our breeding study focused primarily on seedless genotypes. Our study shows that seedless, larger-berry, and muscat-aroma genotypes achieved higher degustation scores in the degustation panels, consistent with the information from the literature.
Researchers have reported that the parents selected in seedless grape breeding can significantly affect the number of seedless hybrid genotypes obtained. Authors have also reported that the female parent primarily affects the seedless genotype rate [9]. In our study, seedless genotypes were obtained from different percentages (0–100%) of combinations, and it was determined that the number of seedless genotypes also varied depending on the pollinator cultivar. Piarulli et al. [10] reported recently that the selected parents used in crosses influenced the number of seedless hybrid genotypes, and they obtained a similar amount of seedless hybrid populations in total, which is consistent with the results of our study. In our study, different numbers of seeded and seedless genotypes were obtained depending on the selected parent combination. These researchers also reported that the selected crossing combinations were important in breeding for disease resistance. Based on this idea, we included a V. labrusca genotype and the ‘Kishmish Vatkana’ cultivar as parents in our study (Figure 5).

Figure 5.
Photos of the hybrid genotypes that stand out as a result of the study.
It has also been reported that the stenospermocarpy trait can be affected by environmental conditions in some years [45]. Another similar study on seedless grapes was published by Puglisi et al. [46], who used different parents and two techniques (embryo rescue and gammic propagation). According to the results obtained by the researchers (similar to previous findings), they reported that the parents selected in grape breeding affect the number of seedless hybrid genotypes. Also, among the measured parameters, they noted a positive correlation only between the obtained berries, seeds, and cultivars. Our study also revealed this situation, especially in combinations where the seeded female parent ‘Beyaz Çavuş’ cultivar was used. Although the ‘Beyaz Çavuş’ cultivar, which is a female-flowered variety, generally increases the number of seedless hybrid genotypes, it is also known that these numbers may vary in some years.
Different methods are used to define the genotypes obtained in hybridization studies as seedless. One of these methods is the determination of dry seed weight. Souza Leão and Carvalho [41] accepted hybrid genotypes with dry seed weights between 0 and 25.7 mg as seedless grapes. In our study, we used more sensitive criteria, and accepted genotypes with dry seed weights of 18 mg and below as seedless. In addition, researchers have stated in their studies that parameters related to fruit size (length, weight, and diameter) have a high positive correlation. In our study, consistently with these findings, parameters related to fruit size (length, weight, and diameter) showed a highly positive correlation. According to these results, the larger ones among our hybrid genotypes had increased berry weight, berry length, and berry width values at the same rate.
Studies conducted by different researchers have shown that the total phenol and antioxidant amounts in grape berries might vary depending on both the Vitis species and cultivars. They have also mentioned that the phenolic content and antioxidant activity could be different in fruit parts (peel, pulp, and seeds) [47,48,49]. It was also reported that cultivars or hybrids with darker-coloured berries might have higher total phenolic contents than those with lighter-coloured fruit [50]. In our study, grape berries were homogenized and crushed without being separated into different parts. We tried to determine the average total phenolic contents and antioxidant activity in the berries. Since most of the hybrid genotypes used were seedless, the total contents in the skin and pulp were determined in these genotypes. Our results confirmed that the seedless hybrid genotypes 18BK-02 and 18BK-11, having dark-coloured berry skin, were in the group with the highest values, in line with the cited literature. However, unlike the literature, the seedless hybrid genotypes 22BY-052 and 12KS-09, with light fruit skin colour, were also included in the group with the highest values. This situation indicates that it would not be correct to expect a rich phenolic content based solely on the colour of the berry skin. In another study, in line with these results, Ananga et al. [51] reported that dark colours in fruits were associated with a high anthocyanin content. Additionally, they stated negative and significant correlations between skin colour (L*, a* and b*), flesh colour (L* and m/L), and total antioxidant content.
Whether the hybrid genotypes obtained in breeding studies have quality characteristics suitable for consumer demands must be evaluated comprehensively [52]. Based on the results, hybrid grapes will move on to the next evaluation stages as potential cultivar candidates. Crespo et al. [12] used the same criteria and methods as those used in our study to evaluate hybrid grapes and their quality traits. The same methods were used to identify the prominent traits in sensory, physicochemical quality, and primary bioactive compound analyses. These researchers reported that the developed hybrid genotypes had superior traits in comparison with their parents in many aspects (wildly different colours of fruit flesh and phenolic compounds). In the same study, researchers also examined the correlation between all quality parameters, as in our study. The correlation analyses reported a highly positive correlation, especially in the data related to fruit size and berry skin (L*, a*, and b*), as observed in our study. However, unlike our study, they also reported that larger fruits have higher fruit flesh hardness. They reported that although most selected parents had soft fruit flesh, large-berry hybrids had harder fruit flesh.
According to all quality criteria evaluations, especially the scores obtained from the degustation panel, 14 hybrid genotypes showed superior characteristics. Among these, the seedless hybrid genotypes 22BY-097, 14KS-03, 18BK-02, and 22BY-044, originating from different parents, were the highest-scoring hybrid genotypes. The main factors contributing to the highest score of the 22 BY-097 genotype were that it had seedless berries and a muscat aroma. The 14KS-03 genotype received high scores from the panellists due to its seedlessness and low acid content. The 18BK-02 genotype received high scores due to its seedlessness and large berry size. It was also determined that this hybrid genotype was one of the genotypes with the highest phenolic contents. The 22BY-044 genotype also gained the appreciation of panellists, with its seedlessness, big berries, and high brix content.
This study investigated the quality traits of hybrid genotypes obtained from breeding programmes with the goal of meeting rapidly changing consumer demands, especially those focusing on seedless grapes. According to the obtained results, it is understood that the parents selected in breeding programmes can affect both the number of hybrid genotypes obtained and the number of seedless genotypes in the same way. It has also been determined that hybrids that have larger berries (4 gr and above), are seedless, and, if possible, are aromatic have a higher potential to gain more appreciation and become cultivars. These high-quality new cultivars, which were most appreciated in the sensory tests, will provide more income opportunities for table grape growers.
5. Conclusions
In crossbreeding studies, the parental combinations selected greatly affect the improvement of important quality traits, including the quality of the fruit obtained. As in this study, many hybrid grape genotypes have been obtained using different combinations to develop new seedless grape cultivars, and whether they have the desired traits has been extensively investigated. In particular, degustation panel scores given by trained panellists guide potential consumer preferences for these hybrid genotypes. In this study, among the 14 hybrid genotypes that received sufficient scores from the panellists, the seedless hybrid genotypes 22BY-097, 14KS-03, 18BK-02, and 22BY-044, which received the highest scores, have a higher market chance. Another important point is that hybrid genotypes that receive high scores in taste tests should be subjected to yield and quality evaluations in different locations to assess their suitability for registration in the next stage. More reliable data can be obtained through adaptation trials, especially in the Aegean and Mediterranean regions, where table grapes are widely grown. These data will be very useful in the commercialization of new cultivars that are registered at a later stage.
This approach will confirm the accuracy and consistency of the results obtained, enabling the introduction of new cultivars to table grape growers. Grape growers can increase their income by adopting these new cultivars with superior quality and higher marketing chances. Moreover, this will help to ensure that future generations recognize viticulture as a profitable agricultural activity, significantly contributing to the sustainability of table grape cultivation.
Author Contributions
A.A.: conceptualization, methodology, supervision, resources, visualization, project administration, writing—original draft, writing—review and editing; M.A.: conceptualization, methodology, formal analyses, resources; A.Ş.: conceptualization, methodology, project administration; Z.G.: formal analyses, validation; A.U.: formal analyses. All authors have read and agreed to the published version of the manuscript.
Funding
This research has been funded by The Scientific and Technological Research Council of Turkey (TÜBİTAK) as part of the TÜBİTAK 1004 Project (Project Number: 22AG012). Also, this article contains data from Melek AKGÜN’s master’s thesis study.
Data Availability Statement
The data underlying this article were provided with permission. Data will be shared upon request to the corresponding author.
Acknowledgments
The authors express special thanks to colleagues at the Yalova AHCRI for their help during this experiment.
Conflicts of Interest
Author Aydın Uzun was employed by the company Projenia R&D and Consulting Co., Ltd. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- FAOSTAT. Food and Agricultural Organization of United Nations. Available online: https://www.fao.org/faostat/en/#data/QCL (accessed on 1 October 2024).
- Tırkaz, E.; Aşçıoğul, O.; Yılmaz, E.; Almış, N. Current and future outlook of the Turkish dried grape industry. Acta Hortic. 2020, 1276, 1–6. [Google Scholar] [CrossRef]
- Uysal, H.; Karabat, S. Forecasting and evaluation for raisin export in turkey. BIO Web Conf. 2017, 9, 03002. [Google Scholar] [CrossRef]
- Atak, A.; Ergönül, O.; Dilli, Y.; Kesgin, M.; Altındişli, A. Grapevine breeding studies in Türkiye. Acta Hortic. 2023, 1370, 145–152. [Google Scholar] [CrossRef]
- Alston, J.M.; Sambucci, O. Grapes in the World Economy. In The Grape Genome; Cantu, D., Walker, M., Eds.; Compendium of Plant Genomes; Springer: Cham, Switzerland, 2019. [Google Scholar] [CrossRef]
- Atak, A. Challenges and strategies for table grape breeding in the face of global climate change. Acta Hortic. 2024, 1401, 33–40. [Google Scholar] [CrossRef]
- OIV. OIV Statistical Report on World Viticulture, 2017, 75008, Paris. Available online: https://www.oiv.int/public/medias/5479/oiv-en-bilan-2017.pdf (accessed on 30 December 2024).
- Söylemezoğlu, G.; Atak, A.; Boz, Y.; Ünal, A.; Sağlam, M. Viticulture Studies in Türkiye. Chron. Hortic. 2016, 52, 27–31. Available online: https://www.ishs.org/system/files/chronica-documents/ch5602.pdf (accessed on 20 January 2025).
- Li, S.; Li, Z.; Zhao, Y.; Zhao, J.; Luo, Q.; Wang, Y. New disease-resistant, seedless grapes are developed using embryo rescue and molecular markers. 3 Biotech 2020, 10, 4. [Google Scholar] [CrossRef]
- Piarulli, L.; Pirolo, C.; Roseti, V.; Bellin, D.; Mascio, I.; La Notte, P.; Montemurro, C.; Miazzi, M.M. Breeding new seedless table grapevines for a more sustainable viticulture in Mediterranean climate. Front. Plant Sci. 2024, 15, 1379642. [Google Scholar] [CrossRef]
- Atak, A. Vitis species for stress tolerance/resistance. Genet. Resour. Crop Evol. 2024, 72, 2425–2444. [Google Scholar] [CrossRef]
- Crespo, P.; Martínez-Zamora, L.; Artés-Hernández, F.; Tornel, M. New table grape hybrids development from teinturier population with enhanced phytochemical quality. Sci. Hortic. 2024, 326, 112756. [Google Scholar] [CrossRef]
- Akkurt, M.; Tahmaz, H.; Veziroğlu, S. Recent Developments in Seedless Grapevine Breeding. S. Afr. J. Enol. Vitic. 2019, 40, 1. [Google Scholar] [CrossRef]
- Weisong, M.; Chengcheng, L.; Dong, T.; Jianying, F. Chinese consumers’ behavior and preference to table grapes: Based on a comparative study of 2009 and 2014. Br. Food J. 2016, 118, 231–246. [Google Scholar] [CrossRef]
- Liu, T.; Wang, J.; Feng, Y.; Mu, W. Chinese consumers’ attribute preference prediction for table grapes. Ital. J. Food Sci. 2023, 35, 147–158. [Google Scholar] [CrossRef]
- Atak, A.; Göksel, Z.; Yılmaz, Y. Changes in Major Phenolic Compounds of Seeds, Skins, and Pulps from Various Vitis spp and the Effect of Powdery and Downy Mildew Diseases on Their Levels in Grape Leaves. Plants 2021, 10, 2554. [Google Scholar] [CrossRef] [PubMed]
- Gratl, V.; Sturm, S.; Zini, E.; Letschka, T.; Stefanini, M.; Vezzulli, S.; Stuppner, H. Comprehensive polyphenolic profiling in promising resistant grapevine hybrids including 17 novel breeds in northern Italy. J. Sci. Food Agric. 2021, 101, 2380–2388. [Google Scholar] [CrossRef]
- Vezzulli, S.; Gramaje, D.; Tello, J.; Gambino, G.; Bettinelli, P.; Pirrello, C.; Schwandner, A.; Schwandner, P.; Angelini, E.; Anfora, G.; et al. Genomic Designing for Biotic Stress Resistant Grapevine; Kole, C., Ed.; Genomic Designing for Biotic Stress Resistant Fruit Crops; Springer: Cham, Switzerland, 2022. [Google Scholar] [CrossRef]
- Chu, Y.; Li, M.; Li, R.; Zhang, K.; Qiu, P.; Yuan, X.; Han, Y.; Liu, X.; Xu, Y.; Liu, G. Embryo Rescue Breeding of New Cold-Resistant, Seedless Grapes. Horticulturae 2023, 9, 992. [Google Scholar] [CrossRef]
- Ebadi, A.; Moghadam, J.E.; Fatahi, R. Evaluation of 22 populations achieved from controlled crossing between some seeded × seedless grapevine cultivars. Sci. Hortic. 2009, 119, 371–376. [Google Scholar] [CrossRef]
- Yamada, M.; Sato, A. Advances in table grape breeding in Japan. Breed Sci. 2016, 66, 34–45. [Google Scholar] [CrossRef]
- Atak, A.; Akkurt, M.; Polat, Z.; Çelik, H.; Kahraman, K.; Akgul, D.; Özer, N.; Söylemezoğlu, G.; Şire Gülbasar, G.; Eibach, R. Susceptibility to downy mildew (Plasmopara viticola) and powdery mildew (Erysiphe necator) of different Vitis cultivars and genotypes. Ciência e Técnica Vitivinícola 2017, 32, 23–32. [Google Scholar] [CrossRef]
- Özer, N.; Uzun, H.İ.; Aktürk, B.; Özer, C.; Akkurt, M.; Aydın, S. Resistance assessment of grapevine leaves to downy mildew with sporulation area scoring. Eur. J. Plant Pathol. 2021, 160, 337–348. [Google Scholar] [CrossRef]
- Ruiz-Garcia, L.; Gago, P.; Martínez-Mora, C.; Santiago, J.L.; Fernádez-López, D.J.; Martinez, M.D.C.; Boso, S. Evaluation and pre-selection of new grapevine genotypes resistant to downy and powdery mildew, obtained by Cross-Breeding programs in Spain. Front. Plant Sci. 2021, 12, 674510. [Google Scholar] [CrossRef]
- Luca, L.P.; Di Guardo, M.; Bennici, S.; Ferlito, F.; Nicolosi, E.; La Malfa, S.; Gentile, A.; Distefano, G. Development of an efficient molecular-marker assisted selection strategy for berry color in grapevine. Sci. Hortic. 2024, 337, 113522. [Google Scholar] [CrossRef]
- Vargas, A.M.; Fernández-Pastor, M.; Castro, F.J.; Martínez, M.A.; Gómez-Cifuentes, A.; Espinosa-Roldán, F.; Cabello, F.; Muñoz-Organero, G.; de Andrés, M.T. Strategy to minimize phenotyping in the selection of new table grape varieties. BIO Web Conf. 2023, 56, 01030. [Google Scholar] [CrossRef]
- Zheng, T.; Cheng, J.H.; Wei, L.Z.; Xiang, J.; Wu, A.J. A New early ripening seedless grape cultivar Tiangong Cuixiangmi. J. Fruit Sci. 2023, 40, 1771–1773. [Google Scholar] [CrossRef]
- Fortes, A.M.; Pais, M.S. Grape (Vitis species). In Nutritional Composition of Fruit Cultivars; Simmonds, M.S.J., Preedy, V.R., Eds.; Academic Press: Cambridge, MA, USA, 2016; pp. 257–286. [Google Scholar] [CrossRef]
- Ferreira, F.; Pinto-Carnide, O.; Arroyo-García, R.; Castro, I. Berry color variation in grapevine as a source of diversity. Plant Physiol. Biochem. 2018, 132, 696–707. [Google Scholar] [CrossRef]
- İşçi, B.; Kacar, E.; Altındişli, A. The Effects of Some Exogenous Applications on Quality in ‘Crimson Seedless’ Grape. Erwerbs Obstbau 2020, 62, 87–100. [Google Scholar] [CrossRef]
- Kupe, M.; Karatas, N.; Unal, M.S.; Ercisli, S.; Baron, M.; Sochor, J. Phenolic Composition and Antioxidant Activity of Peel, Pulp and Seed Extracts of Different Clones of the Turkish Grape Cultivar ‘Karaerik’. Plants 2021, 10, 2154. [Google Scholar] [CrossRef]
- Castillejo, N.; Martínez-Hernández, G.B.; Monaco, K.; Gómez, P.A.; Aguayo, E.; Artés, F.; Artés-Hernández, F. Preservation of bioactive compounds of a green vegetable smoothie using short time-high temperature mild thermal treatment. Food Sci. Technol. Int. 2017, 23, 46–60. [Google Scholar] [CrossRef]
- Ferrara, G.; Mazzeo, A.; Matarrese, A.M.S.; Pacucci, C.; Pacifico, A.; Gambacorta, G.; Faccia, M.; Trani, A.; Gallo, V.; Cafagna, I.; et al. Application of abscisic acid (S-ABA) to ‘Crimson Seedless’ grape berries in a Mediterranean climate: Effects on color, chemical characteristics, metabolic profile, and S-ABA concentration. J. Plant Growth Regul. 2013, 32, 491–505. [Google Scholar]
- Atak, A.; Kahraman, K.A. Breeding studies and new table grapes in Türkiye. E3 J. Agric. Res. Develop. 2012, 2, 80–85. [Google Scholar]
- SAS. JMP 18.1 Statistical Discovery LLC; SAS Institute Inc.: Cary, NC, USA, 1989–2024. [Google Scholar]
- Wang, Z.; Zhou, J.; Xu, X.; Perl, A.; Chen, S.; Ma, H. Adoption of table grape cultivars: An attribute preference study on Chinese grape growers. Sci. Hortic. 2017, 216, 66–75. [Google Scholar] [CrossRef]
- Merdinoglu, D.; Schneider, C.; Prado, E.; Wiedemann-Merdinoglu, S.; Mestre, P. Breeding for durable resistance to downy and powdery mildew in grapevine. OENO One 2018, 52, 203–209. [Google Scholar] [CrossRef]
- Schneider, C.; Onimus, C.; Prado, E.; Dumas, V.; Wiedemann-Merdinoglu, S.; Dorne, M.A.; Lacombe, M.C.; Piron, M.C.; Umar-Faruk, A.; Duchêne, E.; et al. INRA-ResDur: The French grapevine breeding programme for durable resistance to downy and powdery mildew. Acta Hortic. 2019, 1248, 207–214. [Google Scholar] [CrossRef]
- Foria, S.; Monte, C.; Testolin, R.; Di Gaspero, G.; Cipriani, G. Pyramidizing resistance genes in grape: A breeding program for the selection of elite cultivars. Acta Hortic. 1248, 2019, 549–554. [Google Scholar] [CrossRef]
- Maia, J.D.G.; Camargo, U.A.; Tonietto, J.; Zanus, M.C.; Quecini, V.; Ferreira, M.E.; Ritschel, P. Grapevine breeding programs in Brazil. In Grapevine Breeding Programs for the Wine Industry; Elsevier: Amsterdam, The Netherlands, 2015; pp. 247–271. [Google Scholar] [CrossRef]
- Souza Leão, P.C.; Carvalho, J.N. Assessment of table grape progenies and correlation between seedlessness and other agronomic traits. Bras. Frutic. 2023, 45, e-225. [Google Scholar] [CrossRef]
- Zyprian, E.; Eibach, R.; Trapp, O.; Schwander, F.; Töpfer, R. Grapevine breeding under climate change: Applicability of a molecular marker linked to véraison. Vitis 2018, 57, 119–123. [Google Scholar] [CrossRef]
- Volynkin, V.; Polulyah, A.; Klimenko, V.; Likhovskoi, V.; Oleinikov, N.; Levchenko, S.; Pavlova, I.; Zlenko, V.; Kotolovets, Z.; Pytel, I.; et al. Breeding for Ukrainian table grape varieties. Vitis 2015, 54, 157–158. [Google Scholar] [CrossRef]
- Xu, H.Y.; Zhang, G.J.; Yan, A.L.; Sun, L. Table grape breeding at the Beijing Institute of Forestry and Pomology. Acta Hortic. 2015, 1082, 43–46. [Google Scholar] [CrossRef]
- Ponce, M.T.; Agüero, C.B.; Gregori, M.T.; Tizio, R. Factors affecting the development of stenospermic grape (Vitis vinifera) embryos cultured in vitro. Acta Hortic. 2000, 528, 667–672. [Google Scholar] [CrossRef]
- Puglisi, D.; Las Casas, G.; Ferlito, F.; Nicolosi, E.; Di Guardo, M.; Scollo, F.; Saitta, G.; La Malfa, S.; Gentile, A.; Distefano, G. Parents’ selection affects embryo rescue, seed regeneration and the heredity of seedless trait in table grape breeding programs. Agriculture 2022, 12, 1096. [Google Scholar] [CrossRef]
- Pastrana-Bonilla, E.; Akoh, C.C.; Sellappan, S.; Krewer, G. Phenolic content and antioxidant capacity of Muscadine grapes. J. Agric. Food Chem. 2003, 51, 5497–5503. [Google Scholar] [CrossRef]
- Doshi, P.; Adsule, P.; Banargee, K. Phenolic composition and antioxidant activity in grapevine parts and berries (Vitis vinifera L.) cv. Kishmish Chornyi (Sharad Seedless) during maturation. Int.J. Food Sci. Tech. 2006, 41, 1–9. [Google Scholar] [CrossRef]
- Poudel, P.D.; Tamura, H.; Kataoka, I.; Mochioka, R. Phenolic compounds and antioxidant activities of skins and seeds of five wild grapes and two hybrids native to Japan. J. Food Compos. Anal. 2008, 21, 622–625. [Google Scholar] [CrossRef]
- Izcara, S.; Morante-Zarcero, S.; de Andrés, M.T.; Arroyo, T.; Sierra, I. A comparative study of phenolic composition and antioxidant activity in commercial and experimental seedless table grapes cultivated in a Mediterranean climate. Food Meas. 2021, 15, 1916–1930. [Google Scholar] [CrossRef]
- Ananga, A.; Georgiev, V.; Ochieng, J.; Phills, B.; Tsolov, V. Production of Anthocyanins in Grape Cell Cultures: A Potential Source of Raw Material for Pharmaceutical, Food, and Cosmetic Industries [Internet]. In The Mediterranean Genetic Code—Grapevine and Olive; InTechOpen: Rijeka, Croatia, 2013. [Google Scholar] [CrossRef]
- Wang, Z.L.; Yao, F.; Hui, M.; Wu, D.; Wang, Y.; Han, X.; Cao, X.; Li, Y.H.; Li, H.; Wang, H. Fertility analysis of intraspecific hybrids in Vitis vinifera and screening of superior hybrid combinations. Front Plant Sci. 2022, 13, 940540. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).