Transcription Factor RhCUC3 Regulates Petal Numbers in Rose Flowers
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Phytohormone Treatments
2.3. Sequence Analysis of RhCUC3
2.4. RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
2.5. Subcellular Localization
2.6. Virus-Induced Gene Silencing
2.7. Statistical Analysis
3. Results
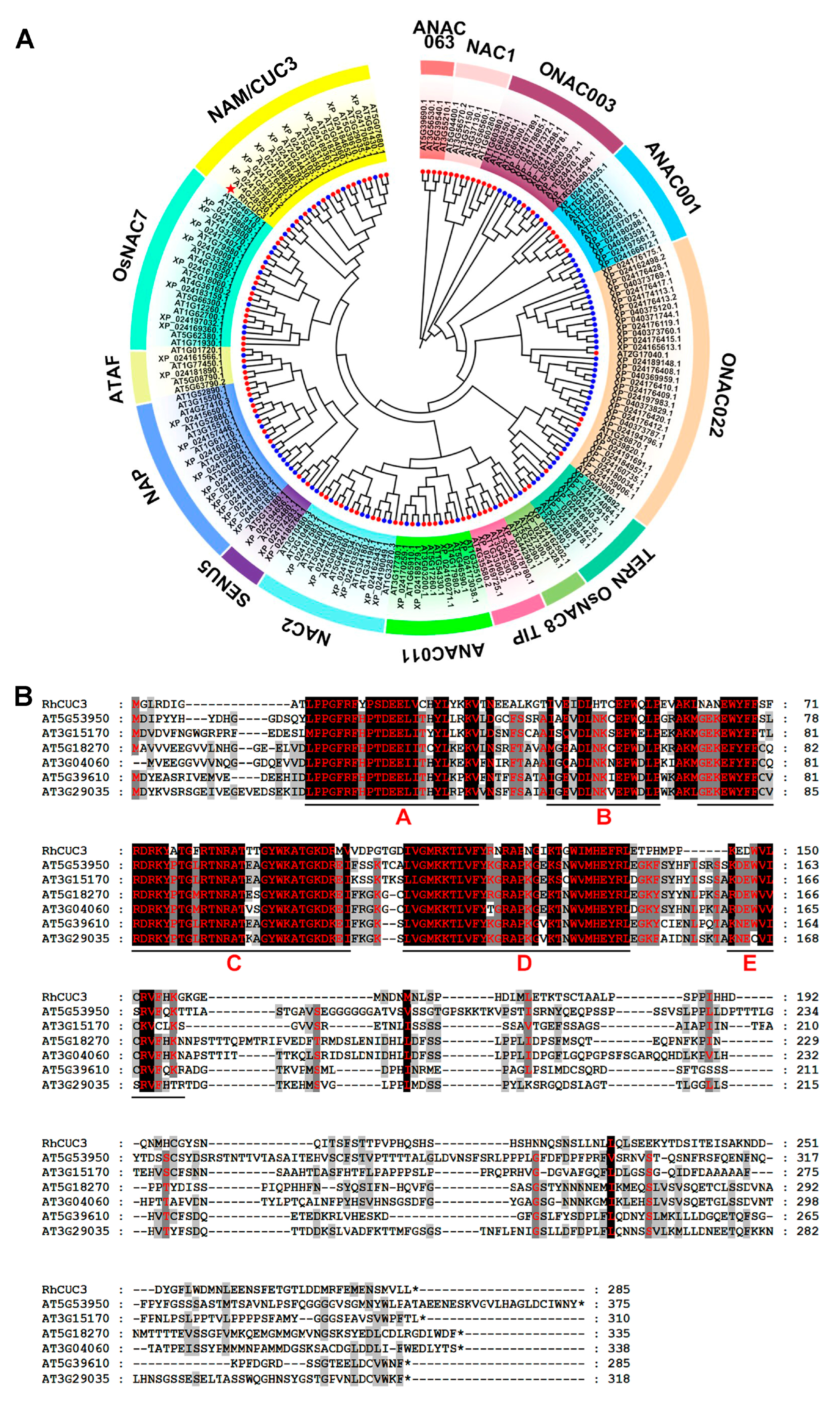
3.1. Isolation and Sequence of RhCUC3
3.2. Expression Analysis of RhCUC3 in Different Developmental Stages and Organs
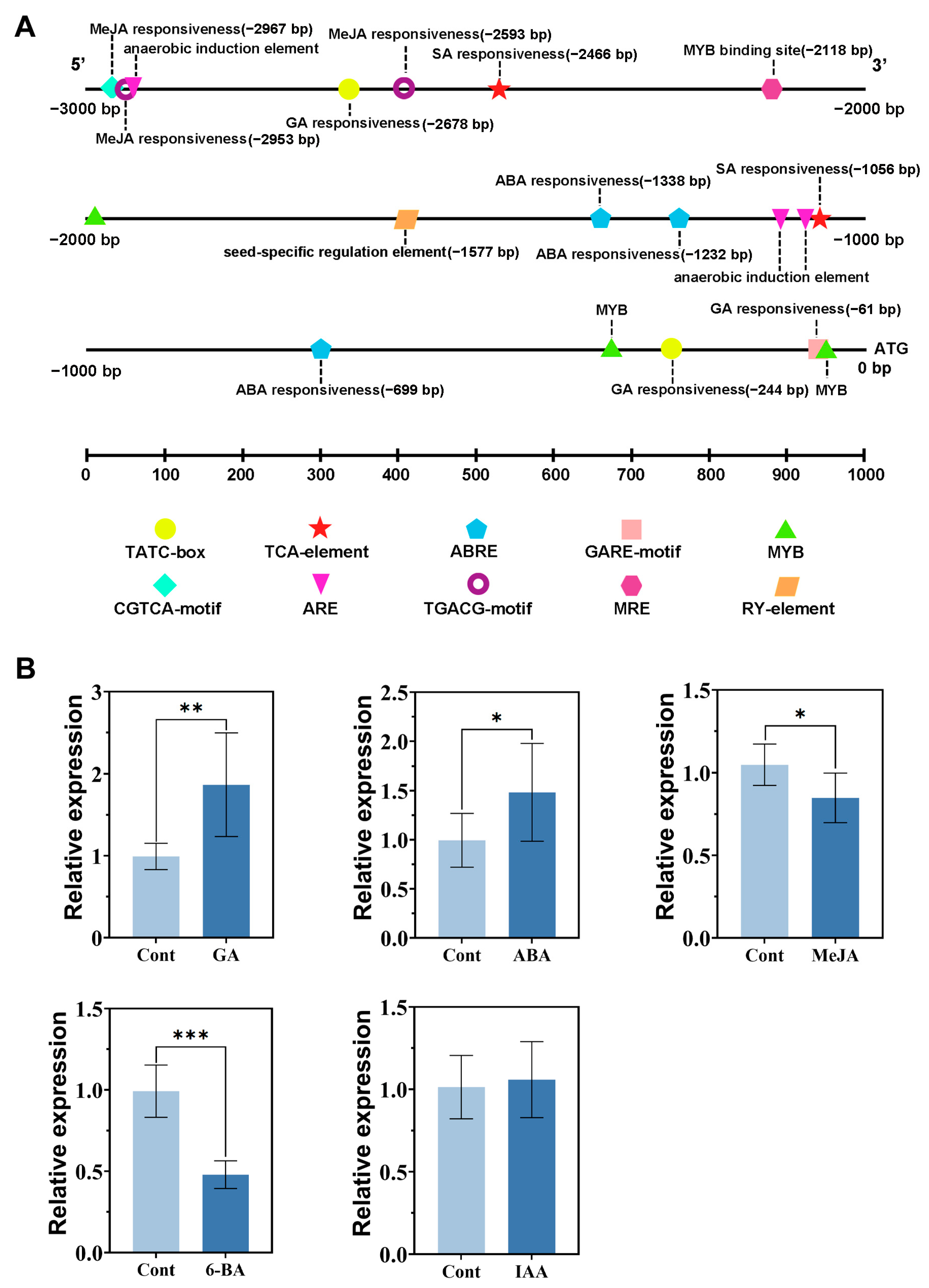
3.3. Expression of RhCUC3 Under Different Phytohormones
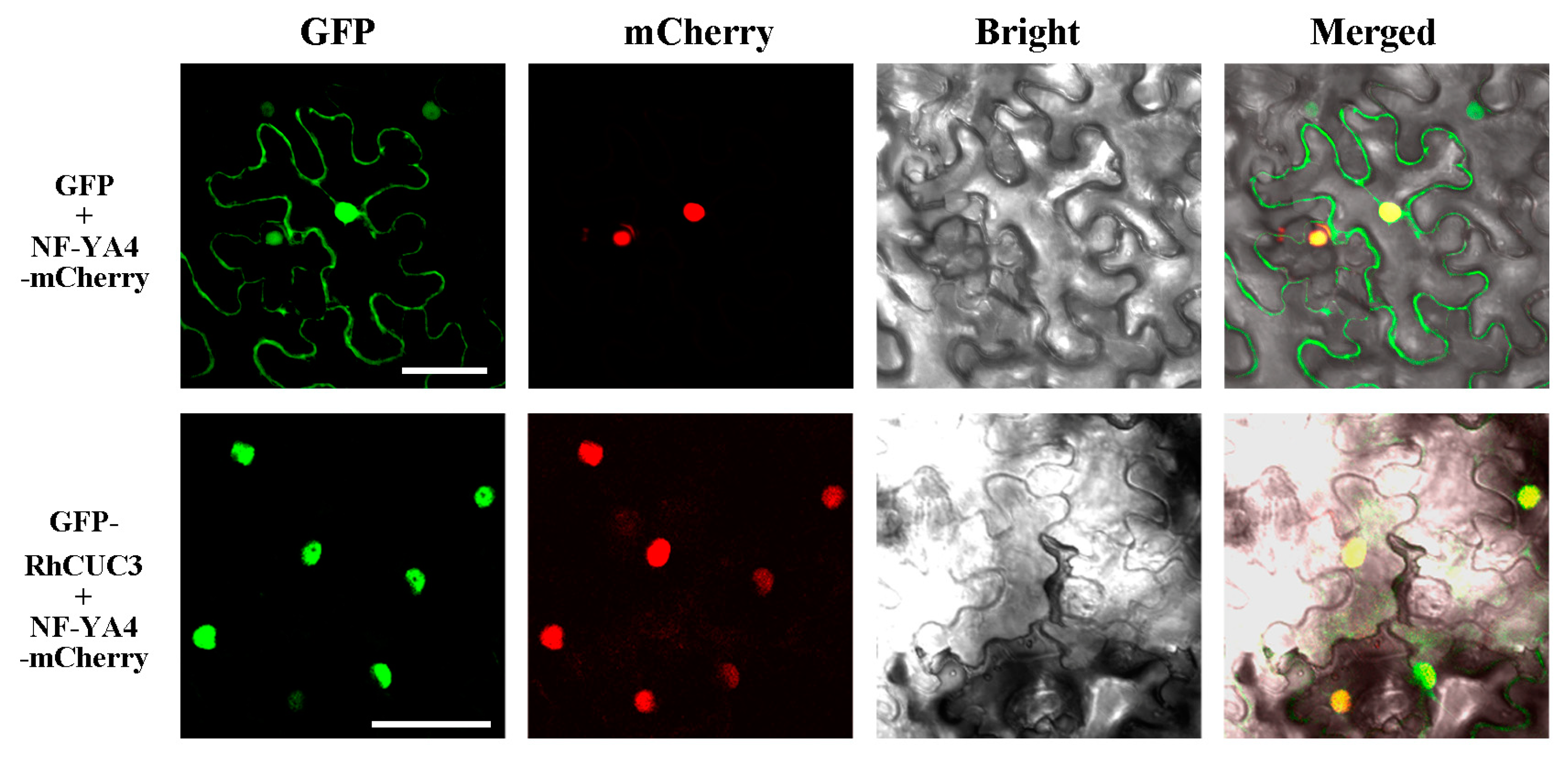
3.4. RhCUC3 Was Localized in the Nucleus
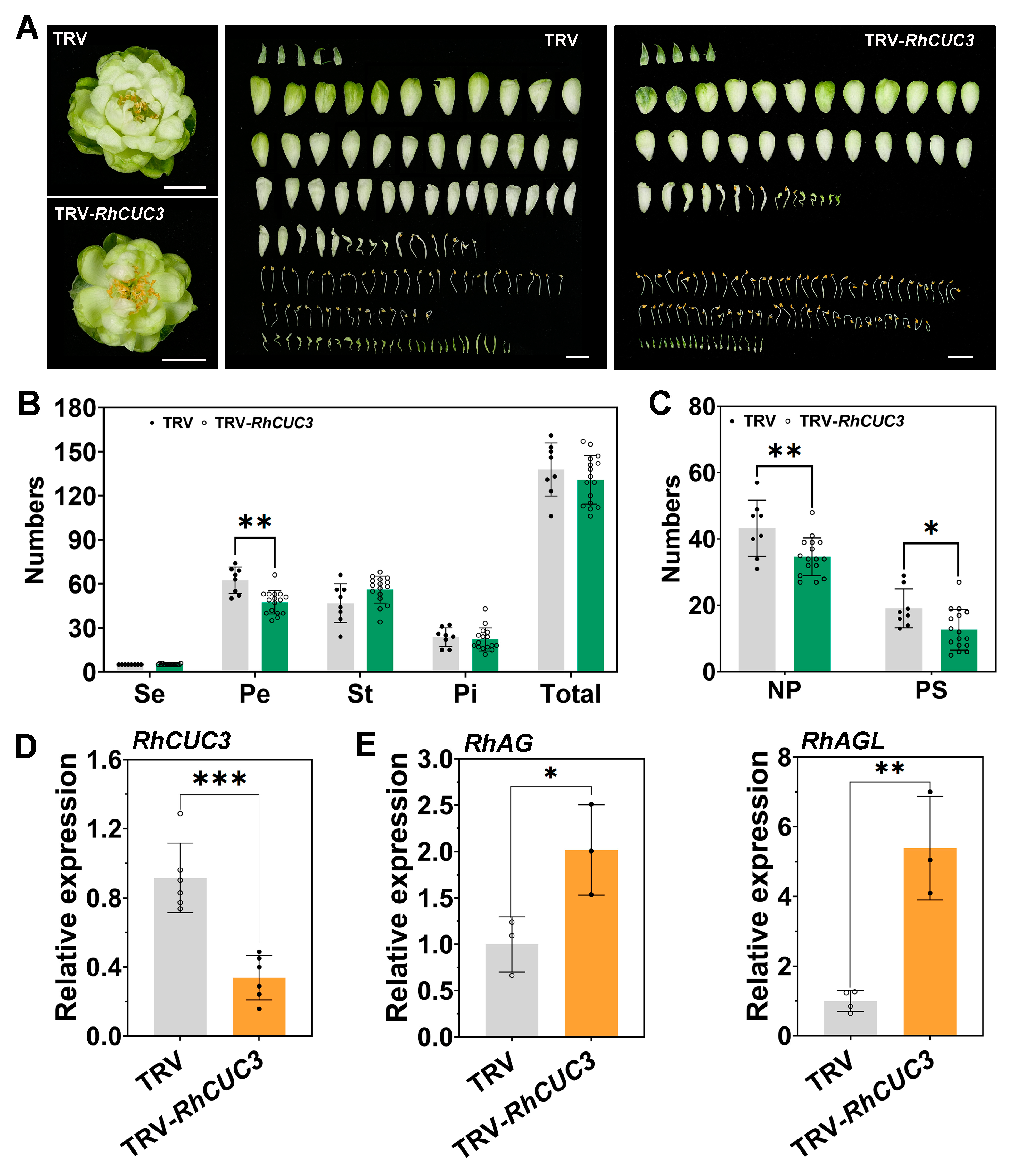
3.5. Silencing of RhCUC3 Decreased the Petal Numbers of Rose Flowers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Meyerowitz, E.M.; Smyth, D.R.; Bowman, J.L. Abnormal flowers and pattern formation in floral development. Development 1989, 106, 209–217. [Google Scholar] [CrossRef]
- Dubois, A.; Raymond, O.; Maene, M.; Baudino, S.; Langlade, N.B.; Boltz, V.; Vergne, P.; Bendahmane, M. Tinkering with the C-Function: A molecular frame for the selection of double flowers in cultivated roses. PLoS ONE 2010, 5, e9288. [Google Scholar] [CrossRef] [PubMed]
- Debener, T. Genetic analysis of horticulturally important morphological and physiological characters in diploid roses. Gartenbauwissenschaft 1999, 64, 14–20. [Google Scholar]
- Morey, D. Observations on the genetics of doubleness in roses. Am. Rose Annu. 1959, 44, 113–116. [Google Scholar]
- Hibrand-Saint Oyant, L.; Crespel, L.; Rajapakse, S.; Zhang, L.; Foucher, F. Genetic linkage maps of rose constructed with new microsatellite markers and locating QTL controlling flowering traits. Tree Genet. Genomes 2007, 4, 11–23. [Google Scholar] [CrossRef]
- Chen, J.; Li, Y.; Li, Y.; Li, Y.; Wang, Y.; Jiang, C.; Choisy, P.; Xu, T.; Cai, Y.; Pei, D.; et al. AUXIN RESPONSE FACTOR 18–HISTONE DEACETYLASE 6 module regulates floral organ identity in rose (Rosa hybrida). Plant Physiol. 2021, 186, 1074–1087. [Google Scholar] [CrossRef]
- Cai, Y.; Wang, L.; Ogutu, C.O.; Yang, Q.; Luo, B.; Liao, L.; Zheng, B.; Zhang, R.; Han, Y. The MADS-box gene PpPI is a key regulator of the double-flower trait in peach. Physiol. Plant. 2021, 173, 2119–2129. [Google Scholar] [CrossRef]
- Peng, L.; Li, Y.; Tan, W.; Wu, S.; Hao, Q.; Tong, N.; Wang, Z.; Liu, Z.; Shu, Q. Combined genome-wide association studies and expression quantitative trait locus analysis uncovers a genetic regulatory network of floral organ number in a tree peony (Paeonia suffruticosa Andrews) breeding population. Hortic. Res. 2023, 10, uhad110. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Li, J.; Fan, Z.; Liu, Z.; Tanaka, T.; Yin, H. Global gene expression defines faded whorl specification of double flower domestication in Camellia. Sci. Rep. 2017, 7, 3197. [Google Scholar] [CrossRef]
- Hu, L.; Zheng, T.; Cai, M.; Pan, H.; Wang, J.; Zhang, Q. Transcriptome analysis during floral organ development provides insights into stamen petaloidy in Lagerstroemia speciosa. Plant Physiol. Biochem. 2019, 142, 510–518. [Google Scholar] [CrossRef]
- Soltis, P.S.; Soltis, D.E.; Kim, S.; Chanderbali, A.; Buzgo, M. Expression of floral regulators in basal angiosperms and the origin and evolution of ABC-Function. Adv. Bot. Res. 2006, 44, 483–506. [Google Scholar]
- Bowman, J.L.; Smyth, D.R.; Meyerowitz, E.M. Genetic interactions among floral homeotic genes of Arabidopsis. Development 1991, 112, 1–20. [Google Scholar] [CrossRef]
- Coen, E.S.; Meyerowitz, E.M. The war of the whorls: Genetic interactions controlling flower development. Nature 1991, 353, 31–37. [Google Scholar] [CrossRef]
- Theißen, G. Development of floral organ identity: Stories from the MADS house. Curr. Opin. Plant Biol. 2001, 4, 75–85. [Google Scholar] [CrossRef]
- Litt, A.; Kramer, E.M. The ABC model and the diversification of floral organ identity. Semin. Cell Dev. Biol. 2010, 21, 129–137. [Google Scholar] [CrossRef]
- Irish, V.F. The flowering of Arabidopsis flower development. Plant J. 2010, 61, 1014–1028. [Google Scholar] [CrossRef]
- Ronse De Craene, L.P. The evolutionary significance of homeosis in flowers: A morphological perspective. Int. J. Plant Sci. 2003, 164, 225–235. [Google Scholar] [CrossRef]
- Lampugnani, E.R.; Kilinc, A.; Smyth, D.R. PETAL LOSS is a boundary gene that inhibits growth between developing sepals in Arabidopsis thaliana. Plant J. 2012, 71, 724–735. [Google Scholar] [CrossRef]
- Ding, L.; Yan, S.; Jiang, L.; Zhao, W.; Ning, K.; Zhao, J.; Liu, X.; Zhang, J.; Wang, Q.; Zhang, X. HANABA TARANU (HAN) bridges meristem and organ primordia boundaries through PINHEAD, JAGGED, BLADE-ON-PETIOLE2 and CYTOKININ OXIDASE 3 during flower development in Arabidopsis. PLoS Genet. 2015, 11, e1005479. [Google Scholar] [CrossRef]
- Thomson, B.; Zheng, B.; Wellmer, F. Floral organogenesis: When knowing your ABCs is not enough. Plant Physiol. 2017, 173, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Quon, T.; Lampugnani, E.R.; Smyth, D.R. PETAL LOSS and ROXY1 interact to limit growth within and between sepals but to promote petal initiation in Arabidopsis thaliana. Front. Plant Sci. 2017, 8, 152. [Google Scholar] [CrossRef]
- Xu, Y.; Prunet, N.; Gan, E.; Wang, Y.; Stewart, D.; Wellmer, F.; Huang, J.; Yamaguchi, N.; Tatsumi, Y.; Kojima, M.; et al. SUPERMAN regulates floral whorl boundaries through control of auxin biosynthesis. EMBO J. 2018, 37, e97499. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Wei, W.; Li, Y.; Kan, L.; Wang, F.; Zhang, X.; Li, F.; Liu, Z.; Kang, C. Conserved and novel roles of miR164-CUC 2 regulatory module in specifying leaf and floral organ morphology in strawberry. New Phytol. 2019, 224, 480–492. [Google Scholar] [CrossRef] [PubMed]
- Aida, M.; Ishida, T.; Fukaki, H.; Fujisawa, H.; Tasaka, M. Genes involved in organ separation in Arabidopsis: An analysis of the cup-shaped cotyledon mutant. Plant Cell 1997, 9, 841–857. [Google Scholar] [CrossRef] [PubMed]
- Berger, Y.; Harpaz-Saad, S.; Brand, A.; Melnik, H.; Sirding, N.; Alvarez, J.P.; Zinder, M.; Samach, A.; Eshed, Y.; Ori, N. The NAC-domain transcription factor GOBLET specifies leaflet boundaries in compound tomato leaves. Development 2009, 136, 823–832. [Google Scholar] [CrossRef]
- Cheng, X.; Peng, J.; Ma, J.; Tang, Y.; Chen, R.; Mysore, K.S.; Wen, J. NO APICAL MERISTEM (MtNAM) regulates floral organ identity and lateral organ separation in Medicago truncatula. New Phytol. 2012, 195, 71–84. [Google Scholar] [CrossRef]
- Baker, C.C.; Sieber, P.; Wellmer, F.; Meyerowitz, E.M. The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Curr. Biol. 2005, 15, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.; López-Giráldez, F.; Townsend, J.P.; Irish, V.F. RBE controls microRNA164 expression to effect floral organogenesis. Development 2012, 139, 2161–2169. [Google Scholar] [CrossRef] [PubMed]
- Takada, S.; Hibara, K.; Ishida, T.; Tasaka, M. The CUP-SHAPED COTYLEDON1 gene of Arabidopsis regulates shoot apical meristem formation. Development 2001, 128, 1127–1135. [Google Scholar] [CrossRef]
- Daimon, Y.; Takabe, K.; Tasaka, M. The CUP-SHAPED COTYLEDON genes promote adventitious shoot formation on calli. Plant Cell Physiol. 2003, 44, 113–121. [Google Scholar] [CrossRef]
- Li, X.G.; Su, Y.H.; Zhao, X.Y.; Li, W.; Gao, X.Q.; Zhang, X.S. Cytokinin overproduction-caused alteration of flower development is partially mediated by CUC2 and CUC3 in Arabidopsis. Gene 2010, 450, 109–120. [Google Scholar] [CrossRef] [PubMed]
- Maugarny, A.; Gonçalves, B.; Arnaud, N.; Laufs, P. CUC transcription factors: To the meristem and beyond. In Plant Transcription Factors; Gonzalez, D.H., Ed.; Academic Press: London, UK, 2016; pp. 229–247. [Google Scholar]
- Vernoux, T.; Kronenberger, J.; Grandjean, O.; Laufs, P.; Traas, J. PIN-FORMED 1 regulates cell fate at the periphery of the shoot apical meristem. Development 2000, 127, 5157–5165. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.L.; Wilson-Sánchez, D.; Bhatia, N.; Rast-Somssich, M.I.; Wu, A.; Vlad, D.; McGuire, L.; Nikolov, L.A.; Laufs, P.; Gan, X.; et al. A CUC1/auxin genetic module links cell polarity to patterned tissue growth and leaf shape diversity in crucifer plants. Proc. Natl. Acad. Sci. USA 2024, 121, e2321877121. [Google Scholar] [CrossRef]
- Wang, J.-R.; Wang, X.; Su, N.; Li, Q.-J.; Zhang, X.-H.; Ma, Y.-P.; Zhao, L.; Ginefra Toni, J.F.; De Craene, L.R. Floral morphology and morphogenesis in Sanguisorba (Rosaceae): Flower diversification despite petal reduction and spatial constraints. Bot. J. Linn. Soc. 2020, 193, 47–63. [Google Scholar] [CrossRef]
- Wang, X.; Gong, J.; Li, Q.; Wang, J.; Ma, Y.; Zhang, X.; Chang, Z.; Wen, J.; Zhao, L. Floral organogenesis of Prunus Laurocerasus and P. Serotina and its significance for the systematics of the genus and androecium diversity in Rosaceae. Botany 2019, 97, 71–84. [Google Scholar] [CrossRef]
- Bendahmane, M.; Dubois, A.; Raymond, O.; Bris, M.L. Genetics and genomics of flower initiation and development in roses. J. Exp. Bot. 2013, 64, 847–857. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Guo, Y.; Wu, Z.; Ren, H.; Jiang, Y.; Ma, N.; Gao, J.; Sun, X. RhAGL24 Regulating Auxin-related gene RhARF18 affects stamen petaloidy in rose. Horticulturae 2022, 8, 407. [Google Scholar] [CrossRef]
- Yang, T.; Wang, Y.; Li, Y.; Liang, S.; Yang, Y.; Huang, Z.; Li, Y.; Gao, J.; Ma, N.; Zhou, X. The transcription factor RhMYB17 regulates the homeotic transformation of floral organs in Rose (Rosa hybrida) under cold stress. J. Exp. Bot. 2024, 75, 2965–2981. [Google Scholar] [CrossRef]
- Noor, S.H.; Ushijima, K.; Murata, A.; Yoshida, K.; Tanabe, M.; Tanigawa, T.; Kubo, Y.; Nakano, R. Double flower formation induced by silencing of C-class MADS-box genes and its variation among petunia cultivars. Sci. Hortic. 2014, 178, 1–7. [Google Scholar] [CrossRef]
- Nakatsuka, T.; Saito, M.; Yamada, E.; Fujita, K.; Yamagishi, N.; Yoshikawa, N.; Nishihara, M. Isolation and characterization of the C-class MADS-box gene involved in the formation of double flowers in Japanese gentian. BMC Plant Biol. 2015, 15, 182. [Google Scholar] [CrossRef] [PubMed]
- Ma, N.; Chen, W.; Fan, T.; Tian, Y.; Zhang, S.; Zeng, D.; Li, Y. Low temperature-induced DNA hypermethylation attenuates expression of RhAG, an AGAMOUS homolog, and increases petal number in rose (Rosa hybrida). BMC Plant Biol 2015, 15, 237. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Tang, A.; Wan, H.; Zhang, T.; Cheng, T.; Wang, J.; Yang, W.; Pan, H.; Zhang, Q. An APETALA2 homolog, RcAP2, regulates the number of rose petals derived from stamens and response to temperature fluctuations. Front. Plant Sci. 2018, 9, 481. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Li, Y.; Wang, Y.; Li, Y.; He, J.; Li, Y.; Du, L.; Gao, Y.; Ma, N.; Gao, J.; et al. Disruption of transcription factor RhMYB123 causes the transformation of stamen to malformed petal in rose (Rosa hybrida). Plant Cell Rep. 2022, 41, 2293–2303. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Meng, Y.; Li, N.; Tian, J.; Gao, J.; Zhang, C. Identification and validation of reference genes for gene expression studies in postharvest rose flower (Rosa hybrida). Sci. Hortic. 2013, 158, 16–21. [Google Scholar] [CrossRef]
- Dai, F.; Zhang, C.; Jiang, X.; Kang, M.; Yin, X.; Lü, P.; Zhang, X.; Zheng, Y.; Gao, J. RhNAC2 and RhEXPA4 are involved in the regulation of dehydration tolerance during the expansion of rose petals. Plant Physiol. 2012, 160, 2064–2082. [Google Scholar] [CrossRef] [PubMed]
- Thomson, B.; Wellmer, F. Molecular regulation of flower development. Curr. Top. Dev. Biol. 2019, 131, 185–210. [Google Scholar]
- Hibara, K.; Karim, M.R.; Takada, S.; Taoka, K.; Furutani, M.; Aida, M.; Tasaka, M. Arabidopsis CUP-SHAPED COTYLEDON3 regulates postembryonic shoot meristem and organ boundary formation. Plant Cell 2006, 18, 2946–2957. [Google Scholar] [CrossRef]
- Ding, A.; Li, S.; Li, W.; Hao, Q.; Wan, X.; Wang, K.; Liu, Q.; Liu, Q.; Jiang, X. RhNAC31, a novel rose NAC transcription factor, enhances tolerance to multiple abiotic stresses in Arabidopsis. Acta Physiol. Plant. 2019, 41, 75. [Google Scholar] [CrossRef]
- Nikovics, K.; Blein, T.; Peaucelle, A.; Ishida, T.; Morin, H.; Aida, M.; Laufs, P. The balance between the MIR164A and CUC2 genes controls leaf margin serration in Arabidopsis. Plant Cell 2006, 18, 2929–2945. [Google Scholar] [CrossRef]
- Lee, B.H.; Jeon, J.O.; Lee, M.M.; Kim, J.H. Genetic interaction between GROWTH-REGULATING FACTOR and CUP-SHAPED COTYLEDON in organ separation. Plant Signal. Behav. 2015, 10, e988071. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jing, W.; Gong, F.; Liu, G.; Deng, Y.; Liu, J.; Yang, W.; Sun, X.; Li, Y.; Gao, J.; Zhou, X.; et al. Petal size is controlled by the MYB73/TPL/HDA19-miR159-CKX6 module regulating cytokinin catabolism in Rosa hybrida. Nat. Commun. 2023, 14, 7106. [Google Scholar] [CrossRef]
- Bartrina, I.; Otto, E.; Strnad, M.; Werner, T.; Schmülling, T. Cytokinin regulates the activity of reproductive meristems, flower organ size, ovule formation, and thus seed yield in Arabidopsis Thaliana. Plant Cell 2011, 23, 69–80. [Google Scholar] [CrossRef]
- Burr, C.A.; Sun, J.; Yamburenko, M.V.; Willoughby, A.; Hodgens, C.; Boeshore, S.L.; Elmore, A.; Atkinson, J.; Nimchuk, Z.L.; Bishopp, A.; et al. The HK5 and HK6 cytokinin receptors mediate diverse developmental pathways in rice. Development 2020, 147, dev191734. [Google Scholar] [CrossRef]
- Sauer, M.; Kleine-Vehn, J. PIN-FORMED and PIN-LIKES auxin transport facilitators. Development 2019, 146, dev168088. [Google Scholar] [CrossRef]
- Hayashi, K.; Nakamura, S.; Fukunaga, S.; Nishimura, T.; Jenness, M.K.; Murphy, A.S.; Motose, H.; Nozaki, H.; Furutani, M.; Aoyama, T. Auxin transport sites are visualized in planta using fluorescent auxin analogs. Proc. Natl. Acad. Sci. USA 2014, 111, 11557–11562. [Google Scholar] [CrossRef] [PubMed]
- Gattolin, S.; Cirilli, M.; Pacheco, I.; Ciacciulli, A.; Da Silva Linge, C.; Mauroux, J.; Lambert, P.; Cammarata, E.; Bassi, D.; Pascal, T.; et al. Deletion of the miR172 target site in a TOE -type gene is a strong candidate variant for dominant double-flower trait in Rosaceae. Plant J. 2018, 96, 358–371. [Google Scholar] [CrossRef]
- Peaucelle, A.; Morin, H.; Traas, J.; Laufs, P. Plants expressing a miR164-resistant CUC2 gene reveal the importance of post-meristematic maintenance of phyllotaxy in Arabidopsis. Development 2007, 134, 1045–1050. [Google Scholar] [CrossRef] [PubMed]
- Chmelnitsky, I.; Azizbekova, N.; Khayat, E.; Zieslin, N. Morphological development of normal and phyllody expressing Rosa hybrida cv. Motrea flowers. Plant Growth Regul. 2002, 37, 215–221. [Google Scholar] [CrossRef]
- Maas, F.M.; Hofman-Eijer, L.B.; Hulsteijn, K. Flower morphogenesis in Rosa hybrida ’Mercedes’ as studied by cryo-scanning electron and light microscopy. Effects on light and shoot position on a branch. Ann. Bot. 1995, 75, 199–205. [Google Scholar] [CrossRef]
- Gendron, J.M.; Liu, J.-S.; Fan, M.; Bai, M.-Y.; Wenkel, S.; Springer, P.S.; Barton, M.K.; Wang, Z.-Y. Brassinosteroids regulate organ boundary formation in the shoot apical meristem of Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 21152–21157. [Google Scholar] [CrossRef]
- Vroemen, C.W.; Mordhorst, A.P.; Albrecht, C.; Kwaaitaal, M.A.C.J.; De Vries, S.C. The CUP-SHAPED COTYLEDON3 gene is required for boundary and shoot meristem formation in Arabidopsis. Plant Cell 2003, 15, 1563–1577. [Google Scholar] [CrossRef] [PubMed]
- Belles-Boix, E.; Hamant, O.; Witiak, S.M.; Morin, H.; Traas, J.; Pautot, V. KNAT6: An Arabidopsis homeobox gene involved in meristem activity and organ separation. Plant Cell 2006, 18, 1900–1907. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Fang, N.; Lu, T.; Tameshige, T.; Nakata, M.T.; Jiang, Y.; Tan, L.; He, H.; Zhang, X.; Huang, Y.; et al. WOX1 controls leaf serration development via temporally restricting BRASSINAZOLE RESISTANT 1 and CUP SHAPED COTYLEDON 3 expression in Arabidopsis. J. Exp. Bot. 2025, 76, 478–492. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, Y.; Zhao, Z.; Shen, Y.; Ding, Z.; Cui, Y.; Chen, W. Transcription Factor RhCUC3 Regulates Petal Numbers in Rose Flowers. Horticulturae 2025, 11, 170. https://doi.org/10.3390/horticulturae11020170
Fang Y, Zhao Z, Shen Y, Ding Z, Cui Y, Chen W. Transcription Factor RhCUC3 Regulates Petal Numbers in Rose Flowers. Horticulturae. 2025; 11(2):170. https://doi.org/10.3390/horticulturae11020170
Chicago/Turabian StyleFang, Yan, Zixin Zhao, Yuanji Shen, Zheyuan Ding, Yongyi Cui, and Wen Chen. 2025. "Transcription Factor RhCUC3 Regulates Petal Numbers in Rose Flowers" Horticulturae 11, no. 2: 170. https://doi.org/10.3390/horticulturae11020170
APA StyleFang, Y., Zhao, Z., Shen, Y., Ding, Z., Cui, Y., & Chen, W. (2025). Transcription Factor RhCUC3 Regulates Petal Numbers in Rose Flowers. Horticulturae, 11(2), 170. https://doi.org/10.3390/horticulturae11020170






