Characterization of LBD Genes in Cymbidium ensifolium with Roles in Floral Development and Fragrance
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Related Treatment Methods
2.2. Identification and Physicochemical Properties Analysis of LBD Genes in C. ensifolium Genome
2.3. Sequence Alignment, Protein Structure, and Phylogenetic Analysis of CeLBD Proteins
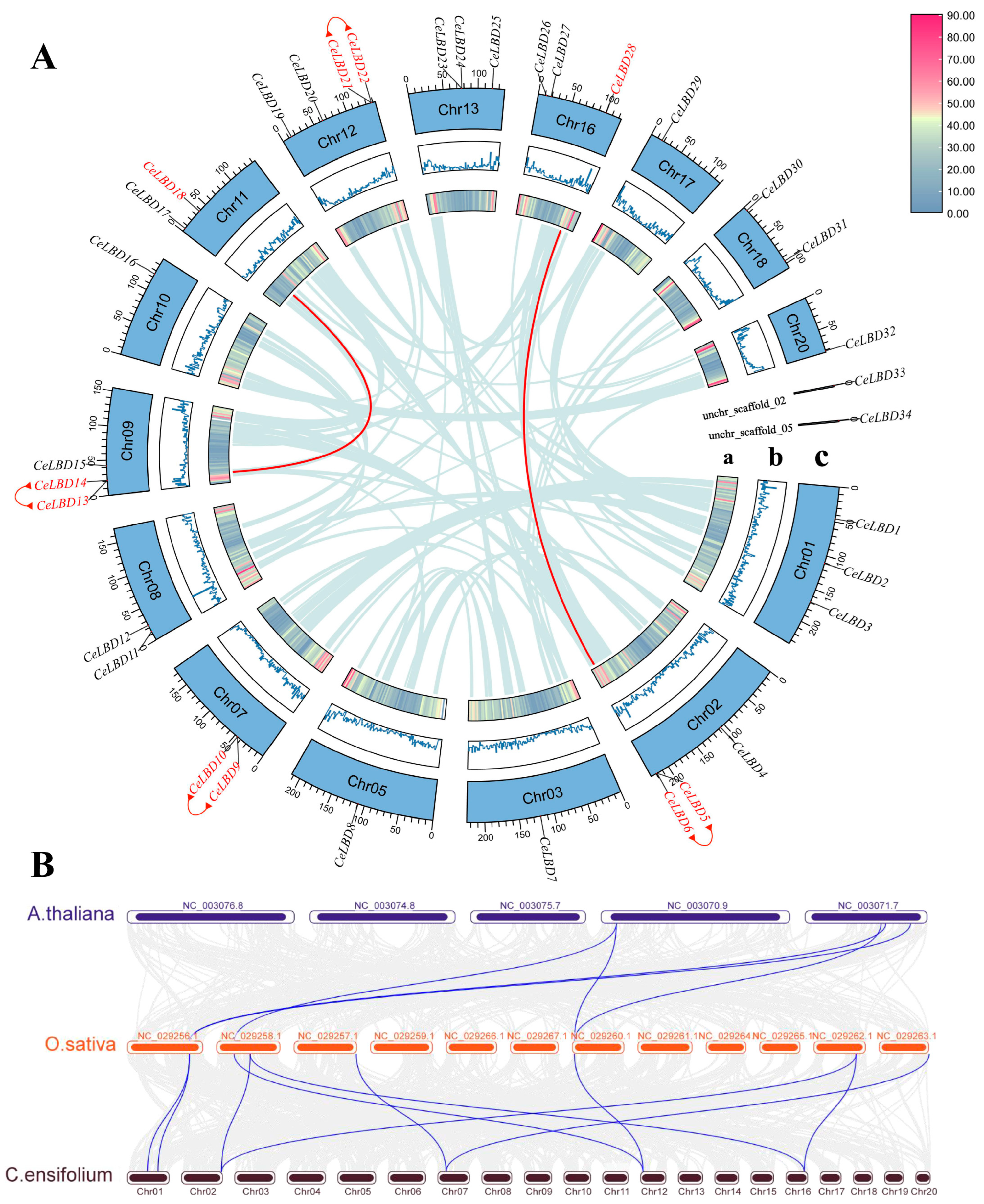
2.4. Chromosomal Distribution and Synteny Analyses
2.5. Prediction of miRNAs Binding Site and Cis-Acting Elements in CeLBDs
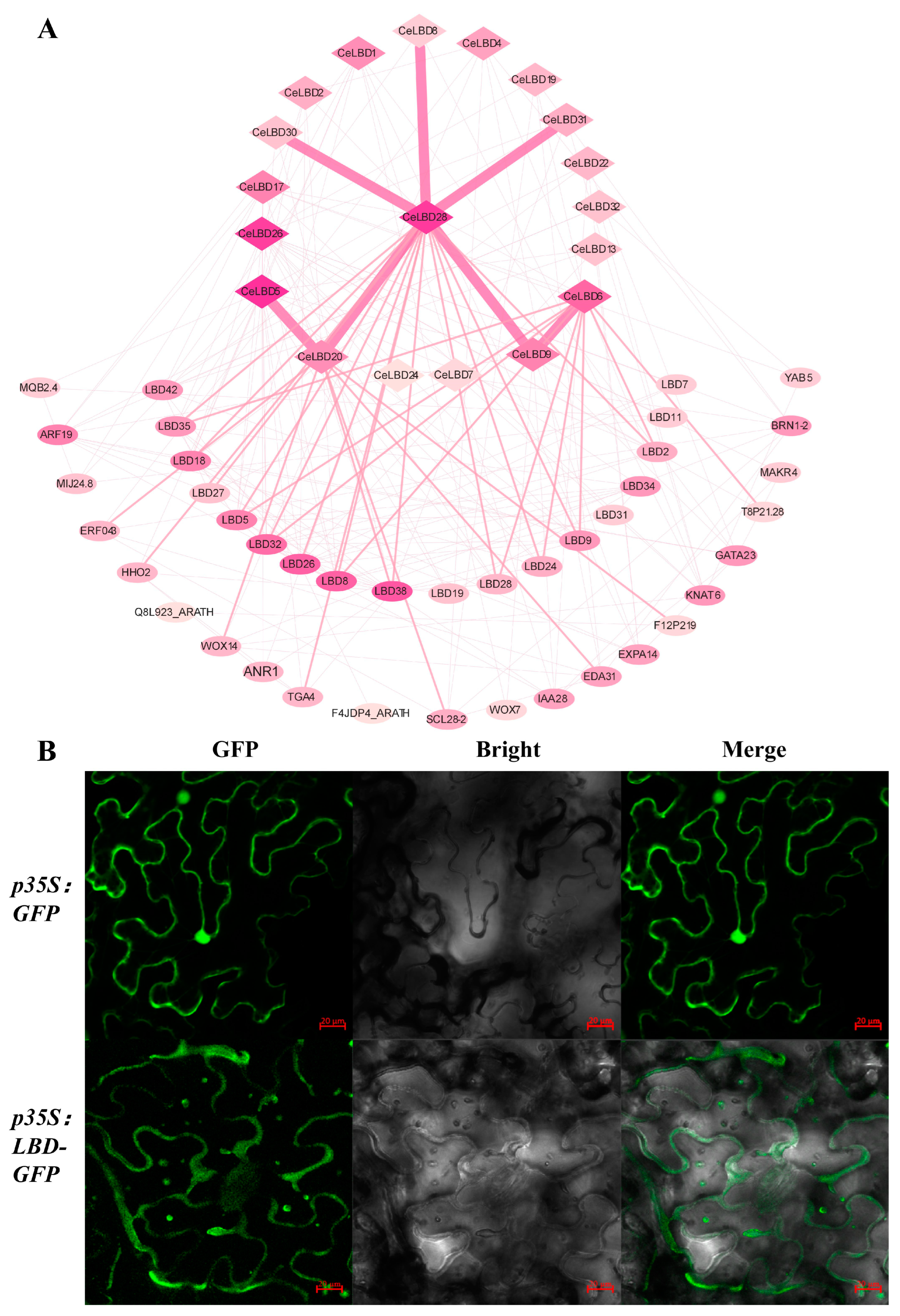
2.6. Protein Interaction Network Analysis of CeLBD Proteins
2.7. CeLBD Gene Expression Patterns from Transcriptomes
2.8. Subcellular Localization of CeLBD
2.9. RNA Extraction, cDNA Synthesis, and RT-qPCR Analysis
3. Results
3.1. Genome-Wide Identification and Physicochemical Properties of CeLBD Genes
3.2. Conserved Domain and Structure in CeLBD Proteins
3.3. CeLBD Structure Variations by Motif Rearrangement and Loss
3.4. CeLBD Location and Synteny in the C. ensifolium Genome
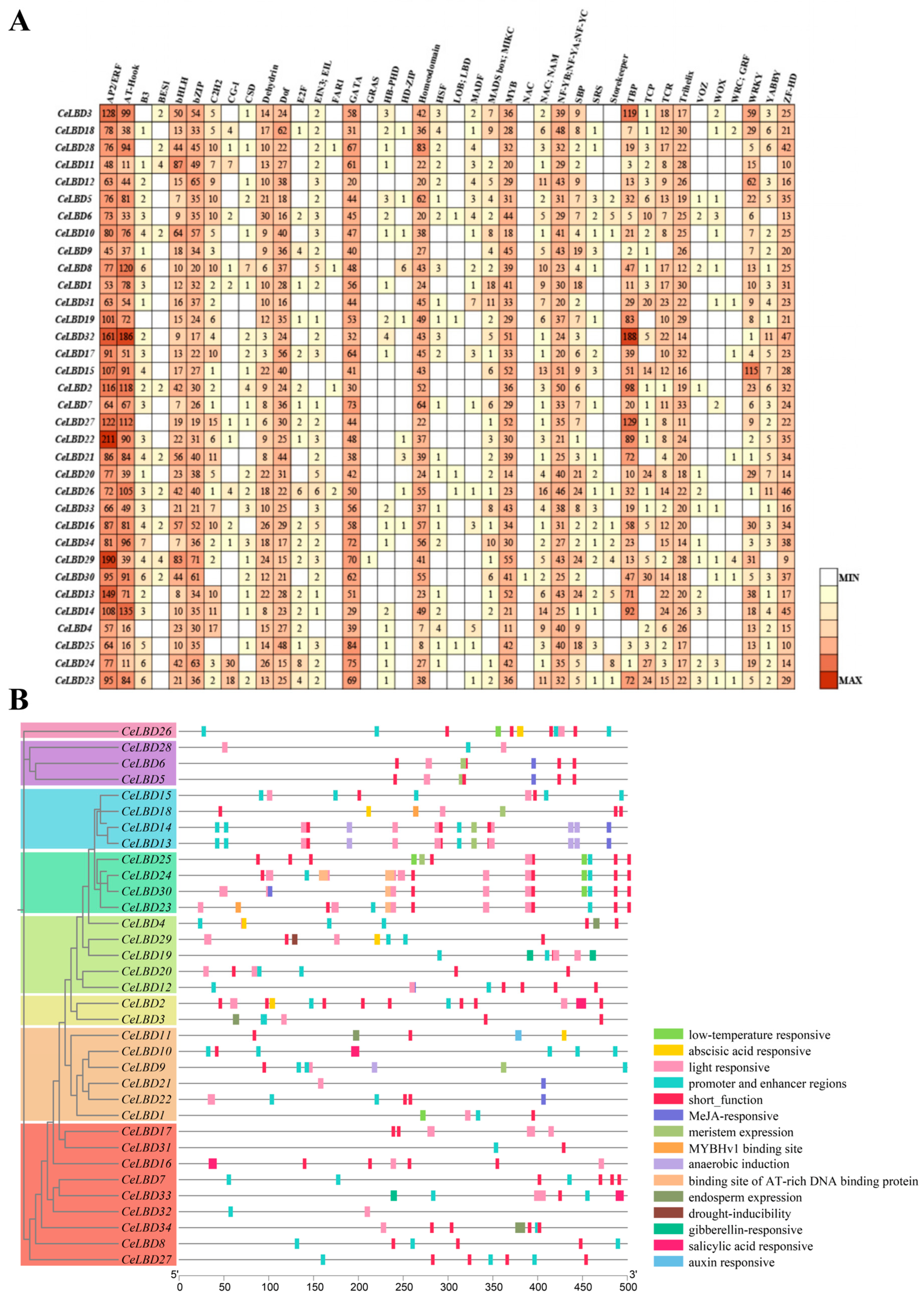
3.5. Analysis of Cis-Regulatory Elements in the Promoter Regions of CeLBD Genes
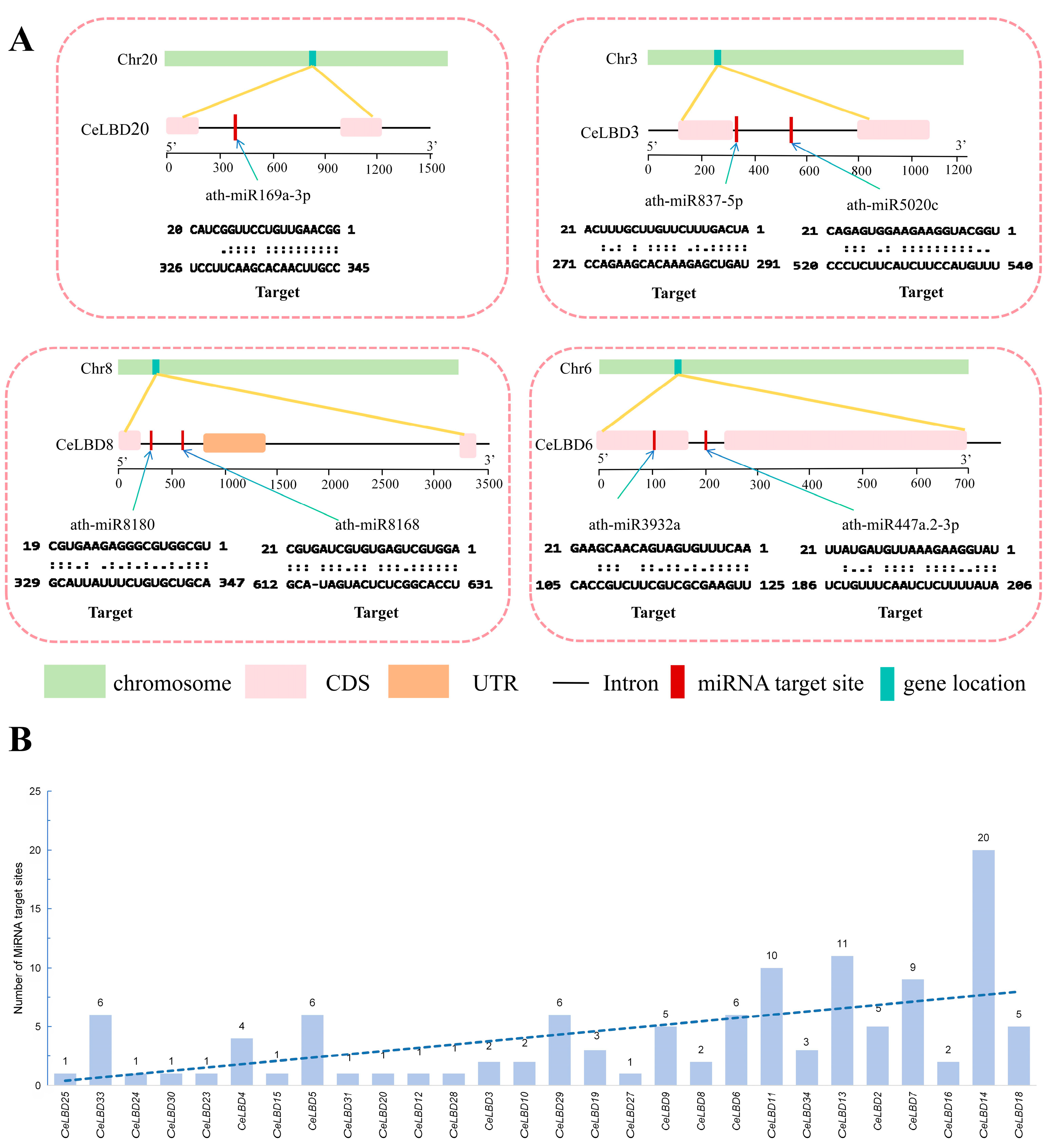
3.6. Prediction of miRNA Targets for 34 CeLBD Genes
3.7. Co-Expression Network Analysis and Subcellular Localization of CeLBD Protein
3.8. The Transcriptional Patterns of the CeLBD Genes and Co-Relation with JA Biosynthesis
3.9. Expression of CeLBD Genes in Response to Hormone Treatments and Abiotic Stress
4. Discussion
4.1. The CeLBD Genes Were Relatively Primitive and Underwent Variation and Functional Differentiation at the Subbranch Level
4.2. The CeLBD Gene Indirectly Regulates the Development of Orchid Flowers and the Release of Fragrance by Responding to Changes in External Abiotic Factors
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shuai, B.; Reynaga-Pena, C.G.; Springer, P.S. The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol. 2002, 129, 747–761. [Google Scholar] [CrossRef] [PubMed]
- Majer, C.; Hochholdinger, F. Defining the boundaries: Structure and function of LOB domain proteins. Trends Plant Sci. 2011, 16, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.W.; Kim, M.J.; Park, M.Y.; Han, K.H.; Kim, J. The conserved proline residue in the LOB domain of LBD18 is critical for DNA-binding and biological function. Mol. Plant 2013, 6, 1722–1725. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Xu, P.; Jing, X.; Chen, L.; Li, L.; Li, X. Decipher the ancestry of the plant-specific LBD gene family. BMC Genom. 2017, 18, 951. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, Z.; Ma, B.; Hou, Q.; Wan, X. Phylogeny and Functions of LOB Domain Proteins in Plants. Int. J. Mol. Sci. 2020, 21, 2278. [Google Scholar] [CrossRef]
- Jhu, M.Y.; Ichihashi, Y.; Farhi, M.; Wong, C.; Sinha, N.R. LATERAL ORGAN BOUNDARIES DOMAIN 25 functions as a key regulator of haustorium development in dodders. Plant Physiol. 2021, 186, 2093–2110. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, J.; Xu, J.; Li, Y.; Guo, L.; Wang, Z.; Zhang, X.; Zhao, B.; Guo, Y.D.; Zhang, N. CRISPR/Cas9 targeted mutagenesis of SlLBD40, a lateral organ boundaries domain transcription factor, enhances drought tolerance in tomato. Plant Sci. 2020, 301, 110683. [Google Scholar] [CrossRef]
- Feng, S.; Shi, J.; Hu, Y.; Li, D.; Guo, L.; Zhao, Z.; Lee, G.S.; Qiao, Y. Genome-Wide Analysis of Soybean Lateral Organ Boundaries Domain Gene Family Reveals the Role in Phytophthora Root and Stem Rot. Front. Plant Sci. 2022, 13, 865165. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Cui, H.; Wang, Z.; Kang, J.; Yang, Q.; Guo, C. Genome-Wide Analysis of the LATERAL ORGAN BOUNDARIES Domain (LBD) Members in Alfalfa and the Involvement of MsLBD48 in Nitrogen Assimilation. Int. J. Mol. Sci. 2023, 24, 4644. [Google Scholar] [CrossRef] [PubMed]
- Rong, M.; Gao, S.X.; Wen, D.; Xu, Y.H.; Wei, J.H. The LOB domain protein, a novel transcription factor with multiple functions: A review. Plant Physiol. Biochem. 2024, 214, 108922. [Google Scholar] [CrossRef]
- Mangeon, A.; Bell, E.M.; Lin, W.C.; Jablonska, B.; Springer, P.S. Misregulation of the LOB domain gene DDA1 suggests possible functions in auxin signalling and photomorphogenesis. J. Exp. Bot. 2011, 62, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Rubin, G.; Tohge, T.; Matsuda, F.; Saito, K.; Scheible, W.R. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell 2009, 21, 3567–3584. [Google Scholar] [CrossRef] [PubMed]
- Albinsky, D.; Kusano, M.; Higuchi, M.; Hayashi, N.; Kobayashi, M.; Fukushima, A.; Mori, M.; Ichikawa, T.; Matsui, K.; Kuroda, H.; et al. Metabolomic screening applied to rice FOX Arabidopsis lines leads to the identification of a gene-changing nitrogen metabolism. Mol. Plant 2010, 3, 125–142. [Google Scholar] [CrossRef]
- Li, C.; Zhu, S.; Zhang, H.; Chen, L.; Cai, M.; Wang, J.; Chai, J.; Wu, F.; Cheng, Z.; Guo, X.; et al. OsLBD37 and OsLBD38, two class II type LBD proteins, are involved in the regulation of heading date by controlling the expression of Ehd1 in rice. Biochem. Biophys. Res. Commun. 2017, 486, 720–725. [Google Scholar] [CrossRef]
- Yu, P.; Gutjahr, C.; Li, C.; Hochholdinger, F. Genetic Control of Lateral Root Formation in Cereals. Trends Plant Sci. 2016, 21, 951–961. [Google Scholar] [CrossRef]
- Zentella, R.; Zhang, Z.L.; Park, M.; Thomas, S.G.; Endo, A.; Murase, K.; Fleet, C.M.; Jikumaru, Y.; Nambara, E.; Kamiya, Y.; et al. Global analysis of della direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 2007, 19, 3037–3057. [Google Scholar] [CrossRef]
- Naito, T.; Yamashino, T.; Kiba, T.; Koizumi, N.; Kojima, M.; Sakakibara, H.; Mizuno, T. A link between cytokinin and ASL9 (ASYMMETRIC LEAVES 2 LIKE 9) that belongs to the AS2/LOB (LATERAL ORGAN BOUNDARIES) family genes in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2007, 71, 1269–1278. [Google Scholar] [CrossRef]
- Geng, L.; Tan, M.; Deng, Q.; Wang, Y.; Zhang, T.; Hu, X.; Ye, M.; Lian, X.; Zhou, D.X.; Zhao, Y. Transcription factors WOX11 and LBD16 function with histone demethylase JMJ706 to control crown root development in rice. Plant Cell 2024, 36, 1777–1790. [Google Scholar] [CrossRef] [PubMed]
- Kirolinko, C.; Hobecker, K.; Cueva, M.; Botto, F.; Christ, A.; Niebel, A.; Ariel, F.; Blanco, F.A.; Crespi, M.; Zanetti, M.E. A lateral organ boundaries domain transcription factor acts downstream of the auxin response factor 2 to control nodulation and root architecture in Medicago truncatula. New Phytol. 2024, 242, 2746–2762. [Google Scholar] [CrossRef]
- Liu, Y.; An, X.H.; Liu, H.; Zhang, T.; Li, X.; Liu, R.; Li, C.; Tian, Y.; You, C.; Wang, X.F. Cloning and functional identification of apple LATERAL ORGAN BOUNDARY DOMAIN 3 (LBD3) transcription factor in the regulation of drought and salt stress. Planta 2024, 259, 125. [Google Scholar] [CrossRef]
- Shi, Y.; Vrebalov, J.; Zheng, H.; Xu, Y.; Yin, X.; Liu, W.; Liu, Z.; Sorensen, I.; Su, G.; Ma, Q.; et al. A tomato LATERAL ORGAN BOUNDARIES transcription factor, SlLOB1, predominantly regulates cell wall and softening components of ripening. Proc. Natl. Acad. Sci. USA 2021, 118, e2102486118. [Google Scholar] [CrossRef] [PubMed]
- Jiao, P.; Wei, X.; Jiang, Z.; Liu, S.; Guan, S.; Ma, Y. ZmLBD2 a maize (Zea mays L.) lateral organ boundaries domain (LBD) transcription factor enhances drought tolerance in transgenic Arabidopsis thaliana. Front. Plant Sci. 2022, 13, 1000149. [Google Scholar] [CrossRef]
- Dong, L.; Manghwar, H. Genome-wide expression analysis of LBD genes in tomato (Solanum lycopersicum L.) under different light conditions. Plant Signal. Behav. 2023, 18, 2290414. [Google Scholar] [CrossRef]
- Liu, W.; Yu, J.; Ge, Y.; Qin, P.; Xu, L. Pivotal role of LBD16 in root and root-like organ initiation. Cell. Mol. Life Sci. 2018, 75, 3329–3338. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Zhang, J.; Jia, H.; Sosso, D.; Li, T.; Frommer, W.B.; Yang, B.; White, F.F.; Wang, N.; Jones, J.B. Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc. Natl. Acad. Sci. USA 2014, 111, E521–E529. [Google Scholar] [CrossRef]
- Xu, C.; Luo, F.; Hochholdinger, F. LOB Domain Proteins: Beyond Lateral Organ Boundaries. Trends Plant Sci. 2016, 21, 159–167. [Google Scholar] [CrossRef]
- Yu, Q.; Hu, S.; Du, J.; Yang, Y.; Sun, X. Genome-wide identification and characterization of the lateral organ boundaries domain gene family in Brassica rapa var. rapa. Plant Divers 2020, 42, 52–60. [Google Scholar] [CrossRef]
- Perez-Escobar, O.A.; Bogarin, D.; Przelomska, N.A.S.; Ackerman, J.D.; Balbuena, J.A.; Bellot, S.; Buhlmann, R.P.; Cabrera, B.; Cano, J.A.; Charitonidou, M.; et al. The origin and speciation of orchids. New Phytol. 2024, 242, 700–716. [Google Scholar] [CrossRef]
- Zheng, R.; Chen, J.; Peng, Y.; Zhu, X.; Niu, M.; Chen, X.; Xie, K.; Huang, R.; Zhan, S.; Su, Q.; et al. General Analysis of Heat Shock Factors in the Cymbidium ensifolium Genome Provided Insights into Their Evolution and Special Roles with Response to Temperature. Int. J. Mol. Sci. 2024, 25, 1002. [Google Scholar] [CrossRef] [PubMed]
- Ba, L.J.; Kuang, J.F.; Chen, J.Y.; Lu, W.J. MaJAZ1 Attenuates the MaLBD5-Mediated Transcriptional Activation of Jasmonate Biosynthesis Gene MaAOC2 in Regulating Cold Tolerance of Banana Fruit. J. Agric. Food Chem. 2016, 64, 738–745. [Google Scholar] [CrossRef]
- Ai, Y.; Li, Z.; Sun, W.H.; Chen, J.; Zhang, D.; Ma, L.; Zhang, Q.H.; Chen, M.K.; Zheng, Q.D.; Liu, J.F.; et al. The Cymbidium genome reveals the evolution of unique morphological traits. Hortic. Res. 2021, 8, 255. [Google Scholar] [CrossRef] [PubMed]
- Mount, D.W. Using the Basic Local Alignment Search Tool (BLAST). Cold Spring Harb. Protoc. 2007, 2007, pdb top17. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; de Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E.; et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Wang, Z.; Qi, B.; Zhang, J.; Yang, H. MEMe: A Mutually Enhanced Modeling Method for Efficient and Effective Human Pose Estimation. Sensors 2022, 22, 632. [Google Scholar] [CrossRef] [PubMed]
- Geourjon, C.; Deleage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Bioinformatics 1995, 11, 681–684. [Google Scholar] [CrossRef]
- Zhao, C.; Liu, T.; Wang, Z. PANDA-3D: Protein function prediction based on AlphaFold models. NAR Genom. Bioinform. 2024, 6, lqae094. [Google Scholar] [CrossRef]
- Janson, G.; Paiardini, A. PyMod 3: A complete suite for structural bioinformatics in PyMOL. Bioinformatics 2021, 37, 1471–1472. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v6: Recent updates to the phylogenetic tree display and annotation tool. Nucleic Acids Res. 2024, 52, W78–W82. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Li, J.; Xu, C.; Wang, L.; Liang, H.; Feng, W.; Cai, Z.; Wang, Y.; Cong, W.; Liu, Y. PSRna: Prediction of small RNA secondary structures based on reverse complementary folding method. J. Bioinform. Comput. Biol. 2016, 14, 1643001. [Google Scholar] [CrossRef] [PubMed]
- Chow, C.N.; Yang, C.W.; Wu, N.Y.; Wang, H.T.; Tseng, K.C.; Chiu, Y.H.; Lee, T.Y.; Chang, W.C. PlantPAN 4.0: Updated database for identifying conserved non-coding sequences and exploring dynamic transcriptional regulation in plant promoters. Nucleic Acids Res. 2024, 52, D1569–D1578. [Google Scholar] [CrossRef]
- Otasek, D.; Morris, J.H.; Boucas, J.; Pico, A.R.; Demchak, B. Cytoscape Automation: Empowering workflow-based network analysis. Genome Biol. 2019, 20, 185. [Google Scholar] [CrossRef] [PubMed]
- Lyu, F.; Han, F.; Ge, C.; Mao, W.; Chen, L.; Hu, H.; Chen, G.; Lang, Q.; Fang, C. OmicStudio: A composable bioinformatics cloud platform with real-time feedback that can generate high-quality graphs for publication. Imeta 2023, 2, e85. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Zhao, K.; Zheng, R.; Chen, J.; Zhu, X.; Xie, K.; Huang, R.; Zhan, S.; Su, Q.; Shen, M.; et al. A Comprehensive Analysis of Auxin Response Factor Gene Family in Melastoma dodecandrum Genome. Int. J. Mol. Sci. 2024, 25, 806. [Google Scholar] [CrossRef]
- Ohashi-Ito, K.; Iwamoto, K.; Fukuda, H. LOB DOMAIN-CONTAINING PROTEIN 15 Positively Regulates Expression of VND7, a Master Regulator of Tracheary Elements. Plant Cell Physiol. 2018, 59, 989–996. [Google Scholar] [CrossRef]
- Bell, E.M.; Lin, W.C.; Husbands, A.Y.; Yu, L.; Jaganatha, V.; Jablonska, B.; Mangeon, A.; Neff, M.M.; Girke, T.; Springer, P.S. Arabidopsis lateral organ boundaries negatively regulates brassinosteroid accumulation to limit growth in organ boundaries. Proc. Natl. Acad. Sci. USA 2012, 109, 21146–21151. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, J.; Ding, T.; Lin, X.; Hu, H.; Ding, Z.; Tian, H. MYB2 and MYB108 regulate lateral root development by interacting with LBD29 in Arabidopsis thaliana. J. Integr. Plant Biol. 2024, 66, 1675–1687. [Google Scholar] [CrossRef]
- Huang, M.; Ma, C.; Yu, R.; Mu, L.; Hou, J.; Yu, Y.; Fan, Y. Concurrent changes in methyl jasmonate emission and the expression of its biosynthesis-related genes in Cymbidium ensifolium flowers. Physiol. Plant. 2015, 153, 503–512. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zhang, W.; Zhang, Y.; Zhang, X.; Lang, D.; Zhang, X. The roles of methyl jasmonate to stress in plants. Funct. Plant Biol. 2019, 46, 197–212. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, J.; Xu, J.; Li, Y.; Lv, H.; Wang, F.; Guo, J.; Lin, T.; Zhao, B.; Li, X.X.; et al. SlMYC2 promotes SlLBD40-mediated cell expansion in tomato fruit development. Plant J. 2024, 118, 1872–1888. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Yuan, S.; Zhou, M.; Yuan, N.; Li, Z.; Hu, Q.; Bethea, F.G., Jr.; Liu, H.; Li, S.; Luo, H. Transgenic creeping bentgrass overexpressing Osa-miR393a exhibits altered plant development and improved multiple stress tolerance. Plant Biotechnol. J. 2019, 17, 233–251. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peng, Y.; Zhan, S.; Tang, F.; Zhao, Y.; Wu, H.; Li, X.; Huang, R.; Su, Q.; Zou, L.-H.; Zhao, K.; et al. Characterization of LBD Genes in Cymbidium ensifolium with Roles in Floral Development and Fragrance. Horticulturae 2025, 11, 117. https://doi.org/10.3390/horticulturae11020117
Peng Y, Zhan S, Tang F, Zhao Y, Wu H, Li X, Huang R, Su Q, Zou L-H, Zhao K, et al. Characterization of LBD Genes in Cymbidium ensifolium with Roles in Floral Development and Fragrance. Horticulturae. 2025; 11(2):117. https://doi.org/10.3390/horticulturae11020117
Chicago/Turabian StylePeng, Yukun, Suying Zhan, Feihong Tang, Yuqing Zhao, Haiyan Wu, Xiangwen Li, Ruiliu Huang, Qiuli Su, Long-Hai Zou, Kai Zhao, and et al. 2025. "Characterization of LBD Genes in Cymbidium ensifolium with Roles in Floral Development and Fragrance" Horticulturae 11, no. 2: 117. https://doi.org/10.3390/horticulturae11020117
APA StylePeng, Y., Zhan, S., Tang, F., Zhao, Y., Wu, H., Li, X., Huang, R., Su, Q., Zou, L.-H., Zhao, K., Liu, Z.-J., & Zhou, Y. (2025). Characterization of LBD Genes in Cymbidium ensifolium with Roles in Floral Development and Fragrance. Horticulturae, 11(2), 117. https://doi.org/10.3390/horticulturae11020117






