Abstract
Luffa is a genus of tropical and subtropical vines in the Cucurbitaceae family, recognized as an important cultivated commercial vegetable. However, the seeds of the luffa species are considered hard-seeded, and the processes governing seed germination remain understudied. The 9-cis-epoxycarotenoid dioxygenase (NCED) genes, which are critical for seed germination, have not been well characterized in Luffa. In this study, we identified four LaNCED genes in Luffa acutangula and four LcNCED genes in Luffa cylindrica, distributed across four chromosomes in each species. Phylogenetic analysis classified these genes into two subgroups. Gene structure and motif composition analyses revealed both similarities and differences among the NCEDs. Cis-element analysis further revealed that these NCEDs may be involved in growth regulation by modulating the phytohormonal network and responding to stress stimuli. Expression profiling of LcNCED genes during seed germination showed a decrease in LcNCED2 levels, coinciding with an increase in α-amylase activity throughout the germination process. Subcellular localization assays demonstrated that LcNCED2 is localized in the chloroplast. Furthermore, transient overexpression of LcNCED2 in tobacco leaves led to a significant increase in ABA content. Our findings provide a comprehensive genomic characterization of the NCED family in Luffa cylindrica and Luffa acutangula and reveal the functional role of LcNCED2 in regulating ABA levels, which may play a critical role in seed germination.
1. Introduction
The growth and development of plants are intricately modulated by interactions among multiple hormones, which are also critical under stress conditions. Abscisic acid (ABA), is one of the well-studied regulators, playing crucial roles in various processes such as seed germination, seedling growth, cuticle formation, fruit development and stress tolerance [1,2]. In plants, ABA biosynthesis occurs via either an indirect or direct pathway, with the indirect pathway being prevalent in most higher plants [3]. In the indirect pathway, the key precursor compound, zeaxanthin, is converted to violaxanthin via an enzymatic reaction catalyzed by zeaxanthin epoxidase (ZEP). Violaxanthin, the immediate precursor to zeaxanthin in the xanthophyll cycle, can subsequently be cleaved into xanthoxin through the catalytic action of 9-cis-epoxycarotenoid dioxygenase (NCED) [4]. The resulting xanthoxin is then transferred from the plastid to the cytosol, where it is further converted to abscisic aldehyde by short-chain alcohol dehydrogenase (SDR/ABA2) [5]. Finally, abscisic aldehyde is oxidized to ABA through the action of aba-aldehyde oxidase (AAO) [6]. Since the ABA synthesis pathway shares common compounds with the xanthophyll cycle, ABA synthesis is often activated together with the xanthophyll cycle, helping plants optimize their response to environmental stimuli [7].
NCED, a pivotal enzyme in the rate-limiting step of ABA synthesis, is encoded by the carotenoid cleavage dioxygenase (CCD) gene family [8]. The first identified NCED gene, denoted as VIVIPAROUS 14 (Vp14), was cloned from an ABA-deficient mutant of maize [8]. The in vitro catalytic activity of the Vp14 protein was observed in a reaction buffer containing ferrous iron, oxygen, ascorbate and detergent [9]. Since the cloning of the Vp14 gene, homologous CCD genes have been identified in various plant species, including Arabidopsis thaliana (Arabidopsis), Glycine max (soybean), Solanum lycopersicum (tomato), Prunus mume (Chinese plum) and Gossypium hirsutum (cotton) [10,11,12,13]. In Arabidopsis, nine CCD gene family members have been identified, five of which are NCED members (AtNCED2, AtNCED3, AtNCED5, AtNCED6 and AtNCED9) that contribute to ABA synthesis [14]. Arabidopsis NCEDs exhibit tissue-specific expression patterns. For instance, AtNCED2 and AtNCED3 are primarily responsible for NCED activity in roots, while AtNCED5, AtNCED6, AtNCED9 and AtNCED3 are expressed in developing seeds [14].
Seed germination marks the transition of plants from dormancy to active growth. This process is finely regulated by phytohormones, including ABA [14]. In Arabidopsis, maternal ABA, derived from NCED expression in vegetative tissues and silique envelopes, accelerate reserve accumulation and seed maturation. Conversely, ABA accumulated in the embryo mediated by the expression of AtNCED6 and AtNCED9 is implicated in seed dormancy [15,16]. Apart from AtNCED6 and AtNCED9, AtNCED5 also plays a role in seed dormancy regulation, working in conjunction with AtNCED3 to contribute to the accumulation of ABA contents in vegetative tissues. The expression of AtNCED5, AtNCED6 and AtNCED9 in seeds appears to function redundantly. The mutation in AtNCED5 alone did not significantly reduce seed dormancy [17]. Consistent results were observed in mutants for AtNCED6 and AtNCED9 [18].
Luffa, also spelled loofah, is commonly known as sponge gourd, ridge gourd or dishcloth gourd. It belongs to the Cucurbitaceae family and is native to the Old-World tropics [19]. Two domesticated and cultivated species, Luffa acutangula (L. acutangula) and Luffa cylindrica (L. cylindrica), are widely grown worldwide. Their fruits are edible when young and serve as a source of fiber when mature. The entire plant of luffa is medicinally important, and more than fifty chemical compounds have been isolated from it [20]. The global demand for luffa is increasing due to its growing economic importance. It has been used worldwide for various purpose, including household cleaning, personal hygiene and skin care products [21]. The main commercial production countries are India, China, Japan and America. China is one of the top producers, with a cultivation area of about 300,000 hectares and an annual production of 7.2 million tons. However, due to their thick seed structure and hard seed coat, luffa seeds exhibit hard seed dormancy. Typically, the seeds are soaked or scarified (i.e., scratched with a knife) before germination [22]. Understanding the germination process and elucidating its underlying mechanisms is crucial for improving the cultivation and production of luffa. While previous studies have identified NCED members in many plant species [23], no information is currently available regarding NCEDs in luffa. The relationship between NCED expression and luffa germination remains unknown. In this study, using HMM-search and local BLAST-P searches, we mined four NCEDs in both L. acutangula and L. cylindrica at the whole genomic level. The physicochemical characteristics, chromosomal distribution, phylogenetic relationships, gene structure, conserved motifs and cis-elements of these identified NCEDs were explored. Furthermore, the expression levels of LcNCED genes during seed germination in L. cylindrica were analyzed. Among these, LcNCED2 was identified as exhibiting a decrease in expression as α-amylase activity increased during germination. Additionally, this gene was found to be localized in the chloroplast and to influence ABA content.
2. Materials and Methods
2.1. Plant Materials and Germination Analysis
Seeds of L. cylindrica cultivar ‘QS−1’ were preserved at the Quzhou Academy of Agricultural and Forestry Sciences. Before germination, the seed coats were carefully scratched. Subsequently, fifty seeds per replicate, with three replicates, were placed in 10 cm diameter plastic germination boxes containing two layers of filter paper moistened with water. The germination boxes were then placed in a growth chamber without light and maintained at a temperature of 28 °C. Germination, defined as the emergence of the radicle, was recorded at 12-h intervals up to the 108th hour.
2.2. Analysis of α-Amylase Activity
Seed samples devoid of seed coats were collected at 24 h, 36 h, 60 h and 72 h post-germination for the determination of α-amylase activity. The α-amylase activity was measured using an α-amylase activity kit (Beijing Solarbio Science & Technology Co. Ltd., Beijing, China) following the method described by Hashemi et al. [24]. Fresh samples (0.2 g) were homogenized with 1 mL distilled water, then transferred to a 1.5 mL centrifuge tube. The tube was incubated at 25 °C for 15 min with intermittent shaking. Subsequently, the tube was centrifuged at 3000× g for 10 min. The resulting supernatant was assayed for α-amylase activity following the protocol provided in the kit.
2.3. Identification of LcNCED and LaNCED Gene Members
The amino acid sequences of 5 reported Arabidopsis NCED proteins and 5 oryza sativa (rice) NCED proteins were downloaded from the Genebank database and used as query sequences. Two local databases, using genomic AA sequence of L. acutangula and L. cylindrica, respectively, were established using TBtool. The reference genomes of L. acutangula and L. cylindrica were sequenced by the authors but have not yet been published. For the genome-wide identification of NCED genes, the query sequences were subjected to local BLAST-P searches with an E value < 1 × 10−5 using TBtools [25]. Additionally, the same sequences were employed to build a Hidden Markov Model (HMM) to predict NCED genes via using HMM search (Hmmer 3.0.). The resulting AA sequences obtained from these two approaches were further verified using NCBI Batch CD-Search (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi, accessed on 19 September 2023) and InterPro (https://www.ebi.ac.uk/interpro/, accessed on 19 September 2023). To ensure credibility, the gene sequences of candidates were further manually re-annotated using Fgenesh (http://www.softberry.com/berry.phtml?topic=fgenesh&group=programs&subgroup=gfind, accessed on 9 January 2025). The online ProtParam tool (https://web.expasy.org/protparam/ accessed on 16 January 2025) was used to calculate the theoretical isoelectric point (pI), molecular weight (MW) and the grand average of hydropathy (GRAVY) index. The subcellular localization of LaNCED and LcNCED proteins were determined using an online tool, Plant-mPLoc [26] (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/, accessed on 1 January 2025).
2.4. Determination of Gene Structure, Conserved Motifs and Potential Cis-Elements
TBtools was utilized for the extraction of coding and genomic DNA sequences of L. acutangula and L. cylindrica. Gene Structure Display Server (GSDS) 2.0 was used to compare the coding sequences with the corresponding genomic DNA sequences and therefore determined the exon-intron arrangements of each NCED [27] (http://gsds.gao-lab.org/, accessed on 9 January 2025). Multiple Expectation Maximization for Motif Elicitation (MEME) Suite 5.5.2 server was employed to identify the conserved motifs of the NCED proteins. The maximum value of the motif was set to 25. To predict the cis-elements, the 1.5 kb promoter fragments upstream from the transcriptional initiation sites for LcNCED and LaNCED genes were retrieved and assessed using the PlantCARE database [28] (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 9 January 2025).
2.5. Chromosome Localization and Phylogenetic Analysis of NCED Proteins
The chromosomal localization of each Luffa NCED were mapped using genomic information and visualized with TBtools. Full-length AA sequences of NCED family proteins from Arabidopsis, rice, L. acutangula, L. cylindrica, Solanum tuberosum, Populus trichocarpa, Triticum aestivum, Zea mays and Sorghum bicolor were aligned using MAFFT 7.0 [29]. The phylogenetic tree was constructed using the MEGAX64 software(MEGA-X), employing the neighbor-joining method with 1000 bootstrap replicates.
2.6. Expression Analysis Performed by Quantitative Real-Time PCR (qRT-PCR)
Total RNA was extracted using the E.Z.N.A. ®Plant RNA Kit (Omega, Norcross, GA, USA). The cDNA synthesis was performed using the HiScript III 1st Strand cDNA Synthesis Kit (Vazyme, Nanjiang, China). For qRT-PCR analysis, a 20-μL reaction system [10 μL TOROGreen® qPCR Master Mix (Toroivd, Shanghai, China), 2 μL cDNA (50 ng/μL), 0.8 μL upstream and downstream primers and 6.4 μL H2O] and a two-step program (95 °C for 1 min; 42 cycles of 95 °C for 10 s and 60 °C for 30 s) was employed. The LcUBQ (Lc_Chr01G03790.1) and LcEF-1α (Lc_Chr13G04430.1) genes were used as internal controls. The relative expression levels were calculated according to the 2−∆∆Ct method with normalization of data to the geometric mean of the internal control genes [30]. The specific primers are listed in Table S1.
2.7. Analysis of Subcellular Localization
The coding region sequence of LcNCED2 (1806 bp), without the terminal codon, was amplified using a cDNA template extracted from L. cylindrica cultivar ‘QS-1’ and fused with GFP by cloning to the pFGC5941 vector. The specific primers for amplification are listed in Table S2. The constructed vector was co-transformed with the 35S::Rubisco-mCherry vector (a chloroplast marker) to the abaxial surface of 4-week-old N. benthamiana leaves using an Agrobacterium-mediated method [31]. Fluorescence was observed 48 h after transformation using a super-resolution microscope (N-STORM, Nikon, Japan).
2.8. Analysis of ABA Content
Tobacco leaves agroinfiltrated with the LcNCED2-GFP constructed vector or an empty vector were collected to measure the ABA content. Collected 0.1 g samples were homogenized with 1 mL cold methanol/water/formic acid (15:4:1, v:v:v). The extraction mix was spiked with 10 μL internal standard d6-ABA (100 ng/mL, OIchemim). Extraction was carried out at 4 °C with shaking for 24 h in darkness. After centrifugation at 12,000 r/min for 5 min, the supernatant was transferred to clean plastic microtubes, followed by evaporation to dryness and dissolved in 100 μL 80% methanol (v:v). The solutions were filtered through a 0.22 μm membrane filter for further analysis [32]. The ABA content was analyzed using an UPLC-ESI-MS/MS system (UPLC, ExionLC™ AD, SCIEX, Framingham, MA, USA; MS, Applied Biosystems 6500 Triple Quadrupole, SCIEX, Framingham, MA, USA). The analytical conditions were as follows: LC: column, Waters ACQUITY UPLC HSS T3 C18 (100 mm × 2.1 mm i.d., 1.8 µm); solvent system, water with 0.04% acetic acid (A), acetonitrile with 0.04% acetic acid (B); gradient program, started at 5% B (0–1 min), increased to 95% B (1–8 min), 95% B (8–9 min), finally ramped back to 5% B (9.1–12 min); flow rate, 0.35 mL/min; temperature, 40 °C; injection volume, 2 μL.
2.9. Statistical Analysis
Statistical analysis was performed using an ANOVA followed by Tukey’s test (p-value at 0.05). Data are shown as the average of three replicates. All statistical analyses were performed using the SPSS package (SPSS 19.0, Chicago, IL, USA).
3. Results
3.1. Identification of the LcNCED and LaNCED Gene Family Members
Genome-wide identification of the NCED gene family based on local BLAST-P searches and HMM searches led to the discovery of 8 members in L. acutangula and L. cylindrica (Table 1, Table S3, Table S4 and Table S5). These 8 genes, classified into two subgroups (group I and group II), were designated as LaNCED2, LaNCED3, LaNCED5, LaNCED6, LcNCED2, LcNCED3, LcNCED5 and LcNCED6. The proteins encoded by these genes exhibited high sequence identity (ranging from 61.5% to 78.8%) to the conserved domain annotated as 9-cis-epoxycarotenoid dioxygenase (PLN02258, Figure S1). LaNCEDs and LcNCEDs show high sequence identity among themselves, with four pairs (LaNCED2/LcNCED2, LaNCED3/LcNCED3, LaNCED5/LcNCED5, LaNCED6/LcNCED6) exceeding 90% (Figure S1). The LaNCED proteins varied in AA numbers from 564 (LaNCED6) to 605 (LaNCED2). The MW ranged from 62,032.84 Da (LaNCED6) to 67,766.82 Da (LaNCED2), and the pIs from 6.22 (LaNCED3) to 7.38 (LaNCED2), while the GRAVY ranged from −0.424 (LaNCED2) to −0.241 (LaNCED6). The physicochemical analysis on LcNCEDs also showed varieties in AA sequences and MW, pI and GRAVY values. LcNCED6 and LcNCED5, with the shortest protein length (561) and the longest protein length (608), have the smallest MW (61,711.64 Da) and the biggest MW (68,127.57 Da), respectively. The pI values of LcNCEDs ranged from 6.13 to 7.08, while the GRAVY ranged from −0.388 to −0.228. According to the predictions conducted by the Plant-mPLoc online tool, LaNCED2, LcNCED2 and LaNCED5 likely localize in both the chloroplast and cytoplasm, while the remaining five members likely localize in the cytoplasm.

Table 1.
The characteristics of NCED genes from L. acutangula and L. cylindrica. Chr, chromosomes; MW, Molecular weight; PI, isoelectric points; GRAVY, grand average of hydropathy; Chl, chloroplast; Cyt, cytoplasm; PL, protein length; PSL, Predictive subcellular localization.
3.2. Chromosomal Distribution and Phylogenetic Analysis
TBtools software (TBtools-II, v 2.149) was employed to visually represent the distribution of LaNCED and LcNCED genes on their corresponding chromosomes (Figure 1). The four LaNCEDs were dispersed among sections of Chr1, 7, 9 and 13, while four LcNCED genes were found to be located on Chr1, 4, 8 and 13.
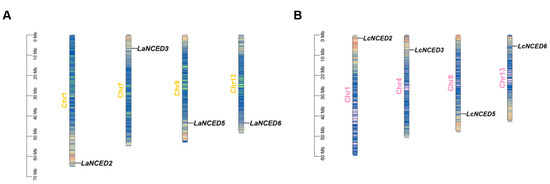
Figure 1.
Chromosomal locations of LaNCED and LcNCED genes. (A) The four LaNCED genes were located on 4 out of the 13 chromosomes of L. acutangula. (B) The four LcNCED genes were located on 4 out of the 13 chromosomes of L. cylindrica. Dark lines indicate the position of NCED genes on chromosomes.
In the phylogenetic analysis involving five eudicots and four monocots, the NCED genes were categorized into five groups labeled as Group I to Group V (Figure 2). LaNCED2, 3, 5, along with LcNCED2, 3, 5, belong to Group I, while LaNCED6 and LcNCED6 are classified under Group II. Notably, NCED proteins from the other three eudicots, including Arabidopsis, Solanum tuberosum and Populus trichocarpa, were all classified under either Group I or Group II. In contrast, proteins from the monocots were grouped under Group III–V. This suggests a relative conservation of NCED genes among dicotyledonous plants, and highlights a differentiation in NCED genes between monocots and eudicots.
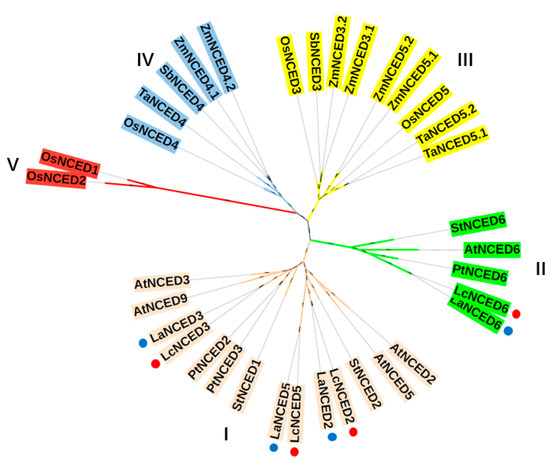
Figure 2.
Phylogenetic analysis of NCED proteins. The tree was divided into 5 groups (I, II, III, IV and V). LaNCED and LcNCED proteins are marked with blue and red dots, respectively.
3.3. Genetic Structure and Conserved Motifs of the LaNCED and LcNCED Gene Families
The Gene Structure Display Server (GSDS2.011) was utilized to explore the exon-intron structures of LaNCEDs and LcNCEDs. As shown in Figure 3A, all LaNCEDs and all LcNCEDs lacked introns. Among the four LcNCEDs, Group I members possessed both a 5’ UTR region and a 3’UTR region. LaNCED2, LaNCED3 and LaNCED6 each have a 5’ UTR region, whereas LaNCED2 and LaNCED3 have a 3’ UTR region. To search the motifs within these LaNCEDs and LcNCEDs, the online tool MEME was employed. A total of 25 motifs with lengths between 6–50 were identified (Figure 3B). Genes encoding NCED2, NCED3, NCED5 and NCED6 in L. acutangula and L. cylindrica exhibited similar compositions. Group I members, encompassing LaNCED2, 3, 5 and LcNCED2, 3, 5, featured 19 motifs. LaNCED6 and LcNCED6, classified under Group II, had 18 motifs. Among the 25 identified motifs, motif 19 was exclusive to Group II members, while motif 17 was consistently located at the end of the protein sequence in all group I members.
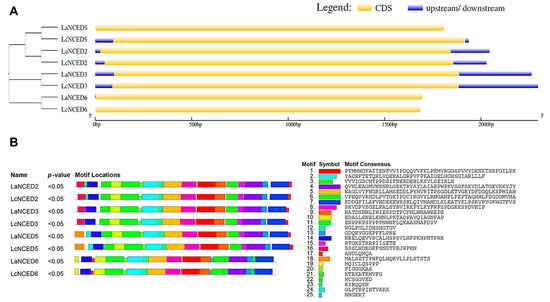
Figure 3.
Gene structures and conserved motifs of LaNCED and LcNCED genes. (A) Gene structures of LaNCED and LcNCED genes. (B) Conserved motifs of LaNCED and LcNCED genes. Conserved motifs were identified using the MEME Suite 5.5.2 server. The maximum value of the motif was set to 25.
3.4. Analysis of Cis-Elements in LaNCED and LcNCED Gene Promoters
Based on the regions of 1500 bp promoter sequences upstream from the transcriptional initiation sites of LaNCED and LcNCED genes, multi cis-elements related to plant development and stress response were identified using PlantCARE (Figure 4, Table S6). Each NCED member possessed light-responsive elements, with their quantity ranking as the highest among all identified elements. Remarkably, a total of 39 ABA responsive cis-elements (ABRE) were identified in all LaNCEDs and all LcNCEDs. Additionally, other cis-elements related to plant hormonal signaling, including MeJA-responsiveness, salicylic acid responsiveness, auxin responsiveness and gibberellin-responsive elements, were observed in specific LaNCEDs and LcNCEDs. Six stress-responsive cis-elements reflecting plant responses to anaerobic induction, defense and stress responsiveness, drought-inducibility, low temperature responsiveness and wound responsiveness were also identified. Moreover, growth-related cis-elements involved in flavonoid biosynthesis, zein metabolism, circadian control and meristem expression were also identified.
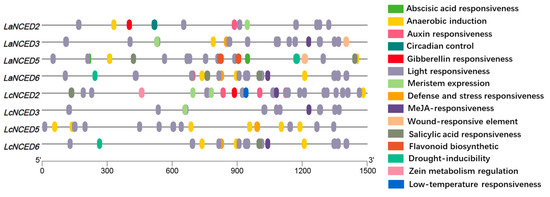
Figure 4.
Cis-elements in the promoters of NCED genes. Different cis-elements with the same or similar functions are present with the same color.
3.5. Expression Patterns of LcNCEDs During the Germination of L. cylindrica
Seeds from the L. cylindrica cultivar ‘QS-1’ were germinated in darkness at 28 °C. As shown in Figure 5A,B, ‘QS-1’ initiated germination at 60 h with a germination percentage of 54%. By 84 h post germination, the germination percentage peaked at 74.6%. α-Amylase, synthesized de novo during seed germination, displayed a positive correlation with the germination rate [33,34]. In alignment with the fluctuation in germination rate, α-amylase activity in ‘QS-1’ increased 81.5% from 24 h to 72 h (Figure 5C). To gain insights into the role of NCED genes during germination in Luffa, we performed qRT-PCR analysis to examine LcNCED expression profiles (Figure 5D). LcNCED5, which belongs to Group I, was not detectable despite attempts to modify primers and reaction procedures. The remaining genes exhibited different expression patterns. The transcript level of LcNCED2 significantly decreased during germination from 24 h to 36 h and remained at a low level between 60 h and 72 h. In contrast, the relative expression level of LcNCED3 dramatically reduced at 36 h but elevated at 60 h, returning to a similar level to that at 24 h by 72 h. In comparison to the expression level at 24 h, LcNCED6 increased 130% at 36 h but subsequently reduced to 47.1% and 31.5% at 60 h and 72 h, respectively.
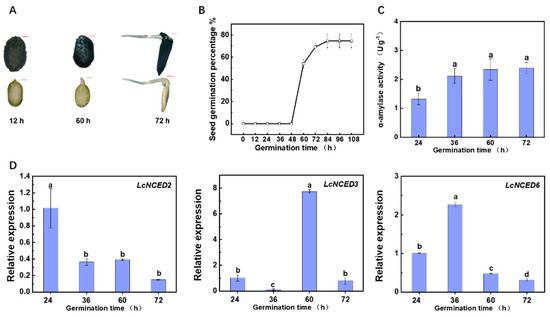
Figure 5.
Germination analysis of seeds from L. cylindrica cultivar ‘QS −1’. (A) The photo of ‘QS −1’ germinated for 12, 60 and 72 h. (B) Seed germination percentage during the germination period. (C) Determination of α-amylase activity. (D) The expression of LcNCEDs in ‘QS −1’. Values represent means of three biological replicates ± SD. Different letters indicate significant differences (p < 0.05) according to Tukey’s test.
3.6. Analysis of Enzyme Activity for LcNCED2
NCED catalyzes the deoxygenation of 9-cis-epoxycarotenoids through a de novo synthesis pathway that occurs in plastids, making NCED activity crucial for regulating ABA levels. Due to the coincidence of its expression peak with the onset of the germination process, we chose LcNCED2 for further analysis. We determined the subcellular localization of LcNCED2 by fusing it with GFP and co-expressing it with 35S::Rubisco-mCherry (a chloroplast marker) in a transient expression system using tobacco (Nicotiana benthamiana) leaves. As Figure 6 shows, the GFP signal of the LcNCED2-GFP protein was highly overlapped with that of mCherry from the chloroplast marker protein, indicating that LcNCED2 is mainly located in the chloroplast. To verify the NCED activity of LcNCED2, we measured the ABA content in tobacco leaves overproducing the LcNCED2-GFP fusion protein compared to the vector control (Figure 7). In LcNCED2-GFP overexpressing plants, ABA levels showed an approximately 12-fold increase compared to the vector control.
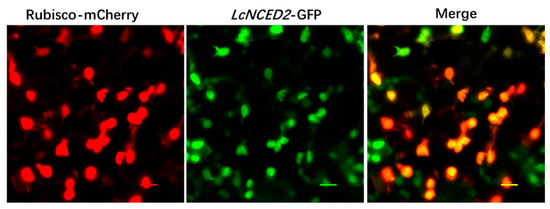
Figure 6.
Subcellular localization of LcNCED2 in chloroplasts. Tobacco (Nicotiana benthamiana) leaves were agroinfiltrated to express the LcNCED-GFP fusion protein for 48 h. The signal for GFP is shown in green. Rubisco-mCherry was used as a chloroplast marker with a red signal.
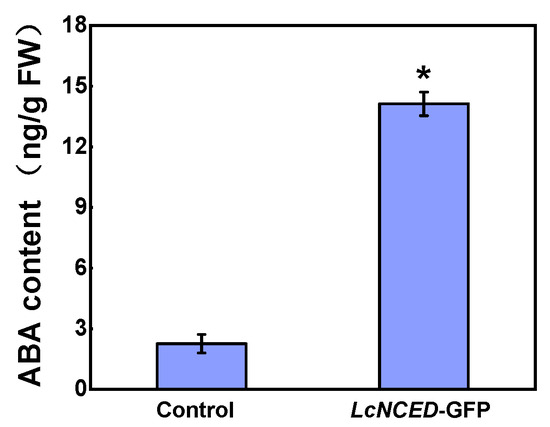
Figure 7.
Determination of ABA content in the tobacco (Nicotiana benthamiana) leaves transiently expressing LcNCED2. Tobacco leaves were agroinfiltrated to express the GFP (control) or LcNCED2-GFP for 48 h. Values represent means of three biological replicates ± SD. * indicates significant differences (p < 0.05) according to Student’s t-test.
4. Discussion
Luffas are globally significant as cultivated vegetables and medicinal plants, playing a substantial role in Cucurbitaceae consumption [35]. However, their seeds are considered hard-seeded due to the hard seed coats and phenolic compounds on the surface, which restrict water uptake into the seed. NCED genes, renowned for their involvement in ABA synthesis, are not only crucial for plant stress responses but also contribute to seed germination processes [1,36]. Although NCED members from various plant species have been identified, no NCED members in the genus Luffa have been reported. In this study, we identified and characterized putative NCED genes in two species of the genus Luffa, namely L. acutangula and L. cylindrica. The exploration encompassed physicochemical characteristics, chromosomal distribution, phylogenetic relationships, gene structure, conserved motifs and cis-elements of the identified NCED genes. Furthermore, we explored the relationship between NCED expression and seed germination activity in the L. cylindrica cultivar ‘QS-1’. LcNCED2, which exhibited a decrease in expression as α-amylase activity increased during germination, was identified. Additionally, this gene was found to be localized in the chloroplast and to influence ABA content.
Using HMMR and BLAST-P searches against the reference genomes of L. acutangula and L. cylindrica, we identified four LaNCED and four LcNCED genes (Table 1), each one fewer in number compared to Arabidopsis. These identified NCEDs varied in AA numbers from 561 (LcNCED6) to 608 (LcNCED5), and were distributed on Chr1, 7, 9 and 13 in L. acutangula and Chr1, 4, 8 and 13 in L. cylindrica (Figure 1). The LaNCED and LcNCED proteins exhibit a high degree of sequence identity, with four pairs demonstrating particularly striking similarity: LaNCED2 with LcNCED2, LaNCED3 with LcNCED3, LaNCED5 with LcNCED5 and LaNCED6 with LcNCED6, all exceeding 94% sequence identity (Supplemental Figure S1). This significant level of similarity suggests a close evolutionary relationship and potentially overlapping functions in ABA biosynthesis. Phylogenetic analysis revealed that NCED proteins from these two species of the genus Luffa and other eudicots, including Arabidopsis, Solanum tuberosum and Populus trichocarpa, were classified under either Group I or Group II, while those from monocots were categorized under Group III–V (Figure 2). This indicates a differentiation of NCED genes between monocots and eudicots. NCEDs that belong to the same group may have similar functions in different species. The LaNCEDs and LcNCEDs classified in the same groups exhibited similar compositions in conserved motifs. Among the identified 25 motifs, only motif 19 appeared in Group II members, while motif 17 was consistently located at the end of the protein sequence in all group I members (Figure 3). The consistency and differences in motif composition may reflect the functional similarities and differences among the NCED members. In Arabidosis, AtNCED6 and AtNCED9 are important for ABA biosynthesis in seeds and have been demonstrated to co-regulate seed germination and dormancy [16,17]. Additionally, these two genes are particularly responsive to drought and play key roles in drought tolerance [37]. AtNCED5, AtNCED6, AtNCED3 and AtNCED2 are expressed in flowers, contributing to the plant’s reproductive success [14]. Therefore, investigating the expression patterns and functions of NCED homologs in Luffa across different tissues and developmental stages would be valuable in future research. NCED-mediated production of xanthoxin in plastids is the rate-limiting step for ABA biosynthesis in plants [4]. The crucial role of ABA in crosstalk with other plant hormonal signaling networks and in regulating adaptation processes in response to stressful environments has been well established [38,39]. In Arabidopsis, each NCED member plays a unique regulatory role in specific environmental responses and developmental processes [14,40]. To explore the putative roles of the LcNCED and LaNCED genes, we searched for cis-elements in the 1.5 kb sequence upstream of their transcriptional initiation site (Figure 4). Among the widespread cis-elements, the light–responsive element was the only one present in each of the LaNCEDs and LcNCEDs. ABRE was second only to the light–responsive element in number and was presented in 7 out of the 8 investigated NCEDs. This reflects the vital role of NCED in the ABA pathway. In addition to ABRE, other elements involved in auxin-, JA-, SA- and GA-responsive were also identified, indicating a putative role of NCED in the crosstalk of ABA and other phytohormone signaling networks. A number of cis-elements related to stress, such as the anaerobic induction element, defense and stress responsive element, drought-inducibility element and low temperature response were also identified. These provide clues for further detailed functional exploration of LaNCEDs and LcNCEDs.
In Arabidopsis, the expressions of AtNCED5, AtNCED6 and AtNCED9 were involved in the regulation of seed dormancy [16,18]. However, little is known about the roles of NCEDs in seed germination in Luffa. This study utilized the ‘QS-1’ seed to explore the putative relationships of the accumulation of NCED transcripts with seed germination. The expression patterns of LcNCED2, LcNCED3 and LcNCED6 showed differences during germination. Among these genes, the transcript of LcNCED2 at 36, 60 and 72 h significantly decreased compared to 24 h. While the relative expression level of LcNCED3 and LcNCED6 were elevated at 60 h and 36 h. α-Amylase is the major enzyme involved in starch mobilization, predominantly synthesized during germination [41]. As expected, the activity of α-amylase increased with germination time, from 1.32 Ug−1 to 2.39 Ug−1 after 72 h. Similar results were also observed in other species [42]. Since the reduced expression level of LcNCED2 was consistent with the increased amylase activity, we tested its NCED activity. Similar to the reported NCEDs, the subcellular localization of LcNCED2 was in the chloroplast [43]. Ectopic expression of this gene caused overproduction of ABA in tobacco leaves. However, further exploration of the specific expression tissues (e.g., endosperm) of this gene will help to clarify its involvement in germination. Furthermore, whether other LcNCED members exhibit NCED activity to produce ABA remains to be confirmed.
5. Conclusions
Each of the four NCED gene members were identified in the genomes of both L. acutangula and L. cylindrica. These NCED genes were distributed across four chromosomes in both L. acutangula and L. cylindrica and were divided into two groups based on their phylogenetic relationship. The consistency and differences in gene structure and motif composition may reflect the functional similarities and differences of the NCED genes. Cis-element analysis revealed the potential involvement of LaNCED and LcNCED genes in light, hormonal and stress responses. Analysis of the expression patterns of NCED genes alongside seed germination revealed a decrease in LcNCED2 levels, coinciding with an increase in α-amylase activity. Subcellular localization assays showed that LcNCED2 is localized in the chloroplast. Furthermore, transient overexpression of LcNCED2 in tobacco leaves resulted in a significant increase in ABA content. In a word, we not only comprehensively identified and characterized LaNCED and LcNCED genes, but also revealed the functional role of LcNCED2 in regulating ABA levels, which may play a critical role in seed germination.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae11020115/s1, Figure S1: Pairwise sequence similarity percentages among LaNCED and LcNCED proteins; Table S1: The specific primers used for qRT-PCR analysis; Table S2: The specific primers used for plasmid construction; Table S3: Protein sequences of identified LaNCED and LcNCED genes. Table S4: cDNA sequences of identified LaNCED and LcNCED genes. Table S5: Genomic sequences of identified LaNCED and LcNCED genes. Table S6: Different cis-elements in the promoter sequences of LaNCED and LcNCED genes.
Author Contributions
Conceptualization, P.F. and Q.G.; methodology, X.L.; validation, X.L.; investigation, J.W. and X.W.; data curation, P.F. and T.Z.; writing—original draft preparation, P.F. and H.M.; writing—review and editing, Q.L.; supervision, Q.G. and Q.L.; funding acquisition, P.F., J.W. and Q.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China—Youth Science Fund, grant number “32202521” and Quzhou Science and Technology Project “2023K083, 2024K063”.
Data Availability Statement
Data analyzed in this study will be available upon request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Brookbank, B.P.; Patel, J.; Gazzarrini, S.; Nambara, E. Role of basal aba in plant growth and development. Genes 2021, 12, 1936. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2019, 62, 25–54. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Cao, S.; Shi, L.; Chen, W.; Yin, X.; Yang, Z. Abscisic acid biosynthesis, metabolism and signaling in ripening fruit. Front. Plant Sci. 2023, 14, 1279031. [Google Scholar] [CrossRef] [PubMed]
- Taylor, I.B.; Alan, B.; Thompson, A.J. Control of abscisic acid synthesis. J. Exp. Bot. 2000, 51, 1563–1574. [Google Scholar] [CrossRef] [PubMed]
- González-Guzmán, M.; Apostolova, N.; Bellés, J.M.; Barrero, J.M.; Piqueras, P.; Ponce, M.R.; Rodriguez, P.L. The short-chain alcohol dehydrogenase ABA2 catalyzes the conversion of xanthoxin to abscisic aldehyde. Plant Cell 2002, 14, 1833–1846. [Google Scholar] [CrossRef]
- González-Guzmán, M.; Rodriguez, L.; Lorenzo-Orts, L.; Pons, C.; Sarrion-Perdigones, A.; Fernandez, M.A. Tomato PYR/PYL/RCAR abscisic acid receptors show high expression in root, differential sensitivity to the abscisic acid agonist quinabactin, and the capability to enhance plant drought resistance. J. Exp. Bot. 2014, 65, 4451–4464. [Google Scholar] [CrossRef]
- Aerts, N.; Hickman, R.; Van Dijken, A.J.H.; Kaufmann, M.; Snoek, B.L.; Pieterse, C.M.J.; Van Wees, S.C.M. Architecture and dynamics of the abscisic acid gene regulatory network. Plant J. 2024, 119, 2538–2563. [Google Scholar] [CrossRef]
- Zhou, Q.; Li, Q.; Li, P.; Zhang, S.; Liu, C.; Jin, J.; Yang, Y. Carotenoid cleavage dioxygenases: Identification, expression, and evolutionary analysis of this gene family in tobacco. Int. J. Mol. Sci. 2019, 20, 5796. [Google Scholar] [CrossRef]
- Schwartz, S.H.; Tan, B.C.; Gage, D.A.; Zeevaart, J.A.; McCarty, D.R. Specific oxidative cleavage of carotenoids by VP14 of maize. Science 1997, 276, 1872–1874. [Google Scholar] [CrossRef]
- Chen, K.; Li, X.; Guo, X.; Yang, L.; Qiu, L.; Liu, W.; Zheng, T. Genome-wide identification and expression profiling of the nced gene family in cold stress response of Prunus mume Siebold & Zucc. Horticulturae 2023, 9, 839. [Google Scholar] [CrossRef]
- Wang, R.K.; Wang, C.E.; Fei, Y.Y.; Gai, J.Y.; Zhao, T.J. Genome-wide identification and transcription analysis of soybean carotenoid oxygenase genes during abiotic stress treatments. Mol. Bio. Rep. 2013, 40, 4737–4745. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.H.; Yu, X.T.; Chen, L.; Zhao, G.; Li, S.Z.; Zhou, H.; Dai, Y.; Sun, N.; Xie, Y.F.; Gao, J.S.; et al. Genome-wide identification and expression analysis of the NCED family in cotton (Gossypium hirsutum L.). PLoS ONE 2021, 16, e0246021. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Lv, R.; Zhang, Y.; Mo, F.; Meng, F.; Cheng, M.; Wang, A. Identification of the NCED gene family in tomato (Solanum lycopersicum) and functional analysis of SlNCED2 in response to drought stress. Sci. Hortic. 2024, 330, 113087. [Google Scholar] [CrossRef]
- Tan, B.C.; Joseph, L.M.; Deng, W.T.; Liu, L.; Li, Q.B.; Cline, K.; McCarty, D.R. Molecular characterization of the Arabidopsis 9-cis epoxycarotenoid dioxygenase gene family. Plant J. 2003, 35, 44–56. [Google Scholar] [CrossRef]
- Ali, F.; Qanmber, G.; Li, F.; Wang, Z. Updated role of ABA in seed maturation, dormancy, and germination. J. Adv. Res. 2022, 35, 199–214. [Google Scholar] [CrossRef]
- Kanno, Y.; Jikumaru, Y.; Hanada, A.E.; Nambara, S.R.; Abrams, Y.; Kamiya, M.S. Comprehensive hormone profiling in developing Arabidopsis seeds: Examination of the site of ABA biosynthesis, ABA transport and hormone interactions. Plant Cell Physiol. 2010, 51, 1988–2001. [Google Scholar] [CrossRef]
- Frey, A.; Effroy, D.; Lefebvre, V.; Seo, M.; Perreau, F.; Berger, A.; Sechet, J.; To, A.; North, H.M.; Marion-Poll, A. Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. Plant J. 2012, 70, 501–512. [Google Scholar] [CrossRef]
- Lefebvre, V.H.; North, A.; Frey, B.; Sotta, M.; Seo, M.; Okamoto, E.; Nambara, A.; Marion, P. Functional analysis of Arabidopsis NCED6 and NCED9 genesindicates that ABA synthesized in the endosperm is involved in the inductionof seed dormancy. Plant J. 2006, 45, 309–319. [Google Scholar] [CrossRef]
- Saeed, A.; Iqbal, M. Loofa (Luffa cylindrica) sponge: Review of development of the biomatrix as a tool for biotechnological applications. Biotechnol. Prog. 2013, 29, 573–600. [Google Scholar] [CrossRef]
- Shendge, P.N.; Belemkar, S. Therapeutic potential of Luffa acutangula: A review on its traditional uses, phytochemistry, Pharmacology and toxicological aspects. Front. Pharmacol. 2018, 9, 1177. [Google Scholar] [CrossRef]
- Awal, A.; Adam, S.; Shamsuri, S.; Yusuf, N.A.; Ibrahim, N.; Elias, E.Z. Luffa gourd production practices from transplanting and direct seeding methods for composite productions. OP Conf. Ser. Earth Environ. Sci. 2021, 685, 12024. [Google Scholar] [CrossRef]
- Chaodumrikul, S.; Kaewsorn, P.; Chulaka, P.; Chanprasert, W. Breaking seed dormancy in smooth loofah (Luffa cylindrica (L.) M. Roem.) using scarification and dry heat treatment. Agric. Nat. Res. 2016, 50, 85–88. [Google Scholar] [CrossRef][Green Version]
- Priya, R.; Siva, R. Analysis of phylogenetic and functional diverge in plant nine-cis epoxycarotenoid dioxygenase gene family. J. Plant Res. 2015, 128, 519–534. [Google Scholar] [CrossRef] [PubMed]
- Hashemi, M.; Mousavi, S.M.; Razavi, S.H.; Shojaosadati, S.A. Comparison of submerged and solid-state fermentation systems effects on the catalytic activity of Bacillus sp. KR-8104 α-amylase at different pH and temperatures. Ind. Crops Prod. 2013, 43, 661–667. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene features visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Løvdal, T.; Lillo, C. Reference gene selection for quantitative real-time PCR normalization in tomato subjected to nitrogen, cold, and light stress. Anal. Biochem. 2009, 387, 238–24228. [Google Scholar] [CrossRef]
- Wang, Z.; Li, X.; Liu, N.; Peng, Q.; Wang, Y.; Fan, B.; Zhu, C.; Chen, Z. A Family of NAI2-interacting proteins in the biogenesis of the ER body and related structures. Plant Physiol. 2019, 180, 212–227. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhou, C.; Yan, X.; Zhang, J.; Xu, J. Simultaneous analysis of ten phytohormones in Sargassum horneri by high-performance liquid chromatography with electrospray ionization tandem mass spectrometry. J. Sep. Sci. 2016, 39, 1804–1813. [Google Scholar] [CrossRef] [PubMed]
- Pujadas, G.; Palau, J. Evolution of alpha-amylases: Architectural features and key residues in the stabilization of the (beta/alpha) (8) scaffold. Mol. Biol. Evol. 2001, 18, 38–54. [Google Scholar] [CrossRef]
- Liu, L.; Xia, W.; Li, H.; Zeng, H.; Wei, B.; Han, S.; Yin, C. Salinity inhibits rice seed germination by reducing alpha-amylase activity via decreased bioactive gibberellin content. Front. Plant Sci. 2018, 9, 275. [Google Scholar] [CrossRef]
- Zhang, L.; Shi, M.; Ji, J.; Hu, X.; Chen, F. Gut microbiota determines the prevention effects of Luffa cylindrica (L.) Roem supplementation against obesity and associated metabolic disorders induced by high-fat diet. FASEB J. 2019, 33, 10339. [Google Scholar] [CrossRef]
- Sano, N.; Marion-Poll, A. ABA metabolism and homeostasis in seed dormancy and germination. Int. J. Mol. Sci. 2021, 22, 5069. [Google Scholar] [CrossRef]
- Ruggiero, B.; Koiwa, H.; Manabe, Y.; Quist, T.M.; Inan, G.; Saccardo, F.; Joly, R.J.; Hasegawa, P.M.; Bressan, R.A.; Maggio, A. Uncoupling the effects of abscisic acid on plant growth and water relations: Analysis of sto1/nced3, an abscisic acid-deficient but salt stress-tolerant mutant in Arabidopsis. Plant Physiol. 2004, 136, 3134–3147. [Google Scholar] [CrossRef]
- Müller, M. Foes or Friends: ABA and ethylene interaction under abiotic stress. Plants 2021, 10, 448. [Google Scholar] [CrossRef]
- Parwez, R.; Aftab, T.; Gill, S.S.; Naeem, M. Abscisic acid signaling and crosstalk with phytohormones in regulation of environmental stress responses. Environ. Exp. Bot. 2022, 199, 104885. [Google Scholar] [CrossRef]
- Martínez-Andújar, C.; Ordiz, M.I.; Huang, Z.; Nonogaki, M.; Beachy, R.N.; Nonogaki, H. Induction of 9-cis-epoxycarotenoid dioxygenase in Arabidopsis thaliana seeds enhances seed dormancy. Proc. Natl. Acad. Sci. USA 2011, 108, 17225–17229. [Google Scholar] [CrossRef]
- Han, C.; Yang, P. Studies on the molecular mechanisms of seed germination. Proteomics 2015, 15, 1671–1679. [Google Scholar] [CrossRef] [PubMed]
- Helland, M.; Wicklund, T.; Narvhus, J. Effect of germination time on α-amylase production and viscosity of maize porridge. Food Res. Inter. 2002, 35, 315–321. [Google Scholar] [CrossRef]
- Lee, Y.; Chen, M.C.; Lin, L.; Chung, M.C.; Leu, W.M. Increased expression of 9-cis-epoxycarotenoid dioxygenase, PtNCED1, associated with inhibited seed germination in a terrestrial orchid, Phaius tankervilliae. Front. Plant Sci. 2018, 9, 1043. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
