Genetic and Haplotype Diversity of Hoplolaimus (Nematoda: Hoplolaimidae) Through Analysis of COI of mtDNA
Abstract
1. Introduction
2. Materials and Methods
2.1. Genetic Diversity, Phylogenetic Analysis, and Genetic Structure
2.2. Neutrality and Population Size Change Test
3. Results
3.1. Haplotype Network
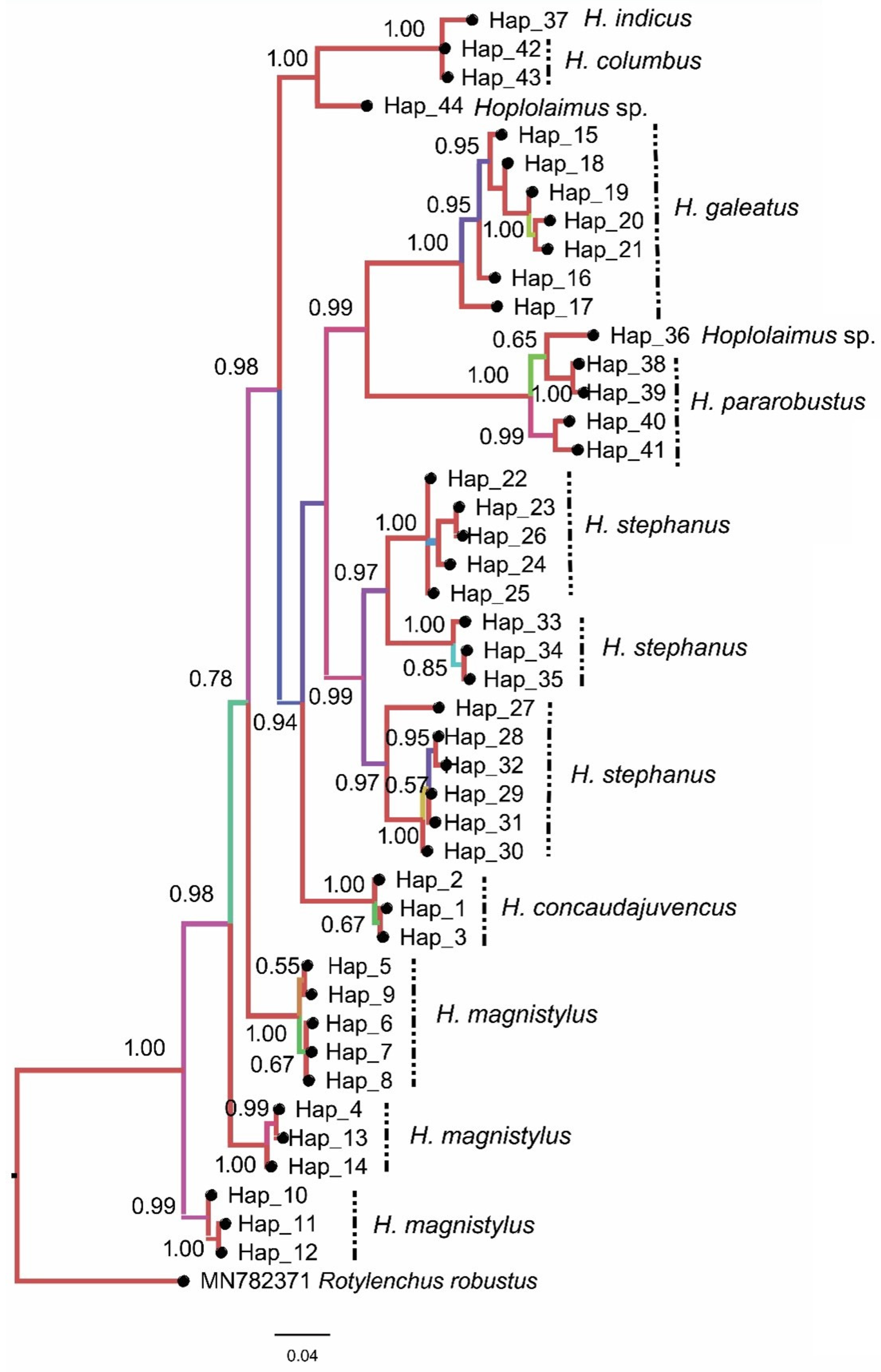
3.2. Phylogenetic Analysis
3.3. Genetic Diversity Estimation
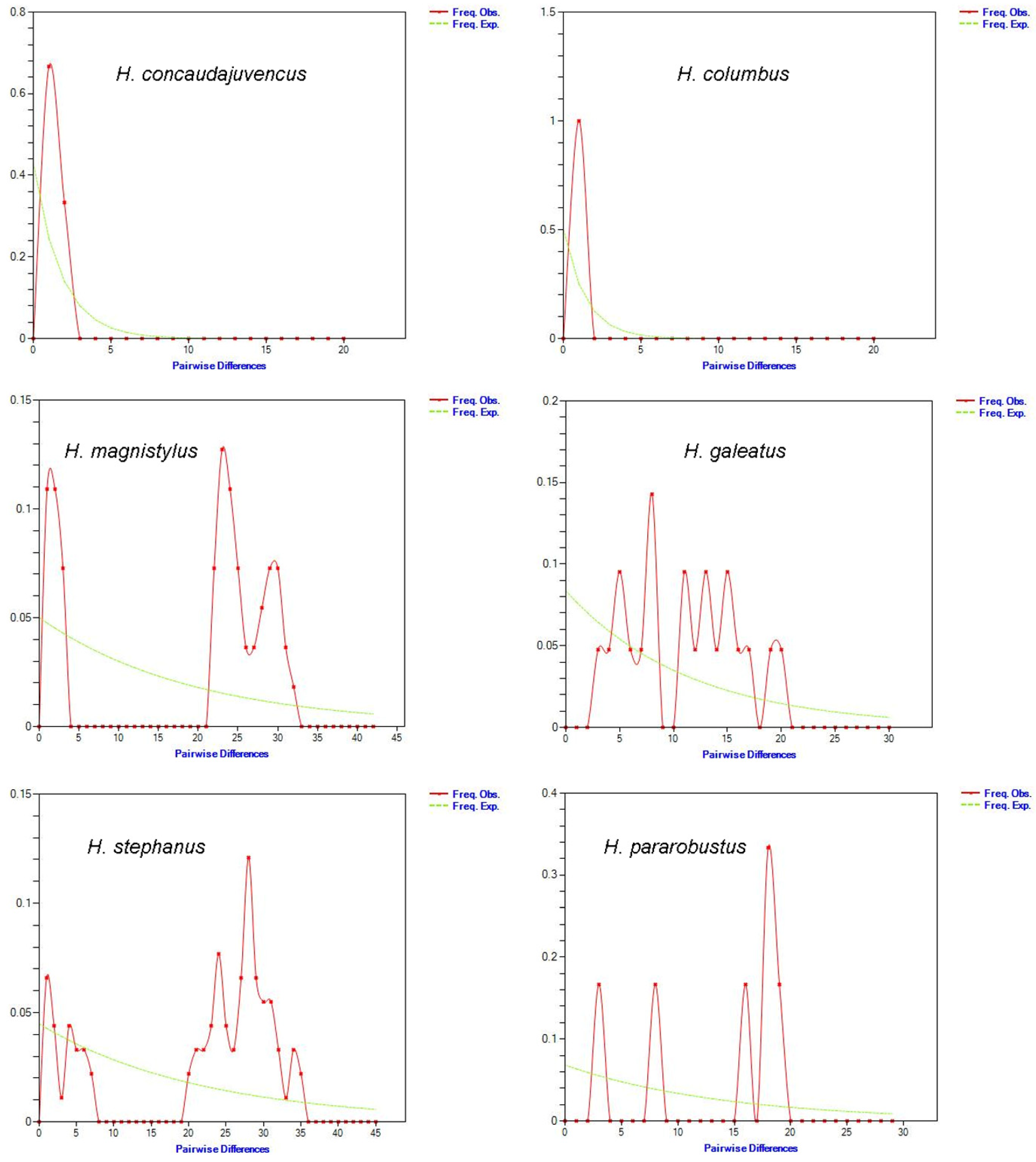
3.4. Demographic History and Neutrality
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shokoohi, E.; Abolafia, J.; Mashela, P.W. Redescription of Hoplolaimus indicus Sher, 1963 (Rhabditida, Hoplolaimidae) from Iran, including the first SEM study of the species. Biologia 2022, 77, 2161–2172. [Google Scholar] [CrossRef]
- Handoo, Z.; Golden, A.M. A key and diagnostic compendium to the species of the genus Hoplolaimus Daday, 1905 (Nematoda: Hoplolaimidae). J. Nematol. 1992, 24, 45–53. [Google Scholar]
- Olajide, E.; Singh, P.R.; Kolombia, Y.A.; Rumbarar, M.K.; Couvreur, M.; Bert, W. Characterization of Hoplolaimus seinhorsti and Hoplolaimus pararobustus (Tylenchina: Hoplolaimidae) from banana, with phylogeny and species delineation in the genus Hoplolaimus. J. Nematol. 2023, 55, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Bae, C.H.; Robbins, R.T.; Szalanski, A.L. Molecular identification of some Hoplolaimus species from the USA based on duplex PCR, multiplex PCR and PCR-RFLP analysis. Nematology 2009, 11, 471–480. [Google Scholar] [CrossRef]
- Bae, C.H.; Szalanski, A.L.; Robbins, R.T. Molecular analysis of the lance nematode, Hoplolaimus spp., using the first internal transcribed spacer and the D2-D3 expansion segments of 28S ribosomal DNA. J. Nematol. 2008, 40, 201–209. [Google Scholar]
- Ma, X.; Agudelo, P.; Mueller, J.D.; Knap, H.T. Molecular characterization and phylogenetic analysis of Hoplolaimus stephanus. J. Nematol. 2011, 43, 25–34. [Google Scholar] [PubMed]
- Holguin, C.M.; Baeza, J.A.; Mueller, J.D.; Agudelo, P. High genetic diversity and geographic subdivision of three lance nematode species (Hoplolaimus spp.) in the United States. Ecol. Evol. 2015, 5, 2929–2944. [Google Scholar] [CrossRef] [PubMed]
- Triantafyllidis, A.; Abatzopoulos, T.; Economidis, P. Genetic differentiation and phylogenetic relationships among Greek Silurus glanis and Silurus aristotelis (Pisces, Siluridae) populations, assessed by PCR–RFLP analysis of mitochondrial DNA segments. Heredity. 1999, 82, 503–509. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Myers, N.; Mittermeier, R.A.; Mittermeier, C.G.; Da Fonseca, G.A.; Kent, J. Biodiversity hotspots for conservation priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef]
- Blouin, M.S.; Yowell, C.A.; Courtney, C.H.; Dame, J.B. Host movement and the genetic structure of populations of parasitic nematodes. Genetics 1995, 141, 1007–1014. [Google Scholar] [CrossRef] [PubMed]
- Gorton, M.J.; Kasl, E.L.; Detwiler, J.T.; Criscione, C.D. Testing local-scale panmixia provides insights into the cryptic ecology, evolution, and epidemiology of metazoan animal parasites. Parasitology 2012, 139, 981–997. [Google Scholar] [CrossRef] [PubMed]
- Huyse, T.; Poulin, R.; Theron, A. Speciation in parasites: A population genetics approach. Trends. Parasitol. 2005, 21, 469–475. [Google Scholar] [CrossRef]
- Kiontke, K.C.; Félix, M.; Ailion, M.; Rockman, M.V.; Braendle, C.; Pénigault, J.B.; Fitch, D.H.A. A phylogeny and molecular barcodes for Caenorhabditis, with numerous new species from rotting fruits. BMC Evol. Biol. 2011, 11, 339. [Google Scholar] [CrossRef] [PubMed]
- Nadler, S.A.; De León, G.P. Integrating molecular and morphological approaches for characterizing parasite cryptic species: Implications for parasitology. Parasitology 2012, 138, 1688–1709. [Google Scholar] [CrossRef]
- Palomares-Rius, J.E.; Cantalapiedra-Navarrete, C.; Castillo, P. Cryptic species in plant-parasitic nematodes. Nematology 2014, 16, 1105–1118. [Google Scholar] [CrossRef]
- Matczyszyn, J.N.; Harris, T.; Powers, K.; Everhart, S.E.; Powers, T.O. Ecological and morphological differentiation among COI haplotype groups in the plant parasitic nematode species Mesocriconema Xenoplax. J. Nematol. 2022, 54, 20220009. [Google Scholar] [CrossRef]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. In Nucleic Acids Symposium Series; Information Retrieval Ltd.: London, UK, 1999; Volume 41, pp. 95–98. [Google Scholar]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Villesen, P. FaBox: An online toolbox for fasta sequences. Mol. Ecol. Notes 2007, 7, 965–968. [Google Scholar] [CrossRef]
- Rozas, J.; Sánchez-Delbarrio, J.C.; Messeguer, X.; Rozas, R. Dnasp, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 2003, 19, 2496–2497. [Google Scholar] [CrossRef]
- Leigh, J.W.; Bryant, D. PopART: Full-feature software for haplotype network construction. Methods Ecol. Evol. 2015, 6, 1110–1116. [Google Scholar] [CrossRef]
- Ronquist, F.; Huelsenbeck, J. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003, 19, 1572–1574. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree v1.3.1. Institute of Evolutionary Biology; University of Edinburgh: Edinburgh, Scotland, 2010; Available online: https://tree.bio.ed.ac.uk/software/figtree/ (accessed on 25 December 2018).
- Excoffier, L.; Lischer, H.E.L. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Biol. Evol. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 1989, 123, 585–595. [Google Scholar] [CrossRef]
- Fu, Y.X. Statistical tests of Neutrality of mutations against population growth, hitchhiking and background selection. Genetics 1997, 147, 915–925. [Google Scholar] [CrossRef]
- Bragard, C.; Baptista, P.; Chatzivassiliou, E.; Di Serio, F.; Gonthier, P.; Jaques Miret, J.A.; Justesen, A.F.; MacLeod, A.; Magnusson, C.S.; Milonas, P.; et al. Pest categorisation of Hoplolaimus galeatus. EFSA J. 2023, 21, e08117. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.R.; Haque, Z.; Ahamad, F.; Shah, M.H. Nematode problems in rice and their sustainable management. In Nematode Diseases of Crops and Their Sustainable Management; Academic Press: Cambridge, MA, USA, 2023; pp. 133–166. [Google Scholar]
- Nei, M. Molecular Evolutionary Genetics; Columbia University Press: New York, NY, USA, 1987. [Google Scholar]
- O’Leary, S.J.; Hollenbeck, C.M.; Vega, R.R.; Portnoy, D.S. Disentangling complex genomic signals to understand population structure of an exploited, estuarine-dependent flatfish. Ecol. Evol. 2021, 11, 13415–13429. [Google Scholar] [CrossRef]
- Vásquez-Aguilar, A.A.; Barbachano-Guerrero, A.; Angulo, D.F.; Jarquín-Díaz, V.H. Phylogeography and population differentiation in Hepatozoon canis (Apicomplexa: Hepatozoidae) reveal expansion and gene flow in world populations. Parasit. Vectors. 2021, 14, 467. [Google Scholar] [CrossRef] [PubMed]
- Zu, Y.G.; Sun, M.; Kang, L. The Theory, Method, and Application of Molecular Ecology; Higher Education Press: Beijing, China, 1999; pp. 30–37. [Google Scholar]
- Eckshtain-Levi, N.; Weisberg, A.J.; Vinatzer, B.A. The population genetic test Tajima’s D identifies genes encoding pathogen-associated molecular patterns and other virulence-related genes in Ralstonia solanacearum. Mol. Plant. Pathol. 2018, 19, 2187–2192. [Google Scholar] [CrossRef]
- Chapuis, M.P.; Loiseau, A.; Michalakis, Y.; Lecoq, M.; Franc, A.; Estoup, A. Outbreaks, gene flow and effective population size in the migratory locust, Locusta migratoria: A regional-scale comparative survey. Mol. Ecol. 2009, 18, 792–800. [Google Scholar] [CrossRef] [PubMed]
- Ray, N.; Currat, M.; Excoffier, L. Intra-deme molecular diversity in spatially expanding populations. Mol. Biol. Evol. 2003, 20, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Rogers, A.R.; Harpending, H. Population growth makes waves in the distribution of pairwise genetic differences. Mol. Biol. Evol. 1992, 9, 552–569. [Google Scholar] [PubMed]
- Cortes-Rodríguez, N.; Hernández-Baños, B.E.; Navarro-Sigüenza, A.G.; Omland, K.E. Geographic variation and genetic structure in the Streak-backed Oriole: Low mitochondrial DNA differentiation reveals recent divergence. Condor 2008, 110, 729–739. [Google Scholar] [CrossRef]
- Apolônio Silva de Oliveira, D.; Decraemer, W.; Moens, T.; Dos Santos, G.A.P.; Derycke, S. Low genetic but high morphological variation over more than 1000 km coastline refutes omnipresence of cryptic diversity in marine nematodes. BMC Evol. Biol. 2017, 17, 71. [Google Scholar] [CrossRef] [PubMed]
- Hada, A.; Singh, D.; Venkata Satyanarayana, K.K.V.; Chatterjee, M.; Phani, V.; Rao, U. Effect of fluensulfone on different functional genes of root-knot nematode Meloidogyne incognita. J. Nematol. 2021, 53, e2021-73. [Google Scholar] [CrossRef]
- Derycke, S.; Remerie, T.; Vierstraete, A.; Backeljau, T.; Vanfleteren, J.; Vincx, M.; Moens, T. Mitochondrial DNA variation and cryptic speciation within the free-living marine nematode Pellioditis marina. Mar. Ecol.-Prog. Ser. 2005, 300, 91–103. [Google Scholar] [CrossRef]
- Ristau, K.; Steinfartz, S.; Traunspurger, W. First evidence of cryptic species diversity and significant population structure in a widespread freshwater nematode morphospecies (Tobrilus gracilis). Mol. Ecol. 2013, 22, 4562–4575. [Google Scholar] [CrossRef] [PubMed]

| Species | n | S | h | Hd | π | Tajima’s D | Fu |
|---|---|---|---|---|---|---|---|
| H. concaudajuvencus | 12 | 2 | 3 | 0.32 | 0.0016 | −0.84971 | −0.725 |
| H. magnistylus | 22 | 40 | 11 | 0.85 | 0.04915 | 0.77362 | 2.616 |
| H. galeatus | 27 | 26 | 7 | 0.67 | 0.03479 | 1.32091 | 6.902 |
| H. stephanus | 52 | 50 | 14 | 0.89 | 0.06746 | 1.71201 | 9.731 ** |
| H. pararobustus | 5 | 22 | 4 | 0.90 | 0.04048 | 0.57841 | 1.890 |
| H. columbus | 52 | 1 | 2 | 0.66 | 0.00026 | −0.87674 | −0.917 |
| Source of Variance | df | Sum of Squares | Variance Component | % Total of Variance | Significance |
|---|---|---|---|---|---|
| Among populations | 7 | 152,012.047 | 868.085 | 86.3 | p < 0.001 |
| Within populations | 166 | 22,889.976 | 137.891 | 13.7 | p < 0.001 |
| Total | 173 | 174,902.023 | 1005.976 | 100 | p < 0.001 |
| Species | H. concaudajuvencus | H. stephanus | H. magnistylus | H. galeatus | H. pararobustus | H. columbus |
|---|---|---|---|---|---|---|
| H. concaudajuvencus | 0 | |||||
| H. stephanus | 0.62 | 0 | ||||
| H. magnistylus | 0.74 | 0.48 | 0 | |||
| H. galeatus | 0.84 | 0.65 | 0.75 | 0 | ||
| H. pararobustus | 0.92 | 0.75 | 0.81 | 0.86 | 0 | |
| H. columbus | 0.97 | 0.89 | 0.94 | 0.95 | 0.95 | 0 |
| Species | Raggedness Index | p-Value |
|---|---|---|
| H. concaudajuvencus | 0.6667 | 0.2357 |
| H. stephanus | 0.0200 | 0.2018 |
| H. magnistylus | 0.0324 | 0.2327 |
| H. galeatus | 0.0635 | 0.1700 |
| H. pararobustus | 0.3333 | 0.2189 |
| H. columbus | 2.0000 | 0.5000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shokoohi, E.; Masoko, P. Genetic and Haplotype Diversity of Hoplolaimus (Nematoda: Hoplolaimidae) Through Analysis of COI of mtDNA. Horticulturae 2025, 11, 113. https://doi.org/10.3390/horticulturae11020113
Shokoohi E, Masoko P. Genetic and Haplotype Diversity of Hoplolaimus (Nematoda: Hoplolaimidae) Through Analysis of COI of mtDNA. Horticulturae. 2025; 11(2):113. https://doi.org/10.3390/horticulturae11020113
Chicago/Turabian StyleShokoohi, Ebrahim, and Peter Masoko. 2025. "Genetic and Haplotype Diversity of Hoplolaimus (Nematoda: Hoplolaimidae) Through Analysis of COI of mtDNA" Horticulturae 11, no. 2: 113. https://doi.org/10.3390/horticulturae11020113
APA StyleShokoohi, E., & Masoko, P. (2025). Genetic and Haplotype Diversity of Hoplolaimus (Nematoda: Hoplolaimidae) Through Analysis of COI of mtDNA. Horticulturae, 11(2), 113. https://doi.org/10.3390/horticulturae11020113







