Combining BSA-Seq, High-Density Genetic Map, and RNA-Seq to Identify Candidate Genes Controlling Embryo Spot Trait in Potato
Abstract
1. Introduction
2. Materials and Methods
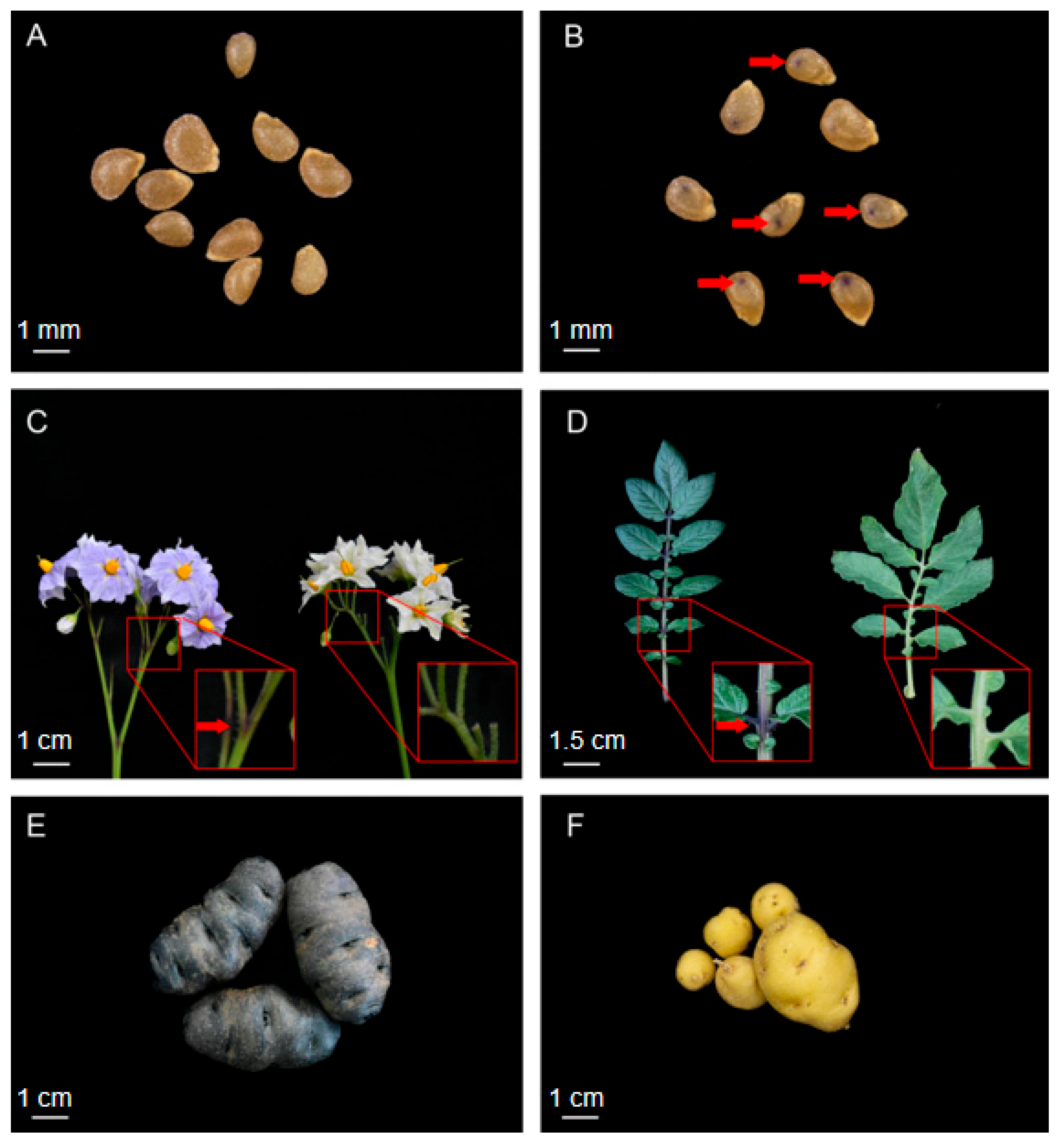
2.1. Experimental Materials and Phenotypic Trait
2.2. DNA Extraction, Library Construction, and Sequencing
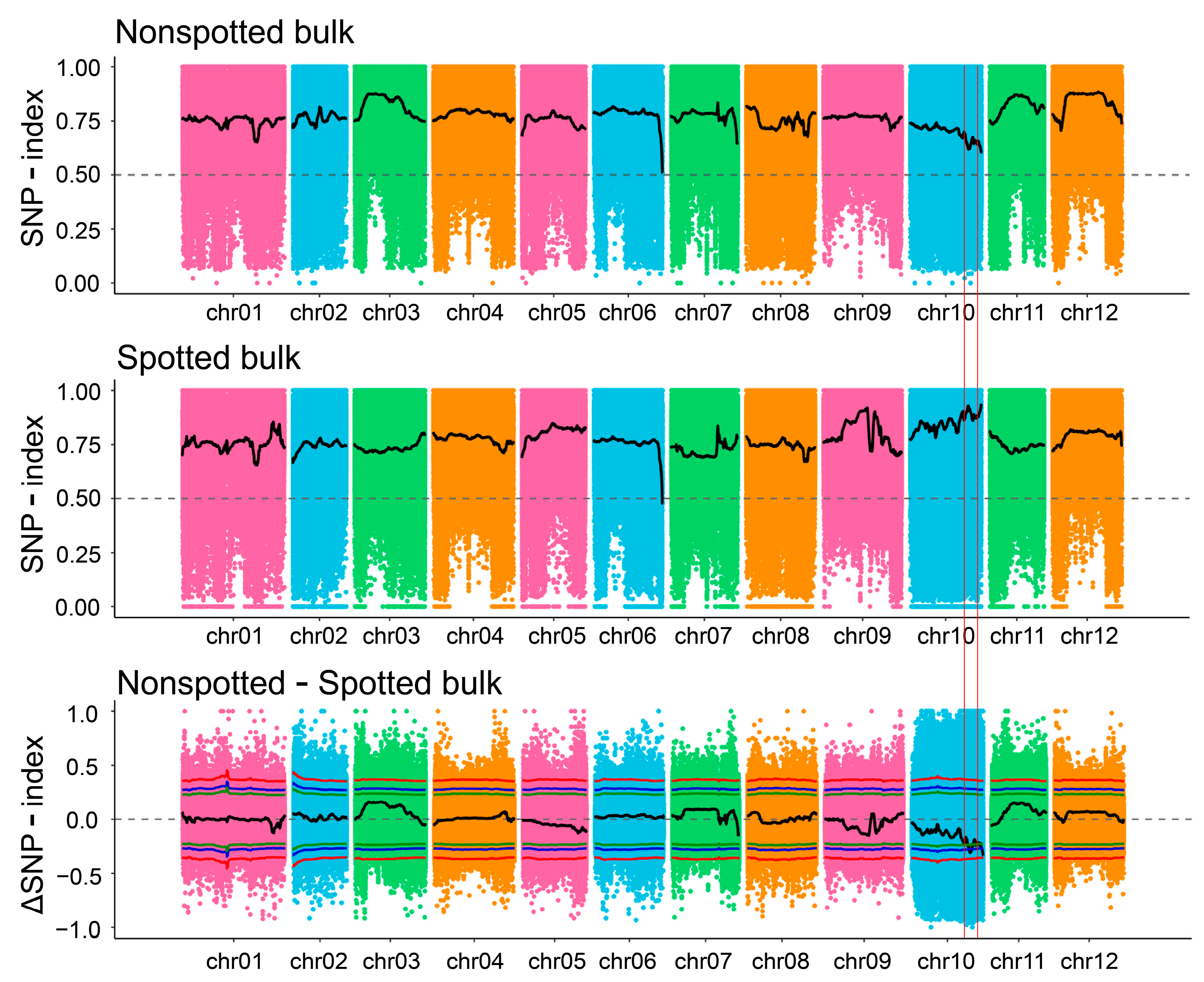
2.3. Analysis of the BSA-Seq Data
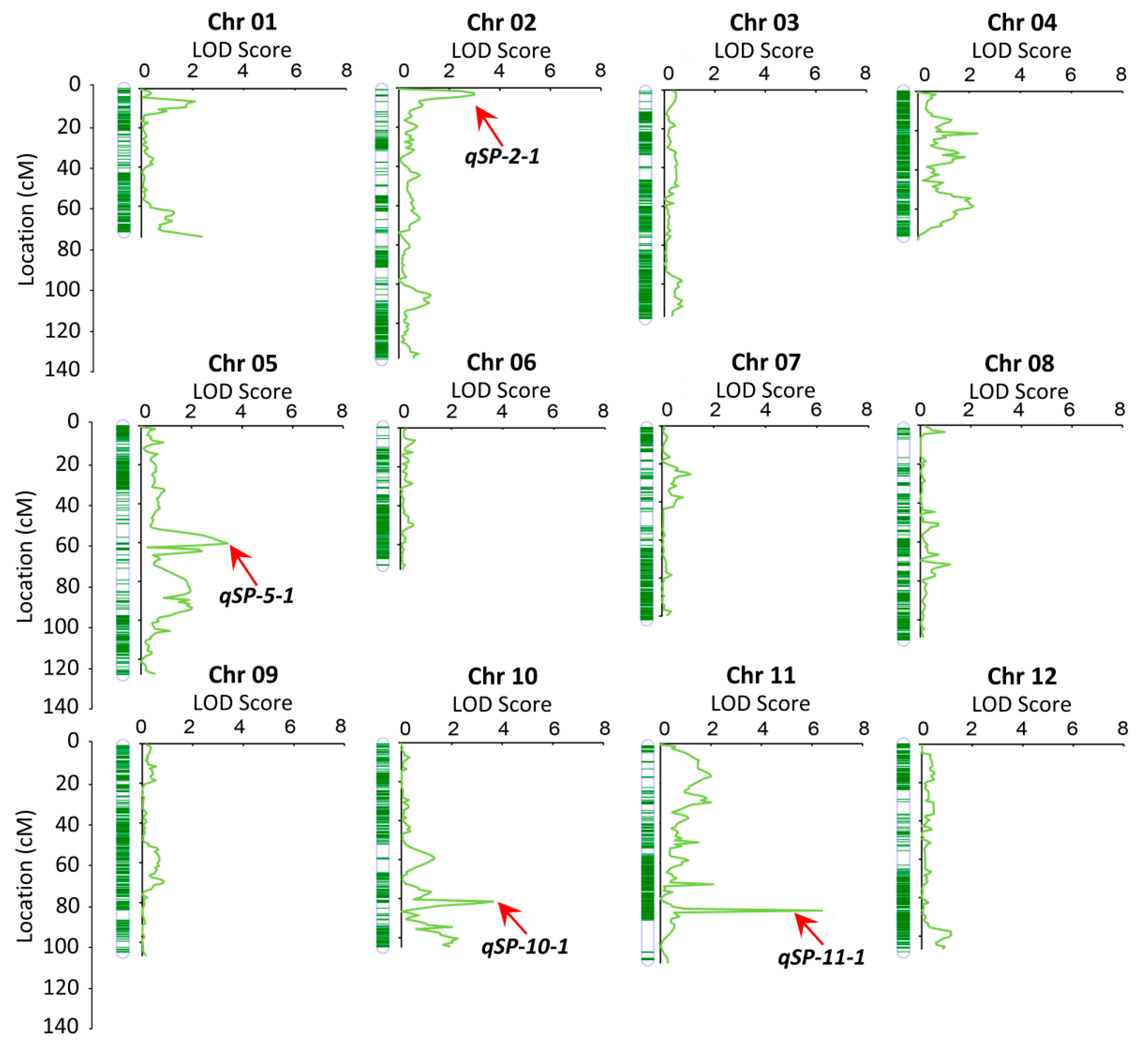
2.4. QTL Mapping
2.5. RNA Sequencing, Transcriptome Assembly, and Data Analysis
2.6. Real-Time qPCR Analysis and Prediction of Conserved Protein Domains of Genes
3. Results
3.1. The Embryo Spot Trait Is Controlled by Two Genetic Loci
3.2. QTL Mapping of Embryo Spot Trait Using High-Density Genetic Map
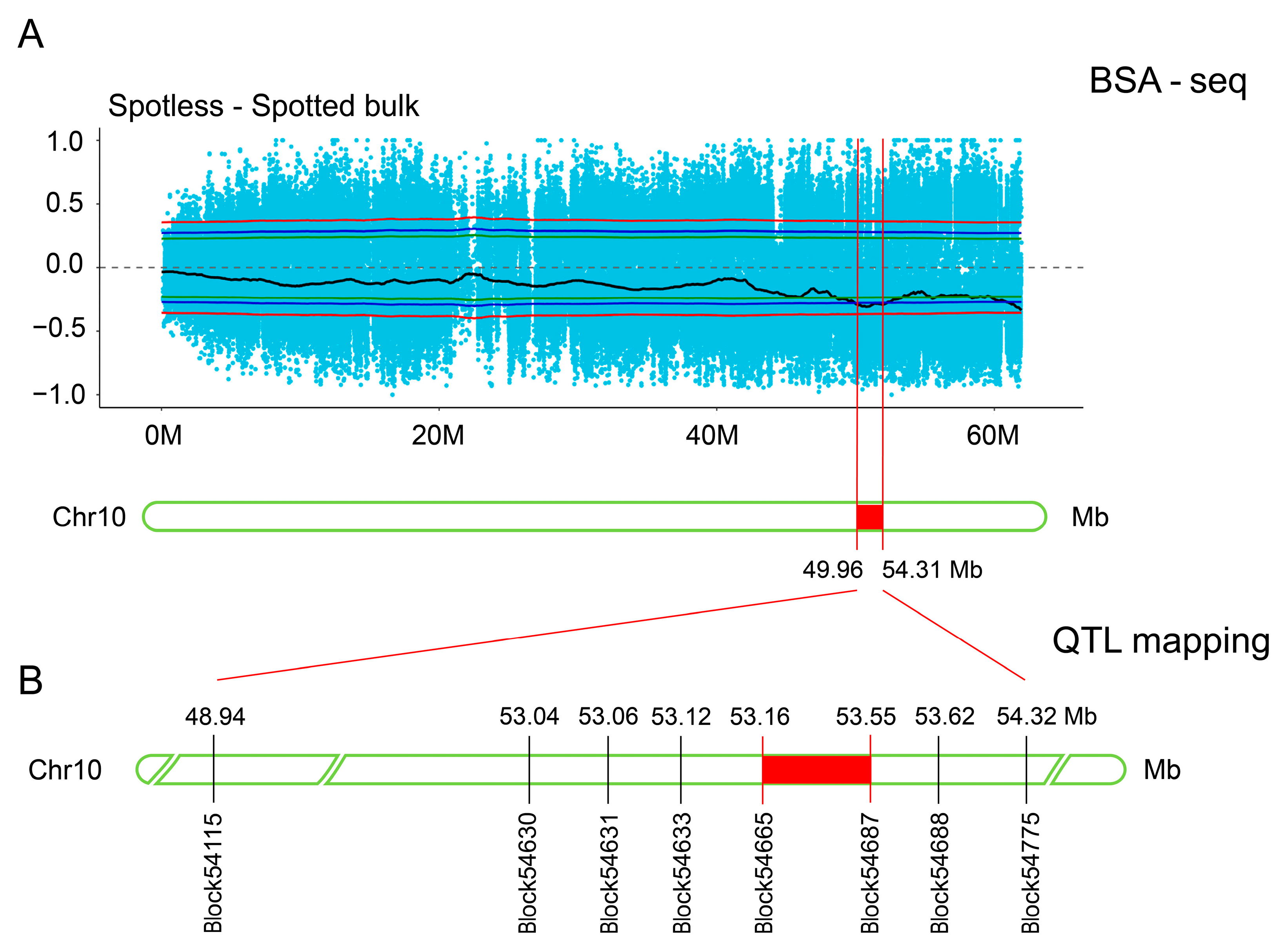
3.3. Further Mapping Analysis by Combining BSA-Seq with QTL Mapping
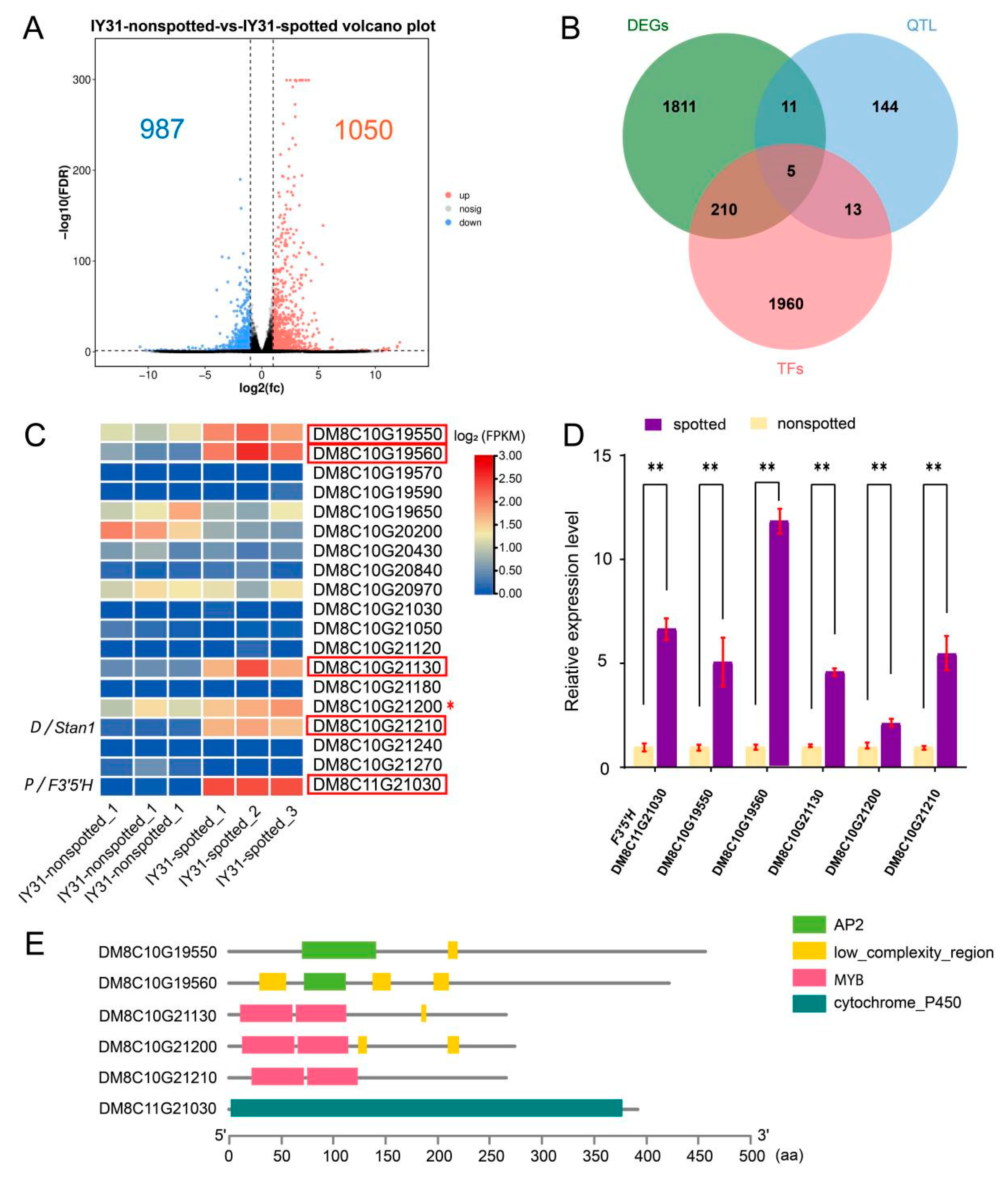
3.4. Identification of Candidate Genes via RNA-Seq
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| BSA-seq | Bulked segregant analysis-sequencing |
| TFs | Transcription factors |
| SP | Spotted |
| NS | Nonspotted |
| ED | Euclidean Distance |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| SNPs | Single nucleotide polymorphisms |
| ICIM | Inclusive Composite Interval Mapping |
| QTLs | Quantitative trait locis |
| PVE | Phenotypic variance |
| DEGs | Differentially expressed genes |
| qRT-PCR | Quantitative real-time PCR |
| BP | Biological process |
References
- Wang, F.; Xia, Z.; Zou, M.; Zhao, L.; Jiang, S.; Zhou, Y.; Zhang, C.; Ma, Y.; Bao, Y.; Sun, H.; et al. The autotetraploid potato genome provides insights into highly heterozygous species. Plant Biotechnol. J. 2022, 20, 1996–2005. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, J.E.; Ramsay, G. Utilisation of the commonwealth potato collection in potato breeding. Euphytica 2005, 146, 9–19. [Google Scholar] [CrossRef]
- Spooner, D.M.; Núñez, J.; Trujillo, G.; Herrera Mdel, R.; Guzmán, F.; Ghislain, M. Extensive simple sequence repeat genotyping of potato landraces supports a major reevaluation of their gene pool structure and classification. Proc. Natl. Acad. Sci. USA 2007, 104, 19398–19403. [Google Scholar] [CrossRef]
- Bethke, P.C.; Halterman, D.A.; Jansky, S. Are we getting better at using wild potato species in light of new tools? Crop Sci. 2017, 57, 1241–1258. [Google Scholar]
- Spooner, D.M.; Ghislain, M.; Simon, R.; Jansky, S.H.; Gavrilenko, T. Systematics, Diversity, Genetics, and Evolution of Wild and Cultivated Potatoes. Bot. Rev. 2014, 80, 283–383. [Google Scholar] [CrossRef]
- Pham, G.M.; Braz, G.T.; Conway, M.; Crisovan, E.; Hamilton, J.P.; Laimbeer, F.P.E.; Manrique-Carpintero, N.; Newton, L.; Douches, D.S.; Jiang, J.; et al. Genome-wide Inference of Somatic Translocation Events During Potato Dihaploid Production. Plant Genome 2019, 12, 180079. [Google Scholar]
- Gebhardt, C. The historical role of species from the Solanaceae plant family in genetic research. Theor. Appl. Genet. 2016, 129, 2281–2294. [Google Scholar] [CrossRef]
- Tang, H.; Qiu, Y.; Wang, W.; Yu, M.; Chang, Y.; Han, Z.; Du, L.; Lin, Z.; Wang, K.; Ye, X. Development of a haploid inducer by editing HvMTL in barley. J. Genet. Genom. 2023, 50, 366–369. [Google Scholar]
- Delzer, B.; Liang, D.; Szwerdszarf, D.; Rodriguez, I.; Mardones, G.; Elumalai, S.; Johnson, F.; Nalapalli, S.; Egger, R.; Burch, E.; et al. Elite, transformable haploid inducers in maize. Crop J. 2024, 12, 314–319. [Google Scholar] [CrossRef]
- Hutten, R.C.B.; Scholberg, E.J.M.M.; Huigen, D.J.; Hermsen, J.G.T.; Jacobsen, E. Analysis of dihaploid induction and production ability and seed parent x pollinator interaction in potato. Euphytica 1993, 72, 61–64. [Google Scholar] [CrossRef]
- Peloquin, S.J.; Gabert, A.C.; Rodomiro, O.J.A.O.B. Nature of ‘Pollinator’ Effect in Potato (Solanum tuberosum L.) Haploid Production. Ann. Bot. 1996, 77, 539–542. [Google Scholar] [CrossRef]
- Breukelen, E.W.M.; Ramanna, M.S.; Hermsen, J.G.T. Parthenogenetic monohaploids (2n=x=12) from Solanum tuberosum L. and S. verrrucosum Schlechtd. and the production of homozygous potato diploids. Euphytica 1977, 26, 263–271. [Google Scholar] [CrossRef]
- Ordoez, B.; Santayana, M.; Aponte, M.; Henry, I.M.; Comai, L.; Eyzaguirre, R.; Lindqvist-Kreuze, H.; Bonierbale, M. PL-4 (CIP596131.4): An Improved Potato Haploid Inducer. Am. J. Potato Res. 2021, 98, 255–262. [Google Scholar] [CrossRef]
- Hermsen, J.G.T.; Verdenius, J.J.E. Selection from Solanum tuberosum group phureja of genotypes combining high-frequency haploid induction with homozygosity for embryo-spot. Euphytica 1973, 22, 244–259. [Google Scholar] [CrossRef]
- Jung, C.S.; Griffiths, H.M.; De Jong, D.M.; Cheng, S.; Bodis, M.; De Jong, W.S. The potato P locus codes for flavonoid 3′,5′-hydroxylase. Theor. Appl. Genet. 2005, 110, 269–275. [Google Scholar] [CrossRef]
- Jong, W.D.D.; De Jong, D.M.; De Jong, H.; Kalazich, J.; Bodis, M. An allele of dihydroflavonol 4-reductase associated with the ability to produce red anthocyanin pigments in potato (Solanum tuberosum L.). Theor. Appl. Genet. 2003, 107, 1375–1383. [Google Scholar] [CrossRef]
- Zhang, Y.; Cheng, S.; De Jong, D.; Griffiths, H.; Halitschke, R.; De Jong, W. The potato R locus codes for dihydroflavonol 4-reductase. Theor. Appl. Genet. 2009, 119, 931–937. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.S.; Griffiths, H.M.; De Jong, D.M.; Cheng, S.; Bodis, M.; Kim, T.S.; De Jong, W.S. The potato developer (D) locus encodes an R2R3 MYB transcription factor that regulates expression of multiple anthocyanin structural genes in tuber skin. Theor. Appl. Genet. 2009, 120, 45–57. [Google Scholar] [CrossRef]
- Du, H.; Zhai, Z.; Pu, J.; Liang, J.; Wang, R.; Zhang, Z.; Wang, P.; Zhu, Y.; Huang, L.; Li, D.; et al. Two tandem R2R3 MYB transcription factor genes cooperatively regulate anthocyanin accumulation in potato tuber flesh. Plant Biotechnol. J. 2025, 23, 1521–1534. [Google Scholar] [CrossRef]
- Laimbeer, F.P.E.; Bargmann, B.O.R.; Holt, S.H.; Pratt, T.; Peterson, B.; Doulis, A.G.; Buell, C.R.; Veilleux, R.E. Characterization of the F Locus Responsible for Floral Anthocyanin Production in Potato. G3 Genes Genomes Genet. 2020, 10, 3871–3879. [Google Scholar] [CrossRef]
- Jong, H.D. Inheritance of anthocyanin pigmentation in the cultivated potato: A critical review. Am. Potato J. 1991, 68, 585–593. [Google Scholar] [CrossRef]
- Dodds, K.S.; Long, D.H. The inheritance of colour in diploid potatoes. J. Genet. 1955, 53, 136–149. [Google Scholar] [CrossRef]
- Dodds, E.S.; Long, D.H. The inheritance oe colour in diploid potatoes II. A three-factor linkage group. J. Genet. 1956, 54, 27–41. [Google Scholar] [CrossRef]
- Van Eck, H.J.; Jacobs, J.M.; van Dijk, J.; Stiekema, W.J.; Jacobsen, E. Identification and mapping of three flower colour loci of potato (S. tuberosum L.) by RFLP analysis. Theor. Appl. Genet. 1993, 86, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Van Eck, H.; Jacobs, J.; van den Berg, P.; Stiekema, W.J.; Heredity, E.J. The inheritance of anthocyanin pigmentation in potato (Solanum tuberosum L.) and mapping of tuber skin colour loci using RFLPs. Heredity 1994, 73, 410–421. [Google Scholar] [CrossRef]
- Endelman, J.B.; Jansky, S.H. Genetic mapping with an inbred line-derived F2 population in potato. Theor. Appl. Genet. 2016, 129, 935–943. [Google Scholar] [CrossRef] [PubMed]
- Sonsungsan, P.; Nganga, M.L.; Lieberman, M.C.; Amundson, K.R.; Stewart, V.; Plaimas, K.; Comai, L.; Henry, I.M. A k-mer-based bulked segregant analysis approach to map seed traits in unphased heterozygous potato genomes. G3 Genes Genomes Genet. 2024, 14, jkae035. [Google Scholar] [CrossRef]
- Wang, P.; Cheng, L.; Pan, J.; Ma, L.; Hu, X.; Zhang, Z.; Li, D.; Zhu, Y.; Chang, S.; Yuan, P. A 6.49-Mb inversion associated with the purple embryo spot trait in potato. Abiotech 2025, 6, 22–32. [Google Scholar] [CrossRef]
- Yang, X.H.; Zhang, L.; Guo, X.; Xu, J.; Zhang, K.; Yang, Y.; Yang, Y.; Jian, Y.; Dong, D.; Huang, S.; et al. The gap-free potato genome assembly reveals large tandem gene clusters of agronomical importance in highly repeated genomic regions. Mol. Plant 2023, 16, 314–317. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Hill, J.T.; Demarest, B.L.; Bisgrove, B.W.; Gorsi, B.; Su, Y.C.; Yost, H.J. MMAPPR: Mutation mapping analysis pipeline for pooled RNA-seq. Genome Res. 2013, 23, 687–697. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Yao, C.; Miao, J.; Li, N.; Ji, F.; Hu, D.; Wang, S.; Zhou, Z.; Dai, K.; Chen, A.; et al. Construction of a High-Density Genetic Map and QTL Mapping Analysis for Yield, Tuber Shape, and Eye Number in Diploid Potato. Agriculture 2025, 15, 2032. [Google Scholar] [CrossRef]
- Lei, M.; Li, H.H.; Zhang, L.Y.; Wang, J.K. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J. 2015, 3, 269–283. [Google Scholar]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data using Real-Time Quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar]
- Riveros-Loaiza, L.M.; Benhur-Cardona, N.; Lopez-Kleine, L.; Soto-Sedano, J.C.; Pinzón, A.M.; Mosquera-Vásquez, T.; Roda, F. Uncovering anthocyanin diversity in potato landraces (Solanum tuberosum L. Phureja) using RNA-seq. PLoS ONE 2022, 17, e0273982. [Google Scholar] [CrossRef]
- Schneeberger, K. Using next-generation sequencing to isolate mutant genes from forward genetic screens. Nat. Rev. Genet. 2014, 15, 662–676. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, H.; Li, H.; Li, N.; Zhang, Y.; Zhang, Q.; Wang, S.; Wang, Q.; Wang, H. Fine-Mapping Quantitative Trait Loci for Body Weight and Abdominal Fat Traits: Effects of Marker Density and Sample Size. Poult. Sci. 2008, 87, 1314–1319. [Google Scholar] [CrossRef]
- Yang, J.; Sun, K.; Li, D.; Luo, L.; Liu, Y.; Huang, M.; Yang, G.; Liu, H.; Wang, H.; Chen, Z.; et al. Identification of stable QTLs and candidate genes involved in anaerobic germination tolerance in rice via high-density genetic mapping and RNA-Seq. BMC Genom. 2019, 20, 355. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zhang, Z.; Zhao, Y.; Guo, D.; Zhao, X.; Gao, W.; Zhang, J.; Song, B. StWRKY13 promotes anthocyanin biosynthesis in potato (Solanum tuberosum) tubers. Funct. Plant Biol. 2021, 49, 102–114. [Google Scholar] [CrossRef] [PubMed]
| Sample | Clean_Reads (bp) | Clean_Base (bp) | Q30 (%) | GC (%) | Mapped (%) | Properly_Mapped (%) | Ave_Depth | Cov_Ratio_1X (%) | Cov_Ratio_5X (%) | Cov_Ratio_10X (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| IVP101 | 304,744,996 | 45,711,749,400 | 85.79 | 35.71 | 99.27 | 89.34 | 56 | 93.95 | 91.08 | 89.12 |
| Y8 | 311,734,376 | 46,587,204,544 | 96.38 | 36.04 | 98.00 | 88.82 | 58 | 91.44 | 87.74 | 85.26 |
| spotted | 307,899,218 | 46,106,384,431 | 96.49 | 34.42 | 98.08 | 86.15 | 57 | 95.46 | 93.17 | 90.96 |
| nonspotted | 344,882,314 | 51,640,905,167 | 96.44 | 34.46 | 99.18 | 87.55 | 65 | 95.57 | 93.45 | 91.39 |
| Chromosome_ID | Chromosome Locations (bp) | Size (Mb) | |
|---|---|---|---|
| Start | End | ||
| chr10 | 49,960,000 | 54,310,000 | 4.3 |
| chr10 | 60,870,000 | 60,910,000 | 0.04 |
| chr10 | 60,950,000 | 61,950,000 | 1 |
| Total | - | - | 5.34 |
| QTL Name | Trait Name | Chromosome | Marker Interval | Physical Location (bp) | LOD a | PVE (%) b | Add c | Dom d |
|---|---|---|---|---|---|---|---|---|
| qSP-2-1 | spotted | 2 | Block4928–Block7296 | 1,004,731–32,809,910 | 2.99 | 5.37 | 0.18 | −0.16 |
| qSP-5-1 | spotted | 5 | Block25984–Block25989 | 26,176,748–26,984,565 | 3.43 | 2.98 | 0.1 | −0.18 |
| qSP-10-1 | spotted | 10 | Block54665–Block54687 | 53,168,851–53,553,482 | 3.63 | 23.85 | 0.13 | 0.07 |
| qSP-11-1 | spotted | 11 | Block59199–Block59202 | 41,280,367–41,823,710 | 6.4 | 18.23 | 0.27 | 0.07 |
| Gene ID | Physical Position (bp) | Nonspotted FPKM_Mean | Spotted FPKM Mean | Log2 (Fold Change) | TF Family | Conserved Domain Accession |
|---|---|---|---|---|---|---|
| DM8C10G19550 | Chr. 10: 51,564,788–51,566,161 | 1.13 | 3.03 | 1.42 | AP2/ERF | smart00380 |
| DM8C10G19560 | Chr. 10: 51,570,542–51,572,171 | 0.40 | 3.77 | 3.23 | AP2/ERF | smart00380 |
| DM8C10G21130 | Chr. 10: 53,332,531–53,334,183 | 0.36 | 2.83 | 2.97 | MYB | PLN03212 |
| DM8C10G21200 | Chr. 10: 53,520,992–53,522,172 | 1.23 | 2.28 | 0.89 | MYB | PLN03212 |
| DM8C10G21210 | Chr. 10: 53,545,170–53,546,332 | 0.08 | 3.84 | 6.62 | MYB | PLN03212 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miao, J.; Yang, M.; Li, N.; Wang, J.; Wang, J.; Zhang, T.; Hu, Z.; Li, Z.; Liu, J.; Li, C.; et al. Combining BSA-Seq, High-Density Genetic Map, and RNA-Seq to Identify Candidate Genes Controlling Embryo Spot Trait in Potato. Horticulturae 2025, 11, 1483. https://doi.org/10.3390/horticulturae11121483
Miao J, Yang M, Li N, Wang J, Wang J, Zhang T, Hu Z, Li Z, Liu J, Li C, et al. Combining BSA-Seq, High-Density Genetic Map, and RNA-Seq to Identify Candidate Genes Controlling Embryo Spot Trait in Potato. Horticulturae. 2025; 11(12):1483. https://doi.org/10.3390/horticulturae11121483
Chicago/Turabian StyleMiao, Jiahao, Min Yang, Nan Li, Jiaji Wang, Jiangqing Wang, Tianzhi Zhang, Zuo Hu, Zhou Li, Jing Liu, Canhui Li, and et al. 2025. "Combining BSA-Seq, High-Density Genetic Map, and RNA-Seq to Identify Candidate Genes Controlling Embryo Spot Trait in Potato" Horticulturae 11, no. 12: 1483. https://doi.org/10.3390/horticulturae11121483
APA StyleMiao, J., Yang, M., Li, N., Wang, J., Wang, J., Zhang, T., Hu, Z., Li, Z., Liu, J., Li, C., & Yang, J. (2025). Combining BSA-Seq, High-Density Genetic Map, and RNA-Seq to Identify Candidate Genes Controlling Embryo Spot Trait in Potato. Horticulturae, 11(12), 1483. https://doi.org/10.3390/horticulturae11121483





