Breeding for Disease Resistance in Cucumber: Current Status, Genetic Insights, and Genomic Resources
Abstract
1. Introduction
2. Genetic Resources
2.1. Germplasm Resources
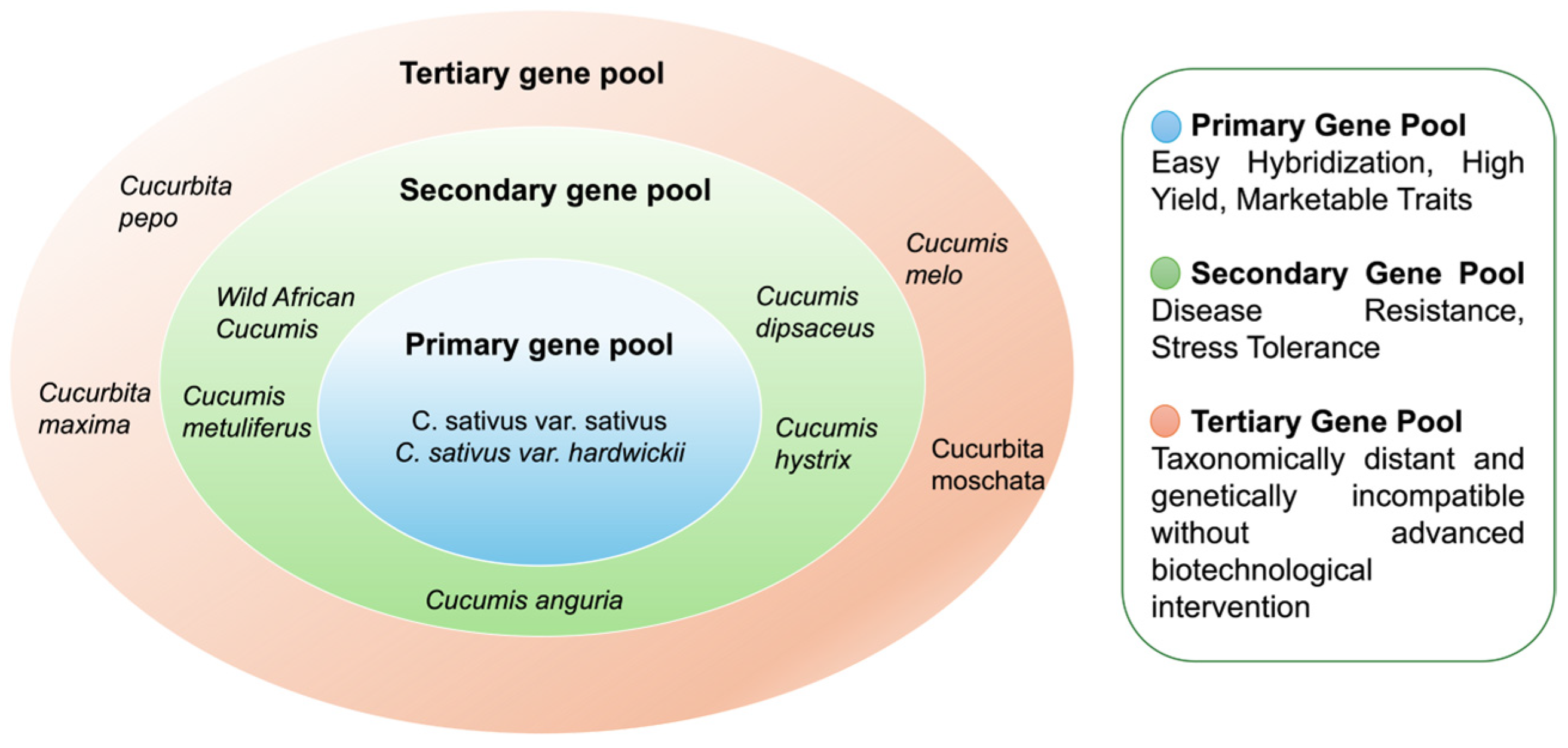
2.2. Cucumber Gene Pool
2.3. Genome Sequencing and Database Resources
3. Host-Pathogen Interaction Mechanisms
4. Major Fungal and Oomycetes Diseases
4.1. Downy Mildew
4.2. Fusarium Wilt
4.3. Scab
4.4. Target Leaf Spot
4.5. Gummy Stem Blight
4.6. Phytophthora Fruit Rot
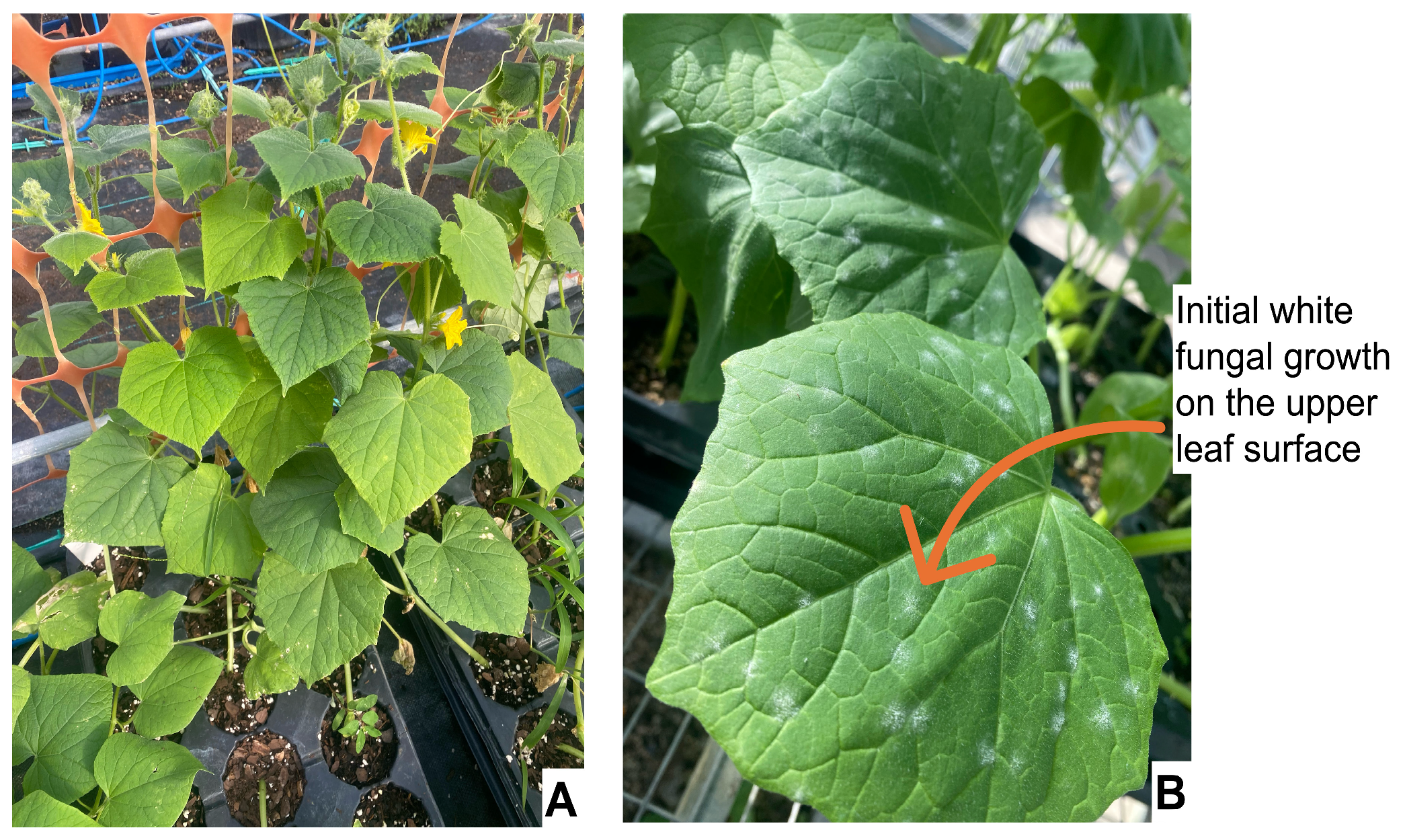
4.7. Powdery Mildew
4.8. Anthracnose
5. Major Viral Diseases
5.1. Cucumber Mosaic Virus (CMV)
5.2. Cucumber Green Mottle Mosaic Virus (CGMMV)
5.3. Watermelon Mosaic Virus (WMV)
5.4. Moroccan Watermelon Mosaic Virus (MWMV) and Zucchini Yellow Fleck Virus (ZYFV)
5.5. Zucchini Yellow Mosaic Virus (ZYMV)
5.6. Papaya Ring Spot Virus (PRSV)
5.7. Cucumber Vein Yellowing Virus (CVYV)
5.8. Melon Yellow Spot Virus (MYSV)
5.9. Cucurbit Yellow Stunting Disorder Virus (CYSDV)
6. Bacterial Disease Resistance
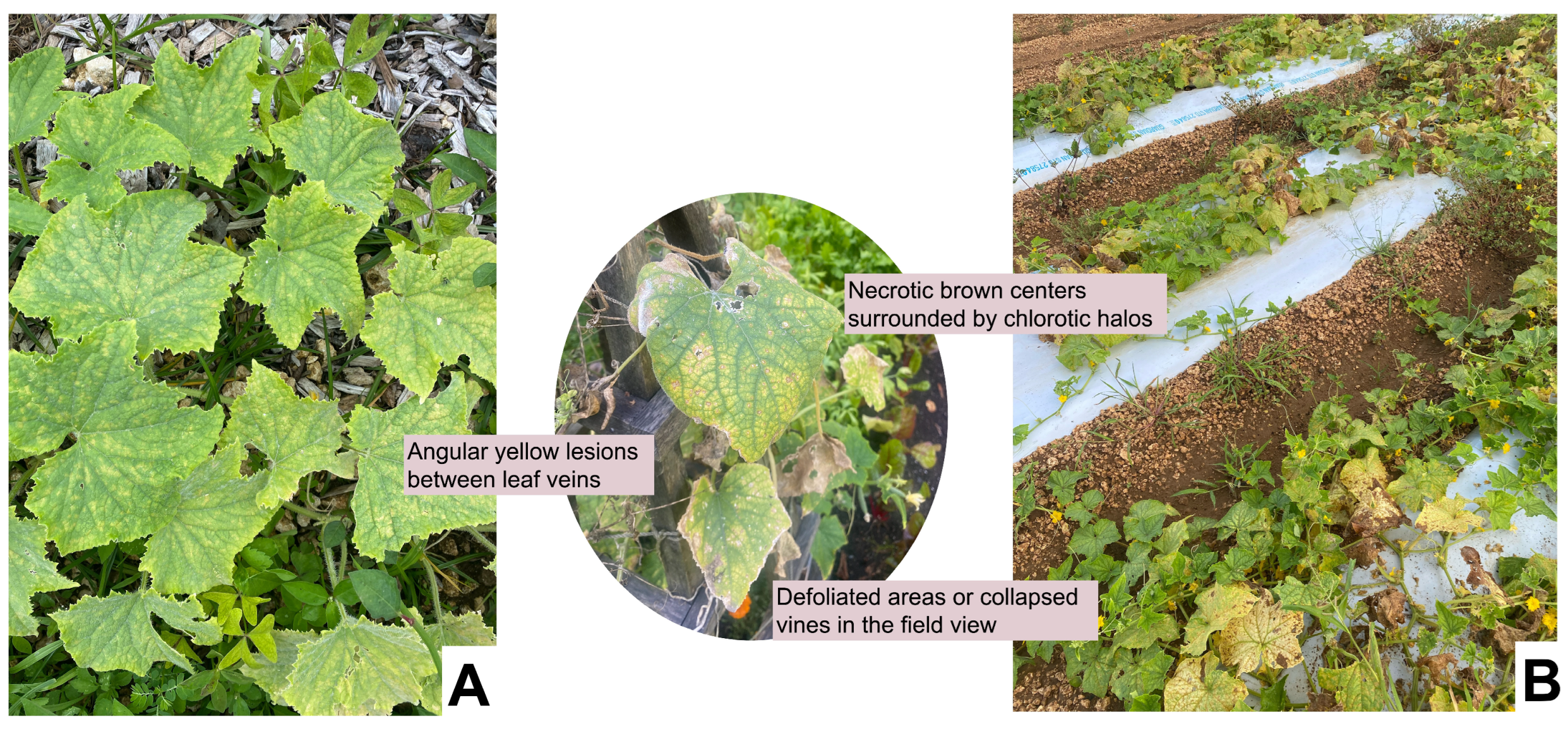
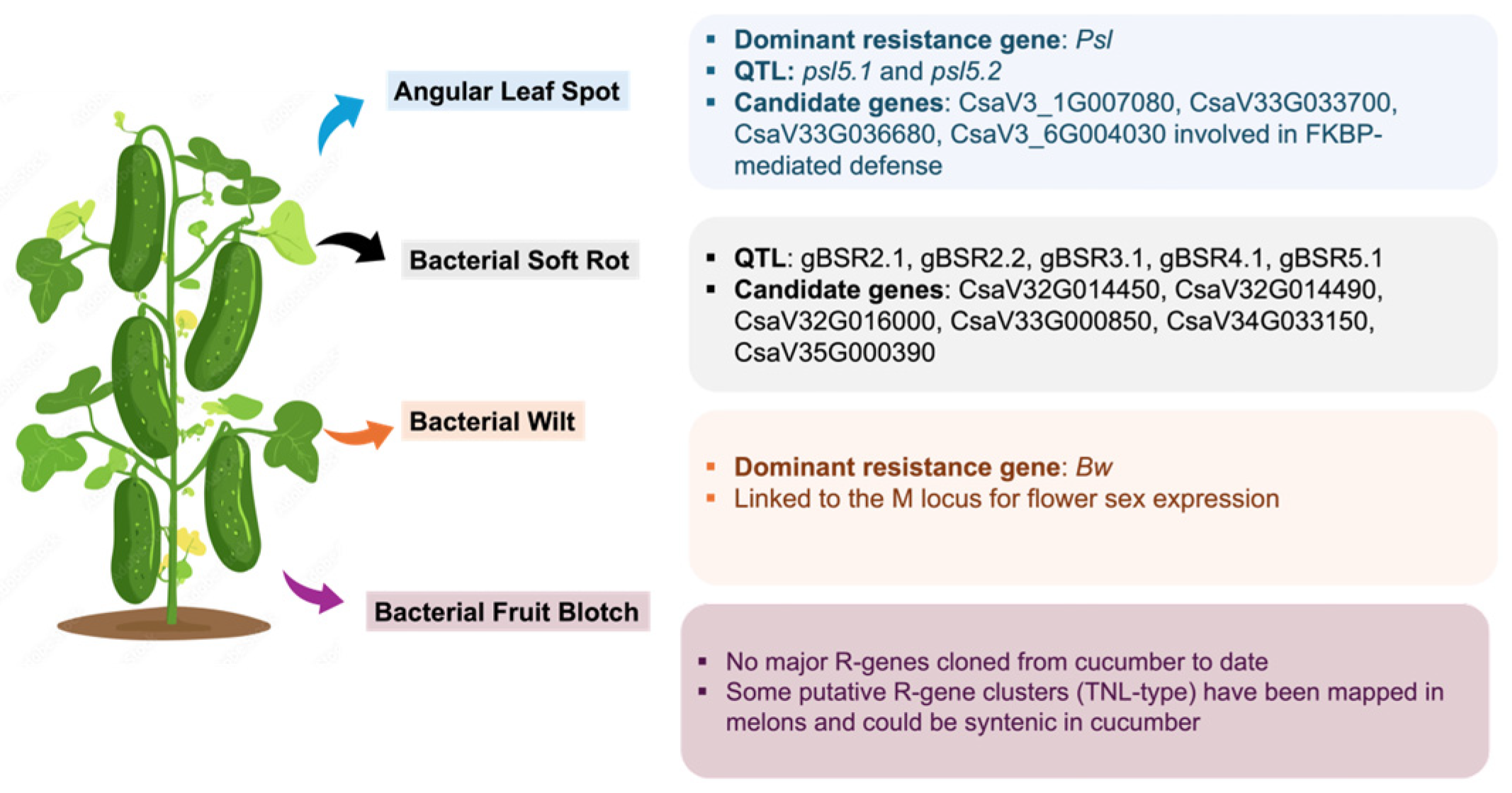
6.1. Angular Leaf Spot
6.2. Bacterial Spot Rot
6.3. Bacterial Wilt and Bacterial Fruit Blotch
7. Future Prospects
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bates, D.M.; Robinson, R.W. Biology and Utilization of the Cucurbitaceae; Cornell University Press: Ithaca, NY, USA, 2019. [Google Scholar]
- Wang, Y.-H.; Joobeur, T.; Dean, R.A.; Staub, J.E. Cucurbits. In Genome Mapping and Molecular Breeding in Plants, Volume 5: Vegetables; Kole, C., Ed.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 315–329. [Google Scholar] [CrossRef]
- Dane, F.; Tsuchiya, T. Chromosome studies in the genus Cucumis. Euphytica 1976, 25, 367–374. [Google Scholar] [CrossRef]
- Whitaker, T.W.; Bemis, W.P. Evolution in the genus Cucurbita. Evolution 1964, 18, 553–559. [Google Scholar] [CrossRef]
- Kirkbride, J.H. Biosystematic Monograph of the Genus Cucumis (Cucurbitaceae): Botanical Identification of Cucumbers and Melons; Parkway Publishers, Inc.: Boone, NC, USA, 1993. [Google Scholar]
- Yang, L.; Koo, D.; Li, Y.; Zhang, X.; Luan, F.; Havey, M.J.; Jiang, J.; Weng, Y. Chromosome rearrangements during domestication of cucumber as revealed by high-density genetic mapping and draft genome assembly. Plant J. 2012, 71, 895–906. [Google Scholar] [CrossRef]
- Pawełkowicz, M.E.; Skarzyńska, A.; Pląder, W.; Przybecki, Z. Genetic and molecular bases of cucumber (Cucumis sativus L.) sex determination. Mol. Breed. 2019, 39, 50. [Google Scholar] [CrossRef]
- Verma, N.; Kumar, R.; Kaur, J.; Thakur, D.S. Floral biology studies in cucumber (Cucumis sativus L.). J. Appl. Hortic. 2021, 23, 371–374. [Google Scholar] [CrossRef]
- Gingras, D.; Gingras, J.; De Oliveira, D. Visits of honeybees (Hymenoptera: Apidae) and their effects on cucumber yields in the field. J. Econ. Entomol. 1999, 92, 435–438. [Google Scholar] [CrossRef]
- Wang, R.; Zeng, J.; Chen, K.; Ding, Q.; Shen, Q.; Wang, M.; Guo, S. Nitrogen improves plant cooling capacity under increased environmental temperature. Plant Soil 2022, 472, 329–344. [Google Scholar] [CrossRef]
- Lu, H.W.; Miao, H.; Tian, G.L.; Wehner, T.C.; Gu, X.F.; Zhang, S.P. Molecular mapping and candidate gene analysis for yellow fruit flesh in cucumber. Mol. Breed. 2015, 35, 64. [Google Scholar] [CrossRef]
- Wang, G.; Qin, Z.; Zhou, X.; Zhao, Z. Genetic analysis and SSR markers of tuberculate trait in Cucumis sativus. Chin. Bull. Bot. 2007, 24, 168. [Google Scholar]
- Kapoor, L.D. CRC Handbook of Ayurvedic Medicinal Plants; CRC Press: Boca Raton, FL, USA, 2018. [Google Scholar] [CrossRef]
- Maggi, F.; Tang, F.H.M.; la Cecilia, D.; McBratney, A. PEST-CHEMGRIDS, global gridded maps of the top 20 crop-specific pesticide application rates from 2015 to 2025. Sci. Data 2019, 6, 170. [Google Scholar] [CrossRef]
- Mordor Intelligence. Cucumber and Gherkins Market Size. Available online: https://www.mordorintelligence.com/industry-reports/cucumber-and-gherkins-market (accessed on 13 October 2025).
- USDA—National Agricultural Statistics Service (NASS). Homepage. Available online: https://www.nass.usda.gov/ (accessed on 13 October 2025).
- Wu, F.; Guan, Z.; Huang, K.-M. The decline of the US cucumber and squash industry: FE1125, 12/2022. EDIS 2022, 2022, 6. [Google Scholar] [CrossRef]
- Savary, S.; Willocquet, L.; Pethybridge, S.J.; Esker, P.; McRoberts, N.; Nelson, A. The global burden of pathogens and pests on major food crops. Nat. Ecol. Evol. 2019, 3, 430–439. [Google Scholar] [CrossRef]
- Bondarenko, S.V.; Stankevych, S.V.; Matsyura, A.V.; Zhukova, L.V.; Zabrodina, I.V.; Rysenko, M.M.; Golovan, L.V.; Romanov, O.V.; Romanova, T.A.; Novosad, K.B.; et al. Major Cucumber Diseases and the Crop Immunity. Ukr. J. Ecol. 2021, 11, 46–54. [Google Scholar] [CrossRef]
- He, Y.; Wei, M.; Yan, Y.; Yu, C.; Cheng, S.; Sun, Y.; Zhu, X.; Wei, L.; Wang, H.; Miao, L. Research advances in genetic mechanisms of major cucumber diseases resistance. Front. Plant Sci. 2022, 13, 862486. [Google Scholar] [CrossRef] [PubMed]
- Arogundade, O.; Ajose, T.; Matthew, J.O.; Osijo, I.A. Current and Emerging Pests and Diseases of Cucumber (Cucumis sativus L.) in Africa. In Cucumber Economic Values and Its Cultivation and Breeding; IntechOpen: London, UK, 2021; p. 179. [Google Scholar]
- Lamichhane, S.; Thapa, S. Advances from conventional to modern plant breeding methodologies. Plant Breed. Biotechnol. 2022, 10, 1–14. [Google Scholar] [CrossRef]
- Razzaq, M.K.; Aleem, M.; Mansoor, S.; Alam Khan, M.; Rauf, S.; Iqbal, S.; Siddique, K.H.M. Omics and CRISPR-Cas9 approaches for molecular insight, functional gene analysis, and stress tolerance development in crops. Int. J. Mol. Sci. 2021, 22, 1292. [Google Scholar] [CrossRef]
- Staub, J.E.; Serquen, F.C.; McCreight, J.D. Genetic diversity in cucumber (Cucumis sativus L.): III. An evaluation of Indian germplasm. Genet. Resour. Crop Evol. 1997, 44, 315–326. [Google Scholar] [CrossRef]
- Meglic, V.; Serquen, F.; Staub, J.E. Genetic diversity in cucumber (Cucumis sativus L.): I. A reevaluation of the U.S. germplasm collection. Genet. Resour. Crop Evol. 1996, 43, 533–546. [Google Scholar] [CrossRef]
- All-Russian Institute of Plant Genetic Resources Named After N.I. Vavilov (VIR). Available online: https://www.vir.nw.ru/ (accessed on 13 October 2025).
- FAO. Forest Genetic Resources No. 22. Available online: https://www.fao.org/4/v3965e/V3965E10.htm (accessed on 13 October 2025).
- USDA-ARS. Germplasm Resources Information Network (GRIN). Available online: https://www.ars-grin.gov/ (accessed on 13 October 2025).
- NIAS [Japan] Genetic Resources—Gene Bank. Available online: https://www.naro.affrc.go.jp/archive/nias/eng/genresources/index.html (accessed on 13 October 2025).
- Crops Research Institute, Guangdong Academy of Agricultural Sciences. Institute Overview. Available online: https://www.gdaas.cn/english/Administration/Research%20Institutes/content/post_987881.html (accessed on 13 October 2025).
- Wehner, T.C.; Robinson, R.W. A brief history of the development of cucumber cultivars in the US. Cucurbit Genet. Coop. Rep. 1991, 14, 2. [Google Scholar]
- Peterson, C.E. Plant introductions in the improvement of vegetable cultivars. HortScience 1975, 10, 575–578. [Google Scholar] [CrossRef]
- Lv, J.; Qi, J.; Shi, Q.; Shen, D.; Zhang, S.; Shao, G.; Li, H.; Sun, Z.; Weng, Y.; Shang, Y.; et al. Genetic diversity and population structure of cucumber (Cucumis sativus L.). PLoS ONE 2012, 7, e46919. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Bao, K.; Reddy, U.K.; Bai, Y.; Hammar, S.A.; Jiao, C.; Wehner, T.C.; Ramírez-Madera, A.O.; Weng, Y.; Grumet, R.; et al. The USDA cucumber (Cucumis sativus L.) collection: Genetic diversity, population structure, genome-wide association studies, and core collection development. Hortic. Res. 2018, 5, 64. [Google Scholar] [CrossRef] [PubMed]
- Bates, D.M.; Merrick, L.C.; Robinson, R.W. (Eds.) Minor cucurbits: Benincasa, Lagenaria, Luffa, Sechium, and Other Genera (Cucurbitaceae). In Evolution of Crop Plants; 1995; Available online: https://worldveg.tind.io/record/47922 (accessed on 13 October 2025).
- van Raamsdonk, L.W.D.; den Nijs, A.P.M.; Jongerius, M.C. Meiotic analyses of Cucumis hybrids and an evolutionary evaluation of the genus Cucumis (Cucurbitaceae). Plant Syst. Evol. 1989, 163, 133–146. [Google Scholar] [CrossRef]
- Suma, A.; Singh, P.K.; Sharma, V.; Pragya; Rana, J.C.; Singh, G.P. Utilization of wild germplasm for vegetable improvement: A review. Curr. Hortic. 2025, 13, 14–25. [Google Scholar]
- Qi, J.; Liu, X.; Shen, D.; Miao, H.; Xie, B.; Li, X.; Zeng, P.; Wang, S.; Shang, Y.; Gu, X.; et al. A genomic variation map provides insights into the genetic basis of cucumber domestication and diversity. Nat. Genet. 2013, 45, 1510–1515. [Google Scholar] [CrossRef]
- Naegele, R.P.; Wehner, T.C. Genetic resources of cucumber. In Genetics and Genomics of Cucurbitaceae; Grumet, R., Katzir, N., Garcia-Mas, J., Eds.; Springer: Cham, Switzerland, 2017; pp. 61–86. [Google Scholar] [CrossRef]
- Chung, S.-M.; Staub, J.E.; Chen, J.-F. Molecular phylogeny of Cucumis species as revealed by consensus chloroplast SSR marker length and sequence variation. Genome 2006, 49, 219–229. [Google Scholar] [CrossRef]
- Staub, J.E.; Chung, S.-M.; Fazio, G. Conformity and genetic relatedness estimation in crop species having a narrow genetic base: The case of cucumber (Cucumis sativus L.). Plant Breed. 2005, 124, 44–53. [Google Scholar] [CrossRef]
- Das, A.; Singh, S.; Islam, Z.; Munshi, A.; Behera, T.; Dutta, S.; Weng, Y.; Dey, S. Current progress in genetic and genomics-aided breeding for stress resistance in cucumber (Cucumis sativus L.). Sci. Hortic. 2022, 300, 111059. [Google Scholar] [CrossRef]
- Sharma, B.D.; Hore, D.K. Indian cucumber germplasm and challenges ahead. Genet. Resour. Crop Evol. 1996, 43, 7–12. [Google Scholar] [CrossRef]
- Walters, S.A.; Wehner, T.C.; Barker, K.R. A single recessive gene for resistance to the root-knot nematode (Meloidogyne javanica) in Cucumis sativus var. hardwickii. J. Hered. 1997, 88, 66–69. [Google Scholar] [CrossRef][Green Version]
- Lebeda, A. Screening of wild Cucumis species against downy mildew (Pseudoperonospora cubensis) isolates from cucumbers. Phytoparasitica 1992, 20, 203–210. [Google Scholar] [CrossRef]
- Chen, J.; Staub, J.E.; Tashiro, Y.; Isshiki, S.; Miyazaki, S. Successful interspecific hybridization between Cucumis sativus L. and C. hystrix Chakr. Euphytica 1997, 96, 413–419. [Google Scholar] [CrossRef]
- Adelberg, J.; Chen, J.; Staub, J.; Skorupska, H.; Rhodes, B. A new synthetic amphidiploid in Cucumis from a C. sativus × C. hystrix F1 interspecific hybrid. In Cucurbitaceae ’98; 1998; Available online: https://open.clemson.edu/ag_pubs/21 (accessed on 13 October 2025).
- Li, H.; Wang, S.; Chai, S.; Yang, Z.; Zhang, Q.; Xin, H.; Xu, Y.; Lin, S.; Chen, X.; Yao, Z.; et al. Graph-based pan-genome reveals structural and sequence variations related to agronomic traits and domestication in cucumber. Nat. Commun. 2022, 13, 682. [Google Scholar] [CrossRef]
- Weng, Y. The cucumber genome. In Genetics and Genomics of Cucurbitaceae; Grumet, R., Katzir, N., Garcia-Mas, J., Eds.; Springer: Cham, Switzerland, 2017; pp. 183–197. [Google Scholar] [CrossRef]
- Shi, L.; Guo, Y.; Dong, C.; Huddleston, J.; Yang, H.; Han, X.; Fu, A.; Li, Q.; Li, N.; Gong, S.; et al. Long-read sequencing and de novo assembly of a Chinese genome. Nat. Commun. 2016, 7, 12065. [Google Scholar] [CrossRef]
- Wóycicki, R.; Witkowicz, J.; Gawroński, P.; Dąbrowska, J.; Lomsadze, A.; Pawełkowicz, M.; Siedlecka, E.; Yagi, K.; Pląder, W.; Seroczyńska, A.; et al. The genome sequence of the North-European cucumber (Cucumis sativus L.) unravels evolutionary adaptation mechanisms in plants. PLoS ONE 2011, 6, e22728. [Google Scholar] [CrossRef]
- Cavagnaro, P.F.; Senalik, D.A.; Yang, L.; Simon, P.W.; Harkins, T.T.; Kodira, C.D.; Huang, S.; Weng, Y. Genome-wide characterization of simple sequence repeats in cucumber (Cucumis sativus L.). BMC Genom. 2010, 11, 569. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.; Zhang, Z.; Lou, Q.; Xia, L.; Li, J.; Li, M.; Zhou, J.; Zhao, X.; Xu, Y.; Li, Q.; et al. Chromosome-scale genome assembly of Cucumis hystrix—A wild species interspecifically cross-compatible with cultivated cucumber. Hortic. Res. 2021, 8, 40. [Google Scholar] [CrossRef] [PubMed]
- Shendure, J.; Mitra, R.D.; Varma, C.; Church, G.M. Advanced sequencing technologies: Methods and goals. Nat. Rev. Genet. 2004, 5, 335–344. [Google Scholar] [CrossRef]
- Turek, S.; Pląder, W.; Hoshi, Y.; Skarzyńska, A.; Pawełkowicz, M. Insight into the organization of the B10v3 cucumber genome by integration of biological and bioinformatic data. Int. J. Mol. Sci. 2023, 24, 4011. [Google Scholar] [CrossRef]
- Seiko, T.; Muto, C.; Shimomura, K.; Yano, R.; Kawazu, Y.; Sugiyama, M.; Kato, K.; Tomooka, N.; Naito, K. Chromosome-level assembly of Cucumis sativus cv. ‘Tokiwa’ as a reference genome of Japanese cucumber. Breed. Sci. 2025, 75, 85–92. [Google Scholar] [CrossRef] [PubMed]
- Guan, J.; Miao, H.; Zhang, Z.; Dong, S.; Zhou, Q.; Liu, X.; Beckles, D.M.; Gu, X.; Huang, S.; Zhang, S. A near-complete cucumber reference genome assembly and Cucumber-DB, a multi-omics database. Mol. Plant 2024, 17, 1178–1182. [Google Scholar] [CrossRef]
- Yu, J.; Wu, S.; Sun, H.; Wang, X.; Tang, X.; Guo, S.; Zhang, Z.; Huang, S.; Xu, Y.; Weng, Y.; et al. CuGenDBv2: An updated database for cucurbit genomics. Nucleic Acids Res. 2023, 51, D1457–D1464. [Google Scholar] [CrossRef]
- Li, Q.; Li, H.; Huang, W.; Xu, Y.; Zhou, Q.; Wang, S.; Ruan, J.; Huang, S.; Zhang, Z. A chromosome-scale genome assembly of cucumber (Cucumis sativus L.). GigaScience 2019, 8, giz072. [Google Scholar] [CrossRef]
- O’Leary, N.A.; Cox, E.; Holmes, J.B.; Anderson, W.R.; Falk, R.; Hem, V.; Tsuchiya, M.T.N.; Schuler, G.D.; Zhang, X.; Torcivia, J.; et al. Exploring and retrieving sequence and metadata for species across the tree of life with NCBI Datasets. Sci. Data 2024, 11, 732. [Google Scholar] [CrossRef] [PubMed]
- Nordberg, H.; Cantor, M.; Dusheyko, S.; Hua, S.; Poliakov, A.; Shabalov, I.; Smirnova, T.; Grigoriev, I.V.; Dubchak, I. The genome portal of the Department of Energy Joint Genome Institute: 2014 updates. Nucleic Acids Res. 2014, 42, D26–D31. [Google Scholar] [CrossRef] [PubMed]
- Horst, R.K. Westcott’s Plant Disease Handbook; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Nazarov, P.A.; Baleev, D.N.; Ivanova, M.I.; Sokolova, L.M.; Karakozova, M.V. Infectious plant diseases: Etiology, current status, problems and prospects in plant protection. Acta Nat. 2020, 12, 46–59. [Google Scholar] [CrossRef]
- Dickman, M.B.; Fluhr, R. Centrality of Host Cell Death in Plant–Microbe Interactions. Annu. Rev. Phytopathol. 2013, 51, 543–570. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Khan, I.U.; Anarjan, M.B.; Hussain, M.; Lee, S. The Mutant STAY-GREEN (Cssgr) in Cucumber Interacts with the CSEP30 Protein to Elicit a Defense Response against Podosphaera xanthii. Mol. Breed. 2024, 44, 67. [Google Scholar] [CrossRef]
- Dey, N.; Roy, U.K.; Aditya, M.; Bhattacharjee, S. Defensive Strategies of ROS in Programmed Cell Death Associated with the Hypersensitive Response in Plant Pathogenesis. Ann. Syst. Biol. 2020, 3, 1–9. [Google Scholar]
- Binyamin, R.; Nadeem, S.M.; Akhtar, S.; Khan, M.Y.; Anjum, R. Beneficial and Pathogenic Plant–Microbe Interactions: A Review. Soil Environ. 2019, 38, 192–202. [Google Scholar] [CrossRef]
- Vela-Corcia, D.; Bautista, R.; de Vicente, A.; Spanu, P.D.; Pérez-García, A. De Novo Analysis of the Epiphytic Transcriptome of the Cucurbit Powdery Mildew Fungus Podosphaera xanthii and Identification of Candidate Secreted Effector Proteins. PLoS ONE 2016, 11, e0163379. [Google Scholar] [CrossRef]
- Mur, L.A.J.; Aubry, S.; Mondhe, M.; Kingston-Smith, A.; Gallagher, J.; Timms-Taravella, E.; James, C.; Papp, I.; Hörtensteiner, S.; Thomas, H.; et al. Accumulation of Chlorophyll Catabolites Photosensitizes the Hypersensitive Response Elicited by Pseudomonas syringae in Arabidopsis. New Phytol. 2010, 188, 161–174. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tan, J.; Wu, Z.; VandenLangenberg, K.; Wehner, T.C.; Wen, C.; Zheng, X.; Owens, K.; Thornton, A.; Bang, H.H. STAYGREEN, STAY HEALTHY: A loss-of-susceptibility mutation in the STAYGREEN gene provides durable, broad-spectrum disease resistances for over 50 years of US cucumber production. New Phytol. 2019, 221, 415–430. [Google Scholar] [CrossRef] [PubMed]
- Keinath, A.P.; Duthie, J.A. Yield and quality reductions in watermelon due to anthracnose, gummy stem blight, and black rot. HortScience 1998, 36, 219–220. [Google Scholar]
- Sitterly, W.R. Breeding for disease resistance in cucurbits. Annu. Rev. Phytopathol. 1972, 10, 471–490. [Google Scholar] [CrossRef]
- Palti, J.; Cohen, Y. Downy mildew of cucurbits (Pseudoperonospora cubensis): The fungus and its hosts, distribution, epidemiology and control. Phytoparasitica 1980, 8, 109–147. [Google Scholar] [CrossRef]
- Forsberg, A.S. Downy mildew—Pseudoperonospora cubensis in Swedish cucumber fields in 1985. Växtskyddsnotiser 1986, 50, 17–19. [Google Scholar]
- Lebeda, A.; Widrlechner, M.P. A set of Cucurbitaceae taxa for differentiation of Pseudoperonospora cubensis pathotypes. J. Plant Dis. Prot. 2003, 110, 337–349. [Google Scholar]
- Lebeda, A.; Schwinn, F.J. The downy mildews—An overview of recent research progress/Falscher Mehltau—Übersicht über neuere Forschungsresultate. Z. Pflanzenkrankh. Pflanzenschutz/J. Plant Dis. Prot. 1994, 101, 225–254. [Google Scholar]
- Thomas, C.E. Downy mildew. In Compendium of Cucurbit Diseases; Zitter, T.A., Ed.; Cornell University Press: Ithaca, NY, USA, 1996; pp. 25–27. [Google Scholar]
- Oerke, E.C.; Steiner, U.; Dehne, H.W.; Lindenthal, M. Thermal imaging of cucumber leaves affected by downy mildew and environmental conditions. J. Exp. Bot. 2006, 57, 2121–2132. [Google Scholar] [CrossRef] [PubMed]
- Kanetis, L.; Holmes, G.J.; Ojiambo, P.S. Survival of Pseudoperonospora cubensis sporangia exposed to solar radiation. Plant Pathol. 2010, 59, 313–323. [Google Scholar] [CrossRef]
- Hahn, M.; Mendgen, K. Signal and nutrient exchange at biotrophic plant–fungus interfaces. Curr. Opin. Plant Biol. 2001, 4, 322–327. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Whisson, S.C.; Boevink, P.C.; Moleleki, L.; Avrova, A.O.; Morales, J.G.; Gilroy, E.M.; Armstrong, M.R.; Grouffaud, S.; van West, P.; Chapman, S.; et al. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 2007, 450, 115–118. [Google Scholar] [CrossRef] [PubMed]
- Fraymouth, J. Haustoria of the Peronosporales. Trans. Br. Mycol. Soc. 1956, 39, 79–107. [Google Scholar] [CrossRef]
- Voglmayr, H.; Riethmüller, A.; Göker, M.; Weiss, M.; Oberwinkler, F. Phylogenetic relationships of Plasmopara, Bremia and other genera of downy mildew pathogens with pyriform haustoria based on Bayesian analysis of partial LSU rDNA sequence data. Mycol. Res. 2004, 108, 1011–1024. [Google Scholar] [CrossRef]
- Heaney, S.P.; Hall, A.A.; Davis, S.A.; Olaya, G. Resistance to fungicides in the QoI-STAR cross-resistance group: Current perspectives. In Brighton Crop Protection Conference—Pests and Diseases; British Crop Protection Council: Brighton, UK, 2000; pp. 755–762. [Google Scholar]
- Jenkins, J.M., Jr. Studies on the inheritance of downy mildew resistance and of other characters in cucumbers. J. Hered. 1946, 37, 267–271. [Google Scholar] [CrossRef]
- Barnes, W.C. The performance of ‘Palmetto’, a new downy mildew resistant cucumber variety. Proc. Am. Soc. Hortic. Sci. 1948, 51, 437–441. [Google Scholar]
- Barnes, W.C.; Epps, W.M. An unreported type of resistance to cucumber downy mildew. Plant Dis. Rep. 1954, 38, 620. [Google Scholar]
- Shimizu, S.; Kanazawa, K.; Kato, A.; Yokota, Y.; Koyama, T. Studies on the breeding of cucumber for resistance to downy mildew and other fruit characters. Engei Shikenjo Hōkoku 1963, 2, 65–81. [Google Scholar]
- van Vliet, G.J.A.; Meysing, W.D. Inheritance of resistance to Pseudoperonospora cubensis Rost. in cucumber (Cucumis sativus L.). Euphytica 1974, 23, 251–255. [Google Scholar] [CrossRef]
- Wang, M.; Liu, W.; Jiang, B.; Peng, Q.; He, X.; Liang, Z.; Lin, Y. Genetic analysis and related gene primary mapping of heat stress tolerance in cucumber using bulked segregant analysis. HortScience 2019, 54, 423–428. [Google Scholar] [CrossRef]
- Doruchowski, R.W.; Lakowska-Ryk, E. Inheritance of resistance to downy mildew (Pseudoperonospora cubensis Berk. & Curt) in Cucumis sativus. In Proceedings of the 5th EUCARPIA Cucurbitaceae Symposium, Skierniewice, Poland, 27–31 July 1992; pp. 66–69. [Google Scholar]
- El-Hafez, A.A.; Shehata, S.A.; El-Din, S.A.B.; El-Doweny, H.H.; Awad, M.M.W. Inheritance of downy mildew disease and its nature of resistance in cucumber. Egypt. J. Appl. Sci. 1990, 5, 1681–1697. [Google Scholar]
- St. Amand, P.C.; Wehner, T.C. Crop loss to 14 diseases in cucumber in North Carolina for 1983 to 1988. Plant Dis. 1991, 75, 15–17. [Google Scholar]
- Call, A.D.; Criswell, A.D.; Wehner, T.C.; Klosinska, U.; Kozik, E.U. Screening cucumber for resistance to downy mildew caused by Pseudoperonospora cubensis (Berk. & Curt.) Rostov. Crop Sci. 2012, 52, 577–592. [Google Scholar] [CrossRef]
- Bommesh, J.C.; Pitchaimuthu, M.; Manjunathagowda, D.C.; Maragal, S.; Ramesh, A.N. Unveiling the downy mildew disease (Pseudoperonospora cubensis Berk. & Curt.) resistance response in cucumber (Cucumis sativus L.). Indian Phytopathol. 2025, 78, 97–104. [Google Scholar] [CrossRef]
- Wang, Y.; VandenLangenberg, K.; Wehner, T.C.; Kraan, P.A.G.; Suelmann, J.; Zheng, X.; Owens, K.; Weng, Y. QTL mapping for downy mildew resistance in cucumber inbred line WI7120 (PI 330628). Theor. Appl. Genet. 2016, 129, 1493–1505. [Google Scholar] [CrossRef]
- Win, K.T.; Vegas, J.; Zhang, C.; Song, K.; Lee, S. QTL mapping for downy mildew resistance in cucumber via bulked segregant analysis using next-generation sequencing and conventional methods. Theor. Appl. Genet. 2017, 130, 199–211. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; He, H.; Zou, Z.; Li, Y. QTL analysis for downy mildew resistance in cucumber inbred line PI 197088. Plant Dis. 2018, 102, 1240–1245. [Google Scholar] [CrossRef] [PubMed]
- Berg, J.A.; Hermans, F.W.K.; Beenders, F.; Lou, L.; Vriezen, W.H.; Visser, R.G.F.; Bai, Y.; Schouten, H.J. Analysis of QTL DM4.1 for downy mildew resistance in cucumber reveals multiple subQTL: A novel RLK as candidate gene for the most important subQTL. Front. Plant Sci. 2020, 11, 569876. [Google Scholar] [CrossRef]
- Bansuli, A.; Sharma, A.; Rana, R.S.; Lata, H.; Thakur, A.; Sharma, A. Mapping quantitative trait loci (QTLs) for resistance to downy mildew and powdery mildew in cucumber (Cucumis sativus L.). J. Plant Biochem. Biotechnol. 2025, 34, 1–15. [Google Scholar] [CrossRef]
- Thakur, S.; Kaur, S.; Adhikari, S.; Sabharwal, P.; Fu, Y.; Meru, G. Turning susceptibility into strength: A new era of durable resistance in plants through genome editing. Plants 2025, 14, 3080. [Google Scholar] [CrossRef]
- Berg, J.A.; Hermans, F.W.K.; Beenders, F.; Abedinpour, H.; Vriezen, W.H.; Visser, R.G.F.; Bai, Y.; Schouten, H.J. The amino acid permease (AAP) genes CsAAP2A and SlAAP5A/B are required for oomycete susceptibility in cucumber and tomato. Mol. Plant Pathol. 2021, 22, 658–672. [Google Scholar] [CrossRef]
- Anarjan, M.B.; Zhang, C.; Begum, S.; Win, K.T.; Li, H.; Wang, C.; Wu, T.; Lee, S. Genetic basis of downy mildew resistance in cucumber: Identification of candidate genes and ethylene-mediated defense mechanism. Res. Sq. 2025. preprint. [Google Scholar] [CrossRef]
- Liu, X.; Lu, H.; Liu, P.; Miao, H.; Bai, Y.; Gu, X.; Zhang, S. Identification of novel loci and candidate genes for cucumber downy mildew resistance using GWAS. Plants 2020, 9, 1659. [Google Scholar] [CrossRef]
- Gao, X.; Guo, P.; Wang, Z.; Chen, C.; Ren, Z. Transcriptome profiling reveals response genes for downy mildew resistance in cucumber. Planta 2021, 253, 112. [Google Scholar] [CrossRef]
- Zhang, P.; Zhu, Y.; Luo, X.; Zhou, S. Comparative proteomic analysis provides insights into the complex responses to Pseudoperonospora cubensis infection of cucumber (Cucumis sativus L.). Sci. Rep. 2019, 9, 9433. [Google Scholar] [CrossRef]
- Wang, Z.; Chang, J.; Han, J.; Yin, M.; Wang, X.; Ren, Z.; Wang, L. Genome-wide reidentification and transcriptome analysis of MADS-box gene family in cucumber. Int. J. Mol. Sci. 2025, 26, 3800. [Google Scholar] [CrossRef]
- Sun, C.; Song, X.; Zheng, J.; Li, X.; Feng, Z.; Yan, L. Comparative proteomic profiles of resistant/susceptible cucumber leaves in response to downy mildew infection. Hortic. Plant J. 2021, 7, 327–340. [Google Scholar] [CrossRef]
- Lai, W.; Zhu, C.; Hu, Z.; Liu, S.; Wu, H.; Zhou, Y. Identification and transcriptional analysis of zinc finger-homeodomain (ZF-HD) family genes in cucumber. Biochem. Genet. 2021, 59, 884–901. [Google Scholar] [CrossRef] [PubMed]
- Yan, S.; Ning, K.; Wang, Z.; Liu, X.; Zhong, Y.; Ding, L.; Zi, H.; Cheng, Z.; Li, X.; Shan, H.; et al. CsIVP functions in vasculature development and downy mildew resistance in cucumber. PLoS Biol. 2020, 18, e3000671. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Li, C.; Tian, H.; Wang, W.; Yang, X.; Beckles, D.M.; Liu, X.; Guan, J.; Gu, X.; Sun, J.; et al. Natural variation in STAYGREEN contributes to low-temperature tolerance in cucumber. J. Integr. Plant Biol. 2023, 65, 2552–2568. [Google Scholar] [CrossRef]
- Wang, H.; Chen, Q.; Feng, W. The emerging role of 2OGDs as candidate targets for engineering crops with broad-spectrum disease resistance. Plants 2024, 13, 1129. [Google Scholar] [CrossRef]
- Tan, J. Genetic Mapping of Downy and Powdery Mildew Resistances and Characterization of CsSGR-Dependent Broad-Spectrum Disease Resistance in Cucumber (Cucumis sativus L.). Ph.D. Thesis, University of Wisconsin, Madison, WI, USA, 2022. [Google Scholar]
- Hegde, R.K.; Bhide, V.P.; Sukapure, R.S.R.S. Fusarium wilt of watermelon and peas in Maharashtra state. Curr. Sci. 1955, 24, 310. [Google Scholar]
- Bhaskaran, R.; Prasad, N.N.; Kothandaraman, R. Occurrence of Fusarium wilt on muskmelon. Madras Agric. J. 1971, 58, 171. [Google Scholar]
- Sarkar, T.; Sengupta, S. (Eds.) Cultivating Progress: New Frontiers in Agricultural Science and Technology, 1st ed.; AkiNik Publications: New Delhi, India, 2025. [Google Scholar] [CrossRef]
- Owen, J.H. Fusarium wilt of cucumber. Phytopathology 1955, 45, 435–439. [Google Scholar]
- Jenkins, S.F., Jr.; Wehner, T.C. Occurrence of Fusarium oxysporum f. sp. cucumerinum on greenhouse-grown Cucumis sativus seed stocks in North Carolina. Plant Dis. 1983, 67, 1024–1025. [Google Scholar] [CrossRef]
- Huang, Z.X.; Zhao, J.H.; Liu, Z.L. Observation on spore germination and growth of cucumber wilt disease fungus and ultrastructural study on fungal colonization in host tissue. Acta Phytopathol. Sin. 1988, 18, 29–36. [Google Scholar]
- Vakalounakis, D.J. Root and stem rot of cucumber caused by Fusarium oxysporum f. sp. radicis-cucumerinum. Plant Dis. 1996, 80, 313–316. [Google Scholar] [CrossRef]
- Liu, D.L.; Yang, R.H.; Ha, Y.H. Research on genetic characteristics of cucumbers resisting Fusarium wilt. Tianjin Agric. Sci. 2003, 9, 33–35. [Google Scholar]
- Wang, Y.J. Studies on Molecular Marker of Fusarium Wilt Resistance Related Gene in Cucumber (Cucumis sativus L.). Master’s Thesis, Northwest A&F University, Yangling, China, 2005; pp. 1–55. [Google Scholar]
- Vakalounakis, D.J. Inheritance and linkage of resistance in cucumber line SMR-18 to races 1 and 2 of Fusarium oxysporum f. sp. cucumerinum. Eur. J. Plant Pathol. 1995, 101, 169–172. [Google Scholar]
- Zhang, S.; Miao, H.; Yang, Y.; Xie, B.; Wang, Y.; Gu, X. A major quantitative trait locus conferring resistance to Fusarium wilt was detected in cucumber by using recombinant inbred lines. Mol. Breed. 2014, 34, 1805–1815. [Google Scholar] [CrossRef]
- Dong, J.; Xu, J.; Xu, X.; Xu, Q.; Chen, X. Inheritance and quantitative trait locus mapping of Fusarium wilt resistance in cucumber. Front. Plant Sci. 2019, 10, 1425. [Google Scholar] [CrossRef] [PubMed]
- Mao, A.; Zhang, Z.; Zhang, L. Analysis on the inheritance of resistance to Fusarium wilt race 4 and cucumber scab and their linkage in cucumber WLS2757. Zhongguo Nongye Kexue 2008, 41, 3382–3388. [Google Scholar] [CrossRef]
- Jaber, E.H.A.; Srour, A.Y.; Zambounis, A.G.; Vakalounakis, D.J.; Doulis, A.G. Identification of SCAR markers linked to the Foc gene governing resistance to Fusarium oxysporum f. sp. cucumerinum in cucumber cv. SMR-18. Eur. J. Plant Pathol. 2020, 157, 845–855. [Google Scholar] [CrossRef]
- Zhang, D.; Meng, K.X.; Hao, Y.H.; Fan, H.Y.; Cui, N.; Wang, S.S.; Song, T.F. Comparative proteomic analysis of cucumber roots infected by Fusarium oxysporum f. sp. cucumerinum Owen. Physiol. Mol. Plant Pathol. 2016, 96, 77–84. [Google Scholar] [CrossRef]
- Du, N.; Shi, L.; Yuan, Y.; Li, B.; Shu, S.; Sun, J.; Guo, S. Proteomic analysis reveals the positive roles of the plant-growth-promoting rhizobacterium NSY50 in the response of cucumber roots to Fusarium oxysporum f. sp. cucumerinum inoculation. Front. Plant Sci. 2016, 7, 1859. [Google Scholar] [CrossRef]
- Bartholomew, E.S.; Black, K.; Feng, Z.; Liu, W.; Shan, N.; Zhang, X.; Wu, L.; Bailey, L.; Zhu, N.; Qi, C.; et al. Comprehensive analysis of the chitinase gene family in cucumber (Cucumis sativus L.): From gene identification and evolution to expression in response to Fusarium oxysporum. Int. J. Mol. Sci. 2019, 20, 5309. [Google Scholar] [CrossRef]
- Zhou, X.; Wu, F. Differentially expressed transcripts from cucumber (Cucumis sativus L.) root upon inoculation with Fusarium oxysporum f. sp. cucumerinum Owen. Physiol. Mol. Plant Pathol. 2009, 74, 142–150. [Google Scholar] [CrossRef]
- Dong, J.; Wang, Y.; Xian, Q.; Chen, X.; Xu, J. Transcriptome analysis reveals ethylene-mediated defense responses to Fusarium oxysporum f. sp. cucumerinum infection in Cucumis sativus L. BMC Plant Biol. 2020, 20, 334. [Google Scholar] [CrossRef]
- Xu, J.; Wang, K.; Xian, Q.; Zhang, N.; Dong, J.; Chen, X. Identification of susceptibility genes for Fusarium oxysporum in cucumber via comparative proteomic analysis. Genes 2021, 12, 1781. [Google Scholar] [CrossRef]
- Bartholomew, E.S.; Xu, S.; Zhang, Y.; Yin, S.; Feng, Z.; Chen, S.; Sun, L.; Yang, S.; Wang, Y.; Liu, P.; et al. A chitinase CsChi23 promoter polymorphism underlies cucumber resistance against Fusarium oxysporum f. sp. cucumerinum. New Phytol. 2022, 236, 1471–1486. [Google Scholar] [CrossRef]
- Arthur, J.C. Spotting of Peaches and Cucumbers; Bulletin No. 19; Purdue University Agricultural Experiment Station: Lafayette, IN, USA, 1889. [Google Scholar]
- Walker, J.C.; Pierson, C.F.; Wiles, A.B. Two new scab-resistant cucumber varieties. Phytopathology 1953, 43, 215–217. [Google Scholar]
- Park, Y.H.; Havey, M.J. Cucumber germplasm resistant to Cladosporium cucumerinum. HortScience 2004, 39, 163–164. [Google Scholar] [CrossRef]
- Andeweg, J.M. The breeding of scab-resistant frame cucumbers in the Netherlands. Euphytica 1956, 5, 185–195. [Google Scholar] [CrossRef]
- Bailey, R.M.; Burgess, I.M. Breeding cucumbers resistant to scab. Proc. Am. Soc. Hortic. Sci. 1934, 32, 474–476. [Google Scholar]
- Bradeen, J.M.; Staub, J.E.; Wye, C.; Antonise, R.; Peleman, J. Towards an expanded and integrated linkage map of cucumber (Cucumis sativus L.). Genome 2001, 44, 111–119. [Google Scholar] [CrossRef]
- Zhang, G.H.; Han, Y.K.; Sun, X.H. Molecular marker linked to the resistant gene of cucumber scab. Sci. Agric. Sin. 2006, 39, 2250–2254. [Google Scholar]
- Sun, X.H.; Du, S.L.; Zhang, G.H. Polymorphism comparison of AFLP and SSR between scab-resistant cucumber materials. Acta Agric. Boreali-Sin. 2006, 21, 105–107. [Google Scholar]
- Zhang, S.P.; Miao, H.; Gu, X.F.; Yang, Y.; Xie, B.; Wang, X.; Huang, S.; Du, Y.; Sun, R.; Wehner, T.C. Genetic mapping of the scab resistance gene in cucumber. J. Am. Soc. Hortic. Sci. 2010, 135, 53–58. [Google Scholar] [CrossRef]
- Kang, H.X.; Weng, Y.Q.; Yang, Y.H.; Zhang, Z.; Zhang, S.; Mao, Z.; Cheng, G.; Gu, X.; Huang, S.; Xie, B. Fine genetic mapping localizes cucumber scab resistance gene Ccu into an R-gene cluster. Theor. Appl. Genet. 2011, 122, 795–803. [Google Scholar] [CrossRef]
- Zhou, Q.; Miao, H.; Li, S.; Zhang, S.; Wang, Y.; Weng, Y.; Zhang, Z.; Huang, S.; Gu, X. A sequencing-based linkage map of cucumber. Mol. Plant 2015, 8, 961–963. [Google Scholar] [CrossRef]
- Vakalounakis, D. Inheritance and genetic linkage of Fusarium wilt (race 1) and scab resistance genes in cucumber (Cucumis sativus). Ann. Appl. Biol. 1993, 122, 359–365. [Google Scholar] [CrossRef]
- Li, Q.-H.; Shen, D.; Li, X.-X.; Wang, H.-P.; Qiu, Y.; Song, J.-P. Genetic analysis of resistance to Cladosporium cucumerinum from different gene sources in cucumber. J. Plant Genet. Resour. 2011, 12, 291–296. [Google Scholar]
- Berkeley, M.J.; Curtis, M.A. Fungi cubenses (Hymenomycetes). Linn. Soc. Trans. 1868, 10, 280–320. [Google Scholar] [CrossRef]
- Yang, S.J.; Gu, X.F.; Zhang, S.P.; Miao, H.; Li, B.J. Research progress on cucumber target leaf spot (Corynespora cassiicola). China Veg. 2012, 5, 1–9. [Google Scholar]
- Pavan, M.A.; Rezende, J.A.M.; Krause-Sakate, R. Doenças das cucurbitáceas. In Manual de Fitopatologia, 4th ed.; Ceres: São Paulo, Brazil, 2016; Volume 2. [Google Scholar]
- Abul-Hayja, Z.; Williams, P.H.; Peterson, C.E. Inheritance of resistance to anthracnose and target leaf spot in cucumbers. Plant Dis. Rep. 1978, 62, 43–45. [Google Scholar]
- Wang, H.; Li, S.; Guan, W. Identification of sources of resistance to cucumber target leaf spot and genetic analysis of resistance. Chin. Cucurbits Veget. 2010, 23, 24–25. [Google Scholar] [CrossRef]
- Fu, H.P.; Wei, J.; Li, S.J.; Yang, R.H.; Guan, W.; Wang, H.Z. EST-SSR markers and artificial inoculation identification of leaf spot resistance in cucumber germplasm. Chin. Hortic. Abstract 2012, 2, 1–3. [Google Scholar]
- Wen, C.; Mao, A.; Dong, C.; Liu, H.; Yu, S.; Guo, Y.-D.; Weng, Y.; Xu, Y. Fine genetic mapping of target leaf spot resistance gene cca-3 in cucumber, Cucumis sativus L. Theor. Appl. Genet. 2015, 128, 2495–2506. [Google Scholar] [CrossRef]
- Xu, Y.; Dong, S.; Xie, X.; Beckles, D.M.; Liu, X.; Guan, J.; Li, C.; Gu, X.; Miao, H.; Zhang, S. A genome-wide association study identifies candidate genes for target leaf spot disease resistance in adult cucumber (Cucumis sativus L.). Front. Plant Sci. 2025, 16, 1542274. [Google Scholar] [CrossRef]
- Liu, D.; Xin, M.; Zhou, X.; Wang, C.; Zhang, Y.; Qin, Z. Expression and functional analysis of the transcription factor-encoding gene CsERF004 in cucumber during Pseudoperonospora cubensis and Corynespora cassiicola infection. BMC Plant Biol. 2017, 17, 96. [Google Scholar] [CrossRef]
- Bureau of Plant Industry, U.S. Department of Agriculture. The Plant Disease Reporter; Bureau of Plant Industry, U.S. Department of Agriculture: Washington, DC, USA, 1978.
- Chiu, W.F. Physiology and pathogenicity of the cucurbit black-rot fungus. J. Agric. Res. 1949, 78, 589–615. [Google Scholar]
- Chester, F.D. Notes on three new or noteworthy diseases of plants. Bull. Torrey Bot. Club 1891, 18, 371–374. [Google Scholar] [CrossRef]
- Keinath, A.P. Diagnostic guide for gummy stem blight and black rot on cucurbits. Plant Health Prog. 2013, 14, 35. [Google Scholar] [CrossRef]
- Choi, I.Y.; Choi, J.N.; Choi, D.C.; Sharma, P.K.; Lee, W.H. Identification and characterization of the causal organism of gummy stem blight in the muskmelon (Cucumis melo L.). Mycobiology 2010, 38, 166–170. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Keinath, A.P. Soil amendment with cabbage residue and crop rotation to reduce gummy stem blight and increase growth and yield of watermelon. Plant Dis. 1996, 80, 564–570. [Google Scholar] [CrossRef]
- Wyszogrodzka, A.J.; Williams, P.H.; Peterson, C.E. Search for resistance to gummy stem blight (Didymella bryoniae) in cucumber (Cucumis sativus L.). Euphytica 1986, 35, 603–613. [Google Scholar] [CrossRef]
- Wehner, T.C.; Shetty, N.V. Screening the cucumber germplasm collection for resistance to gummy stem blight in North Carolina field tests. HortScience 2000, 35, 1132–1140. [Google Scholar] [CrossRef]
- Lou, L.; Wang, H.; Qian, C.; Liu, J.; Bai, Y.; Chen, J. Genetic mapping of gummy stem blight (Didymella bryoniae) resistance genes in Cucumis sativus–hystrix introgression lines. Euphytica 2013, 192, 359–369. [Google Scholar] [CrossRef]
- Liu, S.; Shi, Y.; Miao, H.; Wang, M.; Li, B.; Gu, X.; Zhang, S. Genetic analysis and QTL mapping of resistance to gummy stem blight in Cucumis sativus seedling stage. Plant Dis. 2017, 101, 1145–1152. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, S.; Miao, H.; Shi, Y.; Wang, M.; Wang, Y.; Li, B.; Gu, X. Inheritance and QTL mapping of resistance to gummy stem blight in cucumber stem. Mol. Breed. 2017, 37, 49. [Google Scholar] [CrossRef]
- Hu, Z.; Deng, G.; Mou, H.; Xu, Y.; Chen, L.; Yang, J.; Zhang, M. A re-sequencing-based ultra-dense genetic map reveals a gummy stem blight resistance-associated gene in Cucumis melo. DNA Res. 2018, 25, 1–10. [Google Scholar] [CrossRef]
- Han, J.; Dong, S.; Shi, Y.; Dai, Z.; Miao, H.; Li, B.; Gu, X.; Zhang, S. Genome-wide association study reveals candidate genes for gummy stem blight resistance in cucumber. Hortic. Plant J. 2023, 9, 261–272. [Google Scholar] [CrossRef]
- Cohen, Y.; Coffey, M.D. Systemic fungicides and the control of oomycetes. Annu. Rev. Phytopathol. 1986, 24, 311–338. [Google Scholar] [CrossRef]
- Colle, M.; Straley, E.N.; Makela, S.B.; Hammar, S.A.; Grumet, R. Screening the cucumber plant introduction collection for young fruit resistance to Phytophthora capsici. HortScience 2014, 49, 244–249. [Google Scholar] [CrossRef]
- Kousik, C.S.; Ji, P.; Egel, D.S.; Quesada-Ocampo, L.M. Fungicide rotation programs for managing Phytophthora fruit rot of watermelon in the southeastern United States. Plant Health Prog. 2017, 18, 28–34. [Google Scholar] [CrossRef]
- Quesada-Ocampo, L.M.; Parada-Rojas, C.H.; Hansen, Z.; Vogel, G.; Smart, C.; Hausbeck, M.K.; Carmo, R.M.; Huitema, E.; Naegele, R.P.; Kousik, C.S.; et al. Phytophthora capsici: Recent Progress on Fundamental Biology and Disease Management 100 Years after Its Description. Annu. Rev. Phytopathol. 2023, 61, 185–208. [Google Scholar] [CrossRef]
- Parada-Rojas, C.H.; Granke, L.L.; Naegele, R.P.; Hansen, Z.; Hausbeck, M.K.; Kousik, C.S.; McGrath, M.T.; Smart, C.D.; Quesada-Ocampo, L.M. A Diagnostic Guide for Phytophthora capsici Infecting Vegetable Crops. Plant Health Prog. 2021, 22, 404–414. [Google Scholar] [CrossRef]
- Nazavari, K.; Jamali, F.; Bayat, F.; Modarresi, M. Evaluation of Resistance to Seedling Damping-Off Caused by Phytophthora drechsleri in Cucumber Cultivars under Greenhouse Conditions. Biol. Forum—Int. J. 2016, 8, 54–60. [Google Scholar]
- Favrin, R.J.; Rahe, J.E.; Mauza, B. Pythium spp. Associated with Crown Rot of Cucumbers in British Columbia Greenhouses. Plant Dis. 1988, 72, 683–687. [Google Scholar] [CrossRef]
- Mansfeld, B.N.; Colle, M.; Zhang, C.; Lin, Y.-C.; Grumet, R. Developmentally regulated activation of defense allows for rapid inhibition of infection in age-related resistance to Phytophthora capsici in cucumber fruit. BMC Genom. 2020, 21, 628. [Google Scholar] [CrossRef]
- Lin, Y.-C.; Mansfeld, B.N.; Tang, X.; Colle, M.; Chen, F.; Weng, Y.; Fei, Z.; Grumet, R. Identification of QTL Associated with Resistance to Phytophthora Fruit Rot in Cucumber (Cucumis sativus L.). Front. Plant Sci. 2023, 14, 1281755. [Google Scholar] [CrossRef]
- Zitter, T.A.; Hopkins, D.; Thomas, C.E. (Eds.) Compendium of Cucurbit Diseases; APS Press: St. Paul, MN, USA, 1996. [Google Scholar]
- Kim, S.W.; Lamsal, K.; Park, H.J.; Kim, Y.S.; Lee, Y.S.; Jung, J.H. Effect of Chitosan-Coated Fungicide Against Colletotrichum gloeosporioides and Powdery Mildew. Agric. Life Environ. Sci. 2011, 23, 14–22. [Google Scholar]
- Kooistra, E. Powdery mildew resistance in cucumber. Euphytica 1968, 17, 236–244. [Google Scholar] [CrossRef]
- Block, C.; Reitsma, K. Powdery mildew resistance in the U.S. National Plant Germplasm System cucumber collection. HortScience 2005, 40, 416. [Google Scholar] [CrossRef]
- Shanmugasundaram, S.; Williams, P.H.; Peterson, C.E. Inheritance of resistance to powdery mildew in cucumber. Phytopathology 1971, 61, 1218–1221. [Google Scholar] [CrossRef]
- Clark, R.L. Powdery mildew resistance in plant introductions of cucumber in Iowa. Plant Dis. Rep. 1975, 59, 1024–1028. [Google Scholar]
- Wehner, T.C.; Shetty, N.V.; Clark, R.L. Screening the cucumber germplasm collection for combining ability for yield. HortScience 2000, 35, 1141–1150. [Google Scholar] [CrossRef]
- Morishita, M.; Sugiyama, K.; Saito, T.; Sakata, Y. Powdery mildew resistance in cucumber. Jpn. Agric. Res. Q. 2003, 37, 7–14. [Google Scholar] [CrossRef]
- Liu, L.; Yuan, X.; Cai, R.; Pan, J.; He, H.; Yuan, L.; Guan, Y.; Zhu, L. Quantitative trait loci for resistance to powdery mildew in cucumber under seedling spray inoculation and leaf disc infection. J. Phytopathol. 2008, 156, 691–697. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, M.; Miao, H.; Zhang, S.; Yang, Y.; Xie, B.; Gu, X. QTL mapping of resistance genes to powdery mildew in cucumber (Cucumis sativus L.). Sci. Agric. Sin. 2011, 44, 3584–3593. [Google Scholar]
- Fukino, N.; Yoshioka, Y.; Sugiyama, M.; Sakata, Y.; Matsumoto, S. Identification and validation of powdery mildew (Podosphaera xanthii)-resistant loci in recombinant inbred lines of cucumber (Cucumis sativus L.). Mol. Breed. 2013, 32, 267–277. [Google Scholar] [CrossRef]
- He, X.; Li, Y.; Pandey, S.; Yandell, B.S.; Pathak, M.; Weng, Y. QTL mapping of powdery mildew resistance in WI 2757 cucumber (Cucumis sativus L.). Theor. Appl. Genet. 2013, 126, 2149–2161. [Google Scholar] [CrossRef]
- Yoshioka, Y.; Sakata, Y.; Sugiyama, M.; Fukino, N. Identification of quantitative trait loci for downy mildew resistance in cucumber (Cucumis sativus L.). Euphytica 2014, 198, 265–276. [Google Scholar] [CrossRef]
- Liu, P.N.; Miao, H.; Lu, H.W.; Cui, J.Y.; Tian, G.L.; Wehner, T.C.; Gu, X.F.; Zhang, S.P. Molecular Mapping and Candidate Gene Analysis for Resistance to Powdery Mildew in Cucumis sativus Stem. Genet. Mol. Res. 2017, 16, gmr16039680. [Google Scholar] [CrossRef] [PubMed]
- Nie, J.; He, H.; Peng, J.; Yang, X.; Bie, B.; Zhao, J.; Wang, Y.; Si, L.; Pan, J.-S.; Cai, R. Identification and Fine Mapping of pm5.1: A Recessive Gene for Powdery Mildew Resistance in Cucumber (Cucumis sativus L.). Mol. Breed. 2015, 35, 7. [Google Scholar] [CrossRef]
- Wang, Y.; Bo, K.; Gu, X.; Pan, J.; Li, Y.; Chen, J.; Wen, C.; Ren, Z.; Ren, H.; Chen, X.; et al. Molecularly Tagged Genes and Quantitative Trait Loci in Cucumber with Recommendations for QTL Nomenclature. Hortic. Res. 2020, 7, 3. [Google Scholar] [CrossRef] [PubMed]
- Schouten, H.J.; Krauskopf, J.; Visser, R.G.F.; Bai, Y. Identification of candidate genes required for susceptibility to powdery or downy mildew in cucumber. Euphytica 2014, 200, 475–486. [Google Scholar] [CrossRef]
- Berg, J.A.; Appiano, M.; Martínez, M.S.; Hermans, F.W.; Vriezen, W.H.; Visser, R.G.; Bai, Y.; Schouten, H.J. A transposable element insertion in the susceptibility gene CsaMLO8 results in hypocotyl resistance to powdery mildew in cucumber. BMC Plant Biol. 2015, 15, 243. [Google Scholar] [CrossRef]
- Tek, M.I.; Calis, O.; Fidan, H.; Shah, M.D.; Celik, S.; Wani, S.H. CRISPR/Cas9-based mlo-mediated resistance against Podosphaera xanthii in cucumber (Cucumis sativus L.). Front. Plant Sci. 2022, 13, 1081506. [Google Scholar] [CrossRef]
- Shnaider, Y.; Elad, Y.; Rav-David, D.; Pashkovsky, E.; Leibman, D.; Kravchik, M.; Shtarkman-Cohen, M.; Gal-On, A.; Spiegelman, Z. Development of powdery mildew resistance in cucumber using CRISPR/Cas9-mediated mutagenesis of CsaMLO8. Phytopathology 2023, 113, 786–790. [Google Scholar] [CrossRef]
- Bo, K.; Wei, S.; Wang, W.; Miao, H.; Dong, S.; Zhang, S.; Gu, X. QTL Mapping and Genome-Wide Association Study Reveal Two Novel Loci Associated with Green Flesh Color in Cucumber. BMC Plant Biol. 2019, 19, 243. [Google Scholar] [CrossRef]
- Alavilli, H.; Anarjan, M.B.; Lee, J.J.; Lee, S.; Song, K. Validation of Csaba2 as a powdery mildew susceptibility gene in cucumber. SSRN Prepr. 2025, 141, 5347149. [Google Scholar] [CrossRef]
- Nie, J.; Wang, H.; Zhang, W.; Teng, X.; Yu, C.; Cai, R.; Wu, G. Characterization of lncRNAs and mRNAs Involved in Powdery Mildew Resistance in Cucumber. Phytopathology 2021, 111, 1613–1624. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Shi, Y.; Yu, T.; Xu, X.; Yan, Y.; Qi, X.; Chen, X. Whole-Genome Resequencing of a Cucumber Chromosome Segment Substitution Line and Its Recurrent Parent to Identify Candidate Genes Governing Powdery Mildew Resistance. PLoS ONE 2016, 11, e0164469. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, X.; Zhu, C.; Qin, X.; Xu, J.; Cheng, C.; Lou, Q.; Li, J.; Chen, J. Complete resistance to powdery mildew and partial resistance to downy mildew in a Cucumis hystrix introgression line of cucumber were controlled by a co-localized locus. Theor. Appl. Genet. 2018, 131, 2229–2243. [Google Scholar] [CrossRef]
- Zhong, C. Identification and Functional Analysis of miRNAs Against Powdery Mildew in Cucumber. Master’s Thesis, Yangzhou University, Yangzhou, China, 2020. [Google Scholar] [CrossRef]
- Biles, C.; Abeles, F.B.; Wilson, C. The role of ethylene in anthracnose of cucumber (Cucumis sativus) caused by Colletotrichum lagenarium. Phytopathology 1990, 80, 732. [Google Scholar] [CrossRef]
- Thompson, D.C.; Jenkins, S.F. Effect of temperature, moisture, and cucumber cultivar resistance on lesion size increase and conidial production by Colletotrichum lagenarium. Phytopathology 1985, 75, 828–832. [Google Scholar] [CrossRef]
- Correll, J.C.; Rhoads, D.D.; Guerber, J.C. Analysis of mitochondrial DNA RFLPs, DNA fingerprints, and RAPDs of Colletotrichum orbiculare. Phytopathology 1993, 83, 1199–1204. [Google Scholar] [CrossRef]
- Zitter, T.A. Anthracnose of cucurbits. Extension Fact Sheet/APS Resource, 1987. [Google Scholar]
- Tsay, J.-G.; Chen, R.-S.; Wang, W.-L.; Weng, B.-C. First report of anthracnose on cucurbitaceous crops caused by Glomerella magna in Taiwan. Plant Dis. 2010, 94, 787. [Google Scholar] [CrossRef]
- Palenchar, J.; Treadwell, D.D.; Datnoff, L.E.; Gevens, A.J.; Vallad, G.E. Cucumber Anthracnose in Florida; Plant Pathology Department, Florida Cooperative Extension Service: Gainesville, FL, USA, 2012; p. 266. [Google Scholar]
- Sun, W.M.; Wen, X.L.; Qi, H.X.; Feng, L.N.; Cao, J.; Han, Z.L.; Yang, W.J.; Zhang, M.Y.; Han, B.J.; Meng, T.Y. First report of anthracnose of Atractylodes chinensis caused by Colletotrichum chlorophyti in China. Plant Dis. 2019, 103, 764. [Google Scholar] [CrossRef]
- Wasilwa, L.; Correll, J.; Morelock, T.; McNew, R.E. Reexamination of races of the cucurbit anthracnose pathogen Colletotrichum orbiculare. Phytopathology 1993, 83, 1190. [Google Scholar] [CrossRef]
- Wasilwa, L.; Morelock, T.E.; Correll, J.C. Current status of cucurbit anthracnose. HortScience 1997, 32, 495f. [Google Scholar] [CrossRef]
- Matsuo, H.; Isobe, S.; Shirasawa, K.; Yoshioka, Y. Novel QTLs for cucumber resistance to two Colletotrichum orbiculare strains of different pathogenic races. bioRxiv 2022. preprint 2022.02.28.482428. [Google Scholar] [CrossRef]
- Dutta, S.K. Pathogenicity of Natural Races and Induced Biochemical Mutants of Colletotrichum lagenarium (Ell. & Halst.) and the Inheritance of Resistance in Cucurbits; Kansas State University: Manhattan, KS, USA, 1960. [Google Scholar]
- Correa, E.; Crosby, K.; Malla, S. Optimizing a seedling screening method for anthracnose resistance in watermelon. Plant Health Prog. 2021, 22, 536–543. [Google Scholar] [CrossRef]
- Fitriyah, F.; Matsuo, H.; Isobe, S.; Shirasawa, K.; Naito, K.; Yoshioka, Y. Different control of resistance to two Colletotrichum orbiculare pathogenic races 0 and 1 in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 2024, 137, 127. [Google Scholar] [CrossRef]
- Barnes, W.C.; Epps, W.M. Two types of anthracnose resistance in cucumbers. Plant Dis. Rep. 1952, 36, 479–480. [Google Scholar]
- Robinson, R.W.; Munger, H.M.; Whitaker, T.W.; Bohn, G.W. Genes of the Cucurbitaceae. HortScience 1976, 11, 564–568. [Google Scholar] [CrossRef]
- Pan, J.; Tan, J.; Wang, Y.; Zheng, X.; Owens, K.; Li, D.; Li, Y.; Weng, Y. STAYGREEN (CsSGR) is a candidate for the anthracnose (Colletotrichum orbiculare) resistance locus cla in Gy14 cucumber. Theor. Appl. Genet. 2018, 131, 1577–1587. [Google Scholar] [CrossRef] [PubMed]
- Goode, M.J.; Bowers, J.L. Breeding for anthracnose resistance in cucumber. Phytopathology 1973, 63, 442. [Google Scholar]
- Wehner, T.C.; St. Amand, P.C. Anthracnose resistance of the cucumber germplasm collection in North Carolina field tests. Crop Sci. 1995, 35, 228–236. [Google Scholar] [CrossRef]
- Shim, S.A.; Jang, K.S.; Choi, Y.H.; Kim, J.C.; Kim, H.T.; Choi, G.J. Resistance degree of cucurbit cultivars to Colletotrichum orbiculare. Korean J. Hortic. Sci. Technol. 2013, 31, 371–379. [Google Scholar] [CrossRef]
- Wang, H.Z.; Li, S.J.; Liu, X.F.; Li, P.; Huo, Z.R.; Guan, W. AFLP markers of cucumber anthracnose resistance-related gene. Acta Hortic. Sin. 2007, 34, 213–216. [Google Scholar]
- Wang, Y.-H. Mapping and molecular breeding of monogenic traits. In Genetics, Genomics and Breeding of Cucurbits; Wang, Y.-H., Behera, T.K., Kole, C., Eds.; CRC Press: Boca Raton, FL, USA; Taylor & Francis: Boca Raton, FL, USA, 2012; pp. 225–237. [Google Scholar] [CrossRef]
- Feng, S.; Zhang, J.; Mu, Z.; Wang, Y.; Wen, C.; Wu, T.; Yu, C.; Li, Z.; Wang, H. Recent progress on the molecular breeding of Cucumis sativus L. in China. Theor. Appl. Genet. 2020, 133, 1777–1790. [Google Scholar] [CrossRef]
- El-Sharkawy, E.E.S.; ElSharawy, A.A. The influence of resistance chemical inducers against anthracnose on cucumber and pepper plants. Catrina Int. J. Environ. Sci. 2024, 29, 57–67. [Google Scholar] [CrossRef]
- Jagger, I.C. Breeding and improvement of cucurbits. In Better Plants and Animals; Whitaker, T.W., Ed.; U.S. Department of Agriculture: Washington, DC, USA, 1937; p. 207. [Google Scholar]
- Caranta, C.; Pflieger, S.; Lefebvre, V.; Daubèze, A.M.; Thabuis, A.; Palloix, A. QTLs involved in the restriction of cucumber mosaic virus (CMV) long-distance movement in pepper. Theor. Appl. Genet. 2002, 104, 586–591. [Google Scholar] [CrossRef] [PubMed]
- Roossinck, M.J. Cucumber mosaic virus, a model for RNA virus evolution. Mol. Plant Pathol. 2001, 2, 59–63. [Google Scholar] [CrossRef]
- Gallitelli, D. The ecology of cucumber mosaic virus and sustainable agriculture. Virus Res. 2000, 71, 9–21. [Google Scholar] [CrossRef]
- Price, W.C. Isolation and study of some yellow strains of cucumber mosaic. Phytopathology 1934, 24, 743–761. [Google Scholar]
- Hamza, E.S.; Al-Naggar, A.M.; El-Shabrawi, H.M.; Tolba, I.H. Characteristics of cucumber mosaic virus isolates infecting cucurbits in Egypt. Al-Azhar J. Agric. Res. 2022, 47, 172–184. [Google Scholar] [CrossRef]
- Pan, J.Q. Comprehensive prevention and control technology of cucurbits virus disease. J. Shanghai Veg. 2009, 4, 86. (In Chinese) [Google Scholar]
- Risser, G.; Pitrat, M.; Rode, J.C. Étude de la Résistance du Melon (Cucumis melo L.) au Virus de la Mosaïque du Concombre. Ann. Amelior. Plantes 1977, 27, 509–522. [Google Scholar]
- Wasuwat, S.L.; Walker, J.C. Inheritance of resistance in cucumber to cucumber mosaic virus. Phytopathology 1961, 51, 423–428. [Google Scholar]
- Kooistra, E. The inheritance of resistance to Cucumis virus 1 in cucumber (Cucumis sativus L.). Euphytica 1969, 18, 326–332. [Google Scholar] [CrossRef]
- Marathe, R.; Guan, Z.; Anandalakshmi, R.; Zhao, H.; Dinesh-Kumar, S. Study of the Arabidopsis thaliana resistome in response to cucumber mosaic virus infection using whole genome microarray. Plant Mol. Biol. 2004, 55, 501–520. [Google Scholar] [CrossRef]
- Porter, R.H. The reaction of cucumbers to types of mosaic. Phytopathology 1931, 21, 95–120. [Google Scholar]
- Hunger, H.M.; Newhall, A. Breeding for disease resistance in celery and cucurbits. Yearb. Agric. 1953, 43, 254–259. [Google Scholar]
- Schaible, L.W. Some aspects of resistance to cucumber virus 1 in cucumber and squash. Phytopathology 1954, 44, 904–905. [Google Scholar]
- Havey, M.J. CMV resistance in cucumber—A correction. HortScience 1997, 32, 18. [Google Scholar]
- Wang, Y.J.; Provvidenti, R.; Robinson, R.W. Inheritance of resistance to watermelon mosaic virus 1 in cucumber. HortScience 1984, 19, 587–588. [Google Scholar] [CrossRef]
- Dhillon, N.P.S. New sources of cucumber mosaic virus–tolerant landraces of cucumber identified at the centre of origin (India). Plant Breed. 1992, 109, 172–174. [Google Scholar] [CrossRef]
- Shi, L.; Yang, Y.; Xie, Q.; Miao, H.; Bo, K.; Song, Z.; Wang, Y.; Xie, B.; Zhang, S.; Gu, X. Inheritance and QTL mapping of cucumber mosaic virus resistance in cucumber (Cucumis sativus L.). PLoS ONE 2018, 13, e0200571. [Google Scholar] [CrossRef]
- Amano, M.; Watanabe, N.; Kondo, H.; Suzuki, M.; Kuroda, T.; Yamasaki, S.; Hanada, K. High-resolution mapping of zym, a recessive gene for Zucchini yellow mosaic virus resistance in cucumber. Theor. Appl. Genet. 2013, 126, 2983–2993. [Google Scholar] [CrossRef]
- Zhang, H.Y.; Mao, A.J.; Zhang, F.; Xu, Y.; Wang, Y.J. Mapping of three major virus-resistant genes in cucumber. J. Agric. Biotechnol. 2005, 13, 709–712. [Google Scholar]
- Zhou, J. Genetic Analysis and Gene Mapping of Watermelon mosaic virus (WMV) Resistance in Cucumber. Ph.D. Thesis, The Chinese Academy of Agricultural Sciences, Beijing, China, 2012. [Google Scholar]
- Elsharkawy, M.M.; Elsawy, M.M.; Ismail, I.A. Mechanism of resistance to Cucumber mosaic virus elicited by inoculation with Bacillus subtilis subsp. subtilis. Pest Manag. Sci. 2022, 78, 86–94. [Google Scholar] [CrossRef]
- Atarashi, H.; Tomita, R.; Uehara, K.; Ishibashi, K.; Ishikawa, M.; Nishihara, M.; Matsumura, T. Artificially edited alleles of the eukaryotic translation initiation factor 4E1 gene differentially reduce susceptibility to Cucumber mosaic virus and Potato virus Y in tomato. Front. Microbiol. 2020, 11, 564310. [Google Scholar] [CrossRef]
- Kavosipour, S.; Niazi, A.; Izadpanah, K.; Afsharifar, A.; Yasaie, M. Induction of resistance to Cucumber mosaic virus (CMV) using hairpin construct of 2b gene. Iran. J. Plant Pathol. 2012, 48, 209–219. [Google Scholar]
- Arinaitwe, W.; Wang, T.; Tungadi, T.; Groen, S.C.; Karban, R.; Casteel, C.L.; Carr, J.P. Induction of aphid resistance in tobacco by the Cucumber mosaic virus CMVΔ2b mutant is jasmonate-dependent. Mol. Plant Pathol. 2023, 24, 391–395. [Google Scholar] [CrossRef]
- Tungadi, T.; Donnelly, R.; Qing, L.; Iqbal, J.; Murphy, A.M.; Pate, A.E.; Cunniffe, N.J.; Carr, J.P. Cucumber mosaic virus 2b proteins inhibit virus-induced aphid resistance in tobacco. Mol. Plant Pathol. 2020, 21, 250–257. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.J.; Antoniw, J.F.; Kreuze, J. Virgaviridae: A new family of rod-shaped plant viruses. Arch. Virol. 2009, 154, 1967–1972. [Google Scholar] [CrossRef] [PubMed]
- Dombrovsky, A.; Tran-Nguyen, L.T.T.; Jones, R.A.C. Cucumber green mottle mosaic virus: Rapidly increasing global distribution, etiology, epidemiology, and management. Annu. Rev. Phytopathol. 2017, 55, 201–225. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, Q.; Wang, Y.; Li, J.; Li, H.; Zhang, Z.; Wang, H.; Li, D. First report of cucumber green mottle mosaic virus infecting zucchini (Cucurbita pepo) and wax gourd (Benincasa hispida) in China. Plant Dis. 2023, 107, 4037. [Google Scholar] [CrossRef]
- Ainsworth, G.C. Mosaic disease of cucumber. Ann. Appl. Biol. 1935, 22, 55–67. [Google Scholar] [CrossRef]
- Van Koot, Y.; Van Dorst, H.J.M. Virusziekten van de komkommer in Nederland: With a summary: Virus diseases of cucumber in The Netherlands. Tijdschr. Over Plantenziekten 1959, 65, 257–271. [Google Scholar]
- Liu, H.W.; Luo, L.X.; Li, J.Q.; Liu, P.F.; Chen, X.Y.; Hao, J.J. Pollen and seed transmission of cucumber green mottle mosaic virus in cucumber. Plant Pathol. 2014, 63, 72–77. [Google Scholar] [CrossRef]
- Varveri, C.; Vassilakos, N.; Bem, F. Characterization and detection of cucumber green mottle mosaic virus in Greece. Phytoparasitica 2002, 30, 493–501. [Google Scholar] [CrossRef]
- Kooistra, E. Significance of the non-appearance of visible disease symptoms in cucumber (Cucumis sativus L.) after infection with Cucumis virus 2. Euphytica 1968, 17, 136–140. [Google Scholar] [CrossRef]
- Rajamony, L.; More, T.A.; Seshadri, V.S.; Varma, A. Resistance to cucumber green mottle mosaic virus (CGMMV) in muskmelon. Cucurbit Genet. Coop. 1987, 10, 58–59. [Google Scholar]
- Pan, R.S.; More, T.A. Screening of melon (Cucumis melo L.) germplasm for multiple disease resistance. Euphytica 1996, 88, 125–128. [Google Scholar] [CrossRef]
- Carnide, V.; Do Rosario Barroso, M. Las cucurbitáceas: Bases para su mejora genética. Hortic. Int. 2006, 53, 16–21. [Google Scholar]
- Crespo, O.; Janssen, D.; Robles, C.; Ruiz, L. Resistance to Cucumber green mottle mosaic virus in Cucumis sativus. Euphytica 2018, 214, 201. [Google Scholar] [CrossRef]
- Ellouze, W.; Mishra, V.; Howard, R.J.; Ling, K.-S.; Zhang, W. Preliminary Study on the Control of Cucumber Green Mottle Mosaic Virus in Commercial Greenhouses Using Agricultural Disinfectants and Resistant Cucumber Varieties. Agronomy 2020, 10, 1879. [Google Scholar] [CrossRef]
- Miao, S.; Liang, C.; Li, J.; Baker, B.; Luo, L. Polycistronic Artificial microRNA-Mediated Resistance to Cucumber Green Mottle Mosaic Virus in Cucumber. Int. J. Mol. Sci. 2021, 22, 12237. [Google Scholar] [CrossRef] [PubMed]
- Tewari, J.P. Transmission studies of watermelon mosaic virus by aphids. Entomon 1976, 1, 111–114. [Google Scholar] [CrossRef]
- Yamamoto, T. Infection cycle of watermelon mosaic virus. JARQ 1986, 19, 259–265. [Google Scholar]
- Tian, G.; Miao, H.; Yang, Y.; Zhou, J.; Lu, H.; Wang, Y.; Xie, B.; Zhang, S.; Gu, X. Genetic analysis and fine mapping of Watermelon mosaic virus resistance gene in cucumber. Mol. Breed. 2016, 36, 131. [Google Scholar] [CrossRef]
- Cohen, S.; Gertman, E.; Kedar, N. Inheritance of resistance to Melon mosaic virus in cucumbers. Phytopathology 1971, 61, 253–255. [Google Scholar] [CrossRef]
- Wai, T.; Grumet, R. Inheritance of resistance to Watermelon mosaic virus in the cucumber line TMG-1: Tissue-specific expression and relationship to Zucchini yellow mosaic virus resistance. Theor. Appl. Genet. 1995, 91, 699–706. [Google Scholar] [CrossRef]
- Roggero, P.; Dellavalle, G.; Lisa, V.; Stravato, V.M. First Report of Moroccan Watermelon Mosaic Potyvirus in Zucchini in Italy. Plant Dis. 1998, 82, 351. [Google Scholar] [CrossRef]
- Fischer, H.U.; Lockhart, B.E.L. Serious losses in cucurbits caused by watermelon mosaic virus in Morocco. Plant Dis. Report. 1974, 58, 143–146. [Google Scholar]
- Kabelka, E.; Grumet, R. Inheritance of Resistance to the Moroccan Watermelon Mosaic Virus in the Cucumber Line TMG-1 and Its Relationship to Zucchini Yellow Mosaic Virus Resistance. HortScience 1996, 31, 622e. [Google Scholar] [CrossRef]
- Miras, M.; Truniger, V.; Silva, C.; Verdaguer, N.; Aranda, M.A.; Querol-Audi, J. Structure of eIF4E in complex with an eIF4G peptide supports a universal bipartite binding mode for protein translation. Plant Physiol. 2017, 174, 1476–1491. [Google Scholar] [CrossRef]
- Vovlas, C.; Hiebert, E.; Russo, M. Zucchini yellow fleck virus, a new potyvirus of zucchini squash. Phytopathol. Mediterr. 1981, 20, 123–128. [Google Scholar]
- Avgelis, A.D. Epidemiological studies of Zucchini yellow fleck virus in Crete. Phytopathol. Mediterr. 1985, 24, 208–210. [Google Scholar]
- Antignus, Y.; Levy, D.; Cohen, S. Characterisation of a variant of zucchini yellow fleck virus (ZYFV), a potyvirus causing a wilt disease of melons in Israel. Ann. Appl. Biol. 1995, 126, 111–120. [Google Scholar] [CrossRef]
- Gilbert-Albertini, F.; Lecoq, H. The characterization of a strain of Zucchini yellow fleck virus found in southeastern France. J. Phytopathol. 1994, 140, 375–384. [Google Scholar] [CrossRef]
- Gilbert-Albertini, F.; Pitrat, M.; Lecoq, H. Inheritance of Resistance to Zucchini Yellow Fleck Virus in Cucumis sativus L. HortScience 1995, 30, 336. [Google Scholar] [CrossRef]
- Desbiez, C.; Lecoq, H. Zucchini yellow mosaic virus. Plant Pathol. 1997, 46, 809–829. [Google Scholar] [CrossRef]
- Xu, Y.; Kang, D.; Shi, Z.; Shen, H.; Wehner, T.C. Inheritance of resistance to zucchini yellow mosaic virus and watermelon mosaic virus in watermelon. J. Hered. 2004, 95, 268–273. [Google Scholar] [CrossRef]
- Provvidenti, R. Inheritance of resistance to a strain of zucchini yellow mosaic virus in cucumber. Plant Dis. 1987, 71, 26–29. [Google Scholar] [CrossRef]
- Kabelka, E.; Ullah, Z.; Grumet, R. Multiple alleles for zucchini yellow mosaic virus resistance at the zym locus in cucumber. Theor. Appl. Genet. 1997, 95, 997–1004. [Google Scholar] [CrossRef]
- Purcifull, D.E.; Edwardson, J.R.; Hiebert, E.; Gonsalves, D. Papaya ringspot virus. CMI/AAB Descr. Plant Viruses 1984, 292. [Google Scholar]
- Owolabi, A.; Nwachukwu, C.; Odok, S. Screening cucumber plant introduction accession lines for resistance against cucumber strain of Papaya ringspot virus (PRSV). Int. J. Virol. 2015, 11, 66–76. [Google Scholar] [CrossRef]
- Park, Y.H.; Sensoy, S.; Wye, C.; Antonise, R.; Peleman, J.; Havey, M.J. A genetic map of cucumber composed of RAPDs, RFLPs, AFLPs, and loci conditioning resistance to papaya ringspot and zucchini yellow mosaic viruses. Genome 2000, 43, 1003–1010. [Google Scholar] [CrossRef]
- Monnot, S.; Cantet, M.; Mary-Huard, T.; Moreau, L.; Lowdon, R.; Van Haesendonck, M.; Ricard, A.; Boissot, N. Unravelling cucumber resistance to several viruses via genome-wide association studies highlights resistance hotspots and new QTLs. Hortic. Res. 2022, 9, uhac184. [Google Scholar] [CrossRef]
- Grumet, R.; Kabelka, E.; McQueen, S.; Wai, T.; Humphrey, R. Characterization of sources of resistance to the watermelon strain of papaya ringspot virus in cucumber: Allelism and co-segregation with other potyvirus resistances. Theor. Appl. Genet. 2000, 101, 463–472. [Google Scholar] [CrossRef]
- Lecoq, H.; Desbiez, C.; Delécolle, B.; Cohen, S.; Mansour, A. Cytological and molecular evidence that the whitefly-transmitted cucumber vein yellowing virus is a tentative member of the family Potyviridae. J. Gen. Virol. 2000, 81, 2289–2293. [Google Scholar] [CrossRef]
- Tian, G.; Li, J.; Fu, J.; Yang, Z.; Wang, G.; Ma, Z.; Zhang, H.; Chen, H.; Wang, Y. Genetic analysis and gene mapping of papaya ringspot virus resistance in cucumber. Mol. Breed. 2015, 35, 110. [Google Scholar] [CrossRef]
- Bateson, M.F.; Henderson, J.; Chaleeprom, W.; Gibbs, A.J.; Dale, J.L. Papaya ringspot potyvirus: Isolate variability and the origin of PRSV type P (Australia). J. Gen. Virol. 1994, 75, 3547–3553. [Google Scholar] [CrossRef]
- Attasart, C.; Charoensilp, G.; Kertbundit, S.; Panyim, S.; Juricek, M. Nucleotide sequence of a Thai isolate of Papaya ringspot virus type W. Acta Virol. 2002, 46, 241–246. [Google Scholar]
- Gonsalves, D. Control of papaya ringspot virus in papaya: A case study. Annu. Rev. Phytopathol. 1998, 36, 415–437. [Google Scholar] [CrossRef]
- Ramírez-Madera, A.O.; Havey, M.J. Different haplotypes encode the same protein for independent sources of zucchini yellow mosaic virus resistance in cucumber. HortScience 2017, 52, 1527–1531. [Google Scholar] [CrossRef]
- Tomlinson, J.A. Epidemiology and control of virus diseases of vegetables. Ann. Appl. Biol. 1987, 110, 661–681. [Google Scholar] [CrossRef]
- Wai, T.; Grumet, R. Inheritance of resistance to the watermelon strain of Papaya ringspot virus in the cucumber line TMG-1. HortScience 1995, 30, 338. [Google Scholar] [CrossRef]
- Kabelka, E.; Grumet, R. Inheritance of resistance to the Moroccan watermelon mosaic virus in the cucumber line TMG-1 and cosegregation with zucchini yellow mosaic virus resistance. Euphytica 1997, 95, 237–242. [Google Scholar] [CrossRef]
- Fidan, H.; Çalış, O.; Fidan, M.; Tek, M.İ.; Çelik, S.; Wani, S.H. Knockout of eIF4E using CRISPR/Cas9 for large-scale production of resistant cucumber cultivar against WMV, ZYMV, and PRSV. Front. Plant Sci. 2023, 14, 1143813. [Google Scholar] [CrossRef] [PubMed]
- Mansour, A.; Al-Musa, A. Cucumber vein yellowing virus: Host range and virus–vector relationships. J. Phytopathol. 1993, 137, 73–78. [Google Scholar] [CrossRef]
- Cohen, S.; Nitzany, F.E. A whitefly-transmitted virus of cucurbits in Israel. Phytopathol. Mediterr. 1960, 1, 44–46. [Google Scholar]
- Mansour, A.N.; Hadidi, N. Cucumber vein yellowing virus: Purification and serological studies. Dirasat Agric. Sci. 1999, 26, 8–14. [Google Scholar]
- Picó, B.; Villar, C.; Nuez, F.; Weber, W. Screening Cucumis sativus landraces for resistance to cucumber vein yellowing virus. Plant Breed. 2003, 122, 426–430. [Google Scholar] [CrossRef]
- Nitzany, F.E. Review of vegetable virus diseases in Israel and their control. FAO Plant Prot. Bull. 1975, 23, 148–155. [Google Scholar]
- Picó, B.; Sifres, A.; Martínez-Pérez, E.; Leiva-Brondo, M.; Nuez, F. Genetics of the resistance to CVYV in cucumber. In Modern Variety Breeding for Present and Future Needs, Proceedings of the 18th EUCARPIA General Congress, Valencia, Spain, 9–12 September 2008; Springer: Berlin/Heidelberg, Germany, 2008; pp. 452–456. [Google Scholar]
- Pujol, M.; Martín-Hernández, A.M.; Martínez, C.; Gómez, P.; Janssen, D.; Picó, B.; Ruiz, L. Mapping cucumber vein yellowing virus resistance in cucumber (Cucumis sativus L.) by using BSA-seq analysis. Front. Plant Sci. 2019, 10, 1583. [Google Scholar] [CrossRef]
- Kato, K.; Hanada, K.; Kameya-Iwaki, M. Melon yellow spot virus: A distinct species of the genus Tospovirus isolated from melon. Phytopathology 2000, 90, 422–426. [Google Scholar] [CrossRef]
- Takeuchi, S.; Okuda, M.; Hanada, K.; Kawada, Y.; Kameya, M. Spotted wilt disease of cucumber (Cucumis sativus) caused by melon yellow spot virus. Jpn. J. Phytopathol. 2001, 67, 76–79. [Google Scholar] [CrossRef][Green Version]
- Sugiyama, M.; Okuda, M.; Sakata, Y. Evaluation of resistance to Melon yellow spot virus in a cucumber germplasm collection. Plant Breed. 2009, 128, 209–213. [Google Scholar] [CrossRef]
- Sugiyama, M.; Kawazu, Y.; Fukino, N.; Yoshioka, Y. Mapping of quantitative trait loci for Melon yellow spot virus resistance in cucumber (Cucumis sativus L.). Euphytica 2015, 205, 307–319. [Google Scholar] [CrossRef]
- Tzanetakis, I.E.; Martin, R.R.; Wintermantel, W.M. Epidemiology of criniviruses: An emerging problem in world agriculture. Front. Microbiol. 2013, 4, 119. [Google Scholar] [CrossRef] [PubMed]
- Duffus, J.E. Beet pseudo-yellows virus, transmitted by the greenhouse whitefly (Trialeurodes vaporariorum). Phytopathology 1965, 55, 450–453. [Google Scholar]
- Berdiales, B.; Bernal, J.J.; Sáez, E.; Woudt, B.; Beitia, F.; Rodríguez-Cerezo, E. Occurrence of Cucurbit yellow stunting disorder virus (CYSDV) and Beet pseudo-yellows virus in cucurbit crops in Spain and transmission of CYSDV by two biotypes of Bemisia tabaci. Eur. J. Plant Pathol. 1999, 105, 211–215. [Google Scholar] [CrossRef]
- Liu, L.Z.; Chen, Y.Y.; Zhu, W.M. First report of Cucurbit yellow stunting disorder virus on melon in China. Plant Dis. 2010, 94, 485. [Google Scholar] [CrossRef]
- Guirao, P.; Beitia, F.; Cenis, J.L. Biotype determination of Spanish populations of Bemisia tabaci (Hemiptera: Aleyrodidae). Bull. Entomol. Res. 1997, 87, 587–593. [Google Scholar] [CrossRef]
- Castle, S.; Palumbo, J.; Merten, P.; Cowden, C.; Prabhaker, N. Effects of foliar and systemic insecticides on whitefly transmission and incidence of Cucurbit yellow stunting disorder virus. Pest Manag. Sci. 2017, 73, 1462–1472. [Google Scholar] [CrossRef]
- Marco, C.F.; Aguilar, J.M.; Abad, J.; Gómez-Guillamón, M.L.; Aranda, M.A. Melon resistance to Cucurbit yellow stunting disorder virus is characterized by reduced virus accumulation. Phytopathology 2003, 93, 844–852. [Google Scholar] [CrossRef]
- Eid, S.; Abou-Jawdah, Y.; El-Mohtar, C.; Sobh, H.; Havey, M. Tolerance in Cucumber to Cucurbit yellow stunting disorder virus. Plant Dis. 2006, 90, 645–649. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Aguilar, J.M.; Abad, J.; Aranda, M.A. Resistance to Cucurbit yellow stunting disorder virus in Cucumber. Plant Dis. 2006, 90, 583–586. [Google Scholar] [CrossRef] [PubMed]
- De Ruiter, W.; Hofstede, R.; De Vries, J.; Van den Heuvel, H.; Pitrat, M. Combining QTLs for Resistance to CYSDV and Powdery Mildew in a Single Cucumber Line. In Cucurbitaceae 2008, Proceedings of the IXth EUCARPIA Meeting on Genetics and Breeding of Cucurbitaceae, Avignon, France, 21–24 May 2008; Institut National de la Recherche Agronomique (INRA): Paris, France, 2008; pp. 181–188. [Google Scholar]
- Faber, N.M.; Torres, L.M.; Sanchez, L.O. Method of Breeding CYSDV-Resistant Cucumber Plants. European Patent EP09775344A, 19 December 2008. [Google Scholar]
- Słomnicka, R.; Olczak-Woltman, H.; Korzeniewska, A.; Gozdowski, D.; Niemirowicz-Szczytt, K.; Bartoszewski, G. Genetic Mapping of psl Locus and Quantitative Trait Loci for Angular Leaf Spot Resistance in Cucumber (Cucumis sativus L.). Mol. Breed. 2018, 38, 111. [Google Scholar] [CrossRef]
- Olczak-Woltman, H.; Bartoszewski, G.; Mądry, W.; Niemirowicz-Szczytt, K. Inheritance of Resistance to Angular Leaf Spot (Pseudomonas syringae pv. lachrymans) in Cucumber and Identification of Molecular Markers Linked to Resistance. Plant Pathol. 2009, 58, 145–151. [Google Scholar] [CrossRef]
- Harighi, B. Angular Leaf Spot of Cucumber Caused by Pseudomonas syringae pv. lachrymans in Kurdistan. Plant Dis. 2007, 91, 769. [Google Scholar] [CrossRef]
- Chand, J.N.; Walker, J.C. Inheritance of Resistance to Angular Leaf Spot of Cucumber. Phytopathology 1964, 54, 51–53. [Google Scholar]
- Dessert, J.M.; Baker, L.R.; Fobes, J.F. Inheritance of Reaction to Pseudomonas lachrymans in Pickling Cucumber. Euphytica 1982, 31, 847–855. [Google Scholar] [CrossRef]
- Zhang, K.; Wei, Y.; Njogu, M.K.; Wang, X.; Lou, Q.; Li, J.; Chen, J. Genetic mapping of angular leaf spot resistance to Pseudomonas syringae pv. lachrymans in a Cucumis hystrix introgression line of cucumber. Euphytica 2019, 215, 176. [Google Scholar] [CrossRef]
- Li, L.; Du, C.; Wang, L.; Lai, M.; Fan, H. Exogenous melatonin improves the resistance to cucumber bacterial angular leaf spot caused by Pseudomonas syringae pv. lachrymans. Physiol. Plant. 2022, 174, e13724. [Google Scholar] [CrossRef]
- Yang, D.; Yang, J.; Qiu, M.; Gao, J.; Ma, W.; Cui, R. Genome-wide identification and expression analysis of the cucumber FKBP gene family in response to abiotic and biotic stresses. Genes 2023, 14, 2006. [Google Scholar] [CrossRef]
- Ma, W.; Yang, D.; Qiu, M.; Gao, J.; Cui, R. Genome-wide identification and expression pattern analysis of the trihelix gene family in cucumber. Pak. J. Bot. 2024, 56, 1853–1866. [Google Scholar] [CrossRef]
- Jones, L.R. Bacillus carotovorus n. sp., die Ursache einer weichen Fäulnis der Möhre. Cent. Für Bakteriol. Parasitenkd. Infekt. 1901, 2, 12–21. [Google Scholar]
- Nazerian, E.; Sijam, K.; Zainal Abidin, M.A.; Vadamalai, G. First report of soft rot caused by Pectobacterium carotovorum subsp. carotovorum on cucumber in Malaysia. Plant Dis. 2011, 95, 1474. [Google Scholar] [CrossRef]
- Li, L.; Yuan, L.; Shi, Y.; Xie, X.; Chai, A.; Wang, Q.; Li, B. Comparative genomic analysis of Pectobacterium carotovorum subsp. brasiliense SX309 provides novel insights into its genetic and phenotypic features. BMC Genom. 2019, 20, 486. [Google Scholar] [CrossRef]
- Oulghazi, S.; Pédron, J.; Cigna, J.; Lau, Y.Y.; Moumni, M.; Van Gijsegem, F.; Faure, D. Pectobacterium brasiliense: Genomics, host range and disease management. Microorganisms 2021, 9, 106. [Google Scholar] [CrossRef]
- Yamagishi, H.; Yoshikawa, H.; Yui, S. Leaf morphology and soft rot resistance in offspring of a somatic hybrid between Chinese cabbage and kale (Cruciferae). Euphytica 1990, 47, 215–221. [Google Scholar] [CrossRef]
- Zhang, Y.; Dong, S.; Guan, J.; Liu, X.; Xie, X.; Albornoz, K.; Han, J.; Wang, Z.; Gu, X.; Zhang, S.; et al. Genome-wide association study identifies candidate genes for bacterial soft rot resistance in cucumber seedlings. Hortic. Plant J. 2025, 11, 1152–1165. [Google Scholar] [CrossRef]
- Wang, Y.; Cao, R.; Yang, L.; Duan, X.; Zhang, C.; Yu, X.; Ye, X. Transcriptome analyses revealed the wax and phenylpropanoid biosynthesis pathways related to disease resistance in rootstock-grafted cucumber. Plants 2023, 12, 2963. [Google Scholar] [CrossRef]
- Atiq, M.; Zulfiqar, H.; Rajput, N.A.; Sahi, S.T.; Abbas, W.; Ahmad, S.; Sultan, A.; Usman, M.; Jabbar, A.; Ghaffar, A.; et al. Bacterial wilt of cucumber: An emerging threat to cucumber production in Pakistan. Plant Cell Biotechnol. Mol. Biol. 2022, 23, 54–65. [Google Scholar] [CrossRef]
- Rojas, E.S.; Gleason, M.L.; Batzer, J.C.; Duffy, M. Feasibility of delaying removal of row covers to suppress bacterial wilt of muskmelon (Cucumis melo). Plant Dis. 2011, 95, 729–734. [Google Scholar] [CrossRef]
- Leach, J.G. Observations on cucumber beetles as vectors of cucurbit wilt. Phytopathology 1964, 54, 606–607. [Google Scholar]
- Caudle, J. Control of Erwinia tracheiphila in Cucumis melo. Master’s Thesis, University of Kentucky, Lexington, KY, USA, 2013. [Google Scholar]
- Rand, F.V.; Enlows, E.M.A. Bacterial Wilt of Cucurbits; U.S. Department of Agriculture: Washington, DC, USA, 1920.
- Rojas, E.S.; Gleason, M.L.; Batzer, J.C.; Duffy, M. Bacterial wilt of cucurbits: Resurrecting a classic pathosystem. Plant Dis. 2015, 99, 564–574. [Google Scholar] [CrossRef] [PubMed]
- Nuttall, V.W.; Jasmin, J.J. The inheritance of resistance to bacterial wilt (Erwinia tracheiphila (E.F. Sm.) Holland) in cucumber. Can. J. Plant Sci. 1958, 38, 401–404. [Google Scholar] [CrossRef]
- Lezzoni, A.F.; Peterson, C.E. Linkage of bacterial wilt resistance and sex expression in cucumber. HortScience 1980, 15, 257–258. [Google Scholar] [CrossRef]
- Staub, J.E.; Peterson, C.E. Comparisons between bacterial wilt–resistant and –susceptible gynoecious cucumber lines and F1 progeny. HortScience 1986, 21, 1428–1430. [Google Scholar] [CrossRef]
- Silver, S. Acriflavine resistance: A bacteriophage mutation affecting the uptake of dye by the infected bacterial cells. Proc. Natl. Acad. Sci. USA 1965, 53, 24–30. [Google Scholar] [CrossRef]
- Webb, R.E.; Goth, R.W. A seedborne bacterium isolated from watermelon. Plant Dis. Rep. 1965, 49, 818–821. [Google Scholar]
- Burdman, S.; Walcott, R. Acidovorax citrulli: Generating basic and applied knowledge to tackle a global threat to the cucurbit industry. Mol. Plant Pathol. 2012, 13, 805–815. [Google Scholar] [CrossRef] [PubMed]
- Hopkins, D.; Thompson, C.M. Seed transmission of Acidovorax avenae subsp. citrulli in cucurbits. HortScience 2002, 37, 924. [Google Scholar] [CrossRef]
- Walcott, R.R.; Feng, J.; Gitaitis, R.D. Detection of Acidovorax citrulli in cucurbit seeds. In Detection of Plant-Pathogenic Bacteria in Seed and Other Planting Material, 2nd ed.; The American Phytopathological Society: St. Paul, MN, USA, 2017; pp. 179–187. [Google Scholar] [CrossRef]
- Zhao, T.; Sun, F.; Wang, B.; Hui, W. Pathogen Identification of Hami Melon Bacterial Fruit Blotch. 2001. Available online: https://www.semanticscholar.org/paper/2eca843aeb147bca0f8300c703c5f2ed7c59acbb (accessed on 13 October 2025).
- Martin, H.L.; O’Brien, R.G.; Abbott, D.V. First report of Acidovorax avenae subsp. citrulli as a pathogen of cucumber. Plant Dis. 1999, 83, 965. [Google Scholar] [CrossRef]
- Eckshtain-Levi, N.; Munitz, T.; Živanović, M.; Traore, S.M.; Spröer, C.; Zhao, B.; Welbaum, G.; Walcott, R.; Sikorski, J.; Burdman, S. Comparative analysis of type III–secreted effector genes reflects divergence of Acidovorax citrulli strains into three distinct lineages. Phytopathology 2014, 104, 1152–1162. [Google Scholar] [CrossRef] [PubMed]

| Database Name | Representative Assemblies | Domain Name (url) | Key Features | Sequence Accessibility | Analysis Tools | Reference |
|---|---|---|---|---|---|---|
| Cucumber-DB | CLv4.0 (‘9930’), Cuc64, W4, W8, 9110Gt, Cu2, Cuc37, Cucu80, Gy14, Hx117, Hx14, XTMC | http://www.cucumberdb.com/ | Genome Annotation, Variome, Population Structure, Selection Signals, Expression Atlas, Synteny, CENH3 Tracks | Genome FASTA, GFF3 Annotations, CDS/mRNA/protein FASTA, Transcriptome Data | GetSequence, BLAST, Primer Designer, Differential Expression Analysis, GO/KEGG, IDConverter, Co-expression, Multiple Sequence Alignment, Motif Prediction | Guan et al., 2024 [57] |
| CuGenDB/CuGenDBv2 | 9930 (v2, v3), Gy14 (v1, v2), C. sativus var. hardwickii (PI 183967) | https://cucurbitgenomics.org/ | Comparative Genomics, ESTs/unigenes, Gene Annotations, Expression Profiles, Genetic Maps, Integrated RNA-Seq visualization | Genome FASTA, GFF3 Annotations, CDS/mRNA, EST/unigene | JBrowse, Genome Browser, SyntenyViewer, BLAST, GO & Pathway Enrichment, Gene Functional Classification, RNA-Seq Module (edgeR/DESeq) | Yu et al., 2023 [58] |
| NCBI Assembly/Genomes | 9930_V3, ASM407v2, others | https://www.ncbi.nlm.nih.gov/assembly, accessed on 19 November 2025 | Chromosome-level Assemblies, Gene and Transcript Annotations, Contig/Scaffold Statistics, Taxonomic Lineage | Genome FASTA, GFF3/GTF Annotations, CDS/mRNA/protein, Assembly Reports, Metadata Tables | Genome viewer, BLAST, metadata search | O’Leary NA et al., 2024 [60] |
| JGI Genome Portal | Gy14 v1.0 | https://genome.jgi.doe.gov/ | EST-mapped Genome Assemblies, Gene Structural and Functional Annotations, and Comparative Genomics Data | genome browser; GFF3/GTF annotations, CDS/mRNA/protein | BLAST, JBrowse-based Genome Browser, Biomart, Biomart v13, Synteny | Nordberg et al., 2014 [61] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaur, S.; Thakur, S.; Sabharwal, P.; Meru, G. Breeding for Disease Resistance in Cucumber: Current Status, Genetic Insights, and Genomic Resources. Horticulturae 2025, 11, 1440. https://doi.org/10.3390/horticulturae11121440
Kaur S, Thakur S, Sabharwal P, Meru G. Breeding for Disease Resistance in Cucumber: Current Status, Genetic Insights, and Genomic Resources. Horticulturae. 2025; 11(12):1440. https://doi.org/10.3390/horticulturae11121440
Chicago/Turabian StyleKaur, Simranjot, Shallu Thakur, Prerna Sabharwal, and Geoffrey Meru. 2025. "Breeding for Disease Resistance in Cucumber: Current Status, Genetic Insights, and Genomic Resources" Horticulturae 11, no. 12: 1440. https://doi.org/10.3390/horticulturae11121440
APA StyleKaur, S., Thakur, S., Sabharwal, P., & Meru, G. (2025). Breeding for Disease Resistance in Cucumber: Current Status, Genetic Insights, and Genomic Resources. Horticulturae, 11(12), 1440. https://doi.org/10.3390/horticulturae11121440







