Genome-Wide Identification and Expression Pattern Analysis of CrLBD Family Reveal Their Involvement in Floral Development in Chionanthus retusus
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. Identification of CrLBDs Transcription Factor Family Members in C. retusus
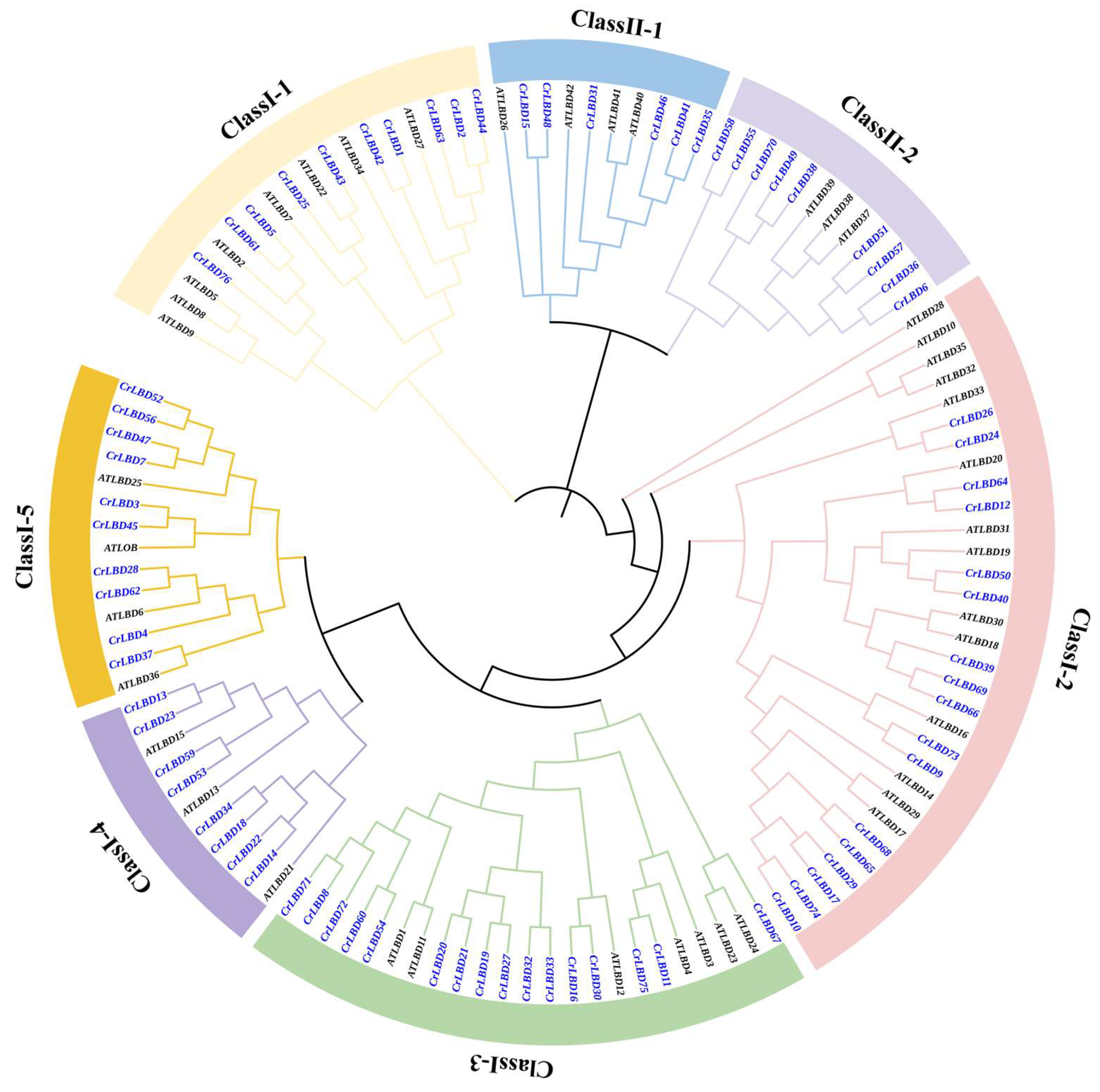
2.3. Phylogenetic Relationship Analysis
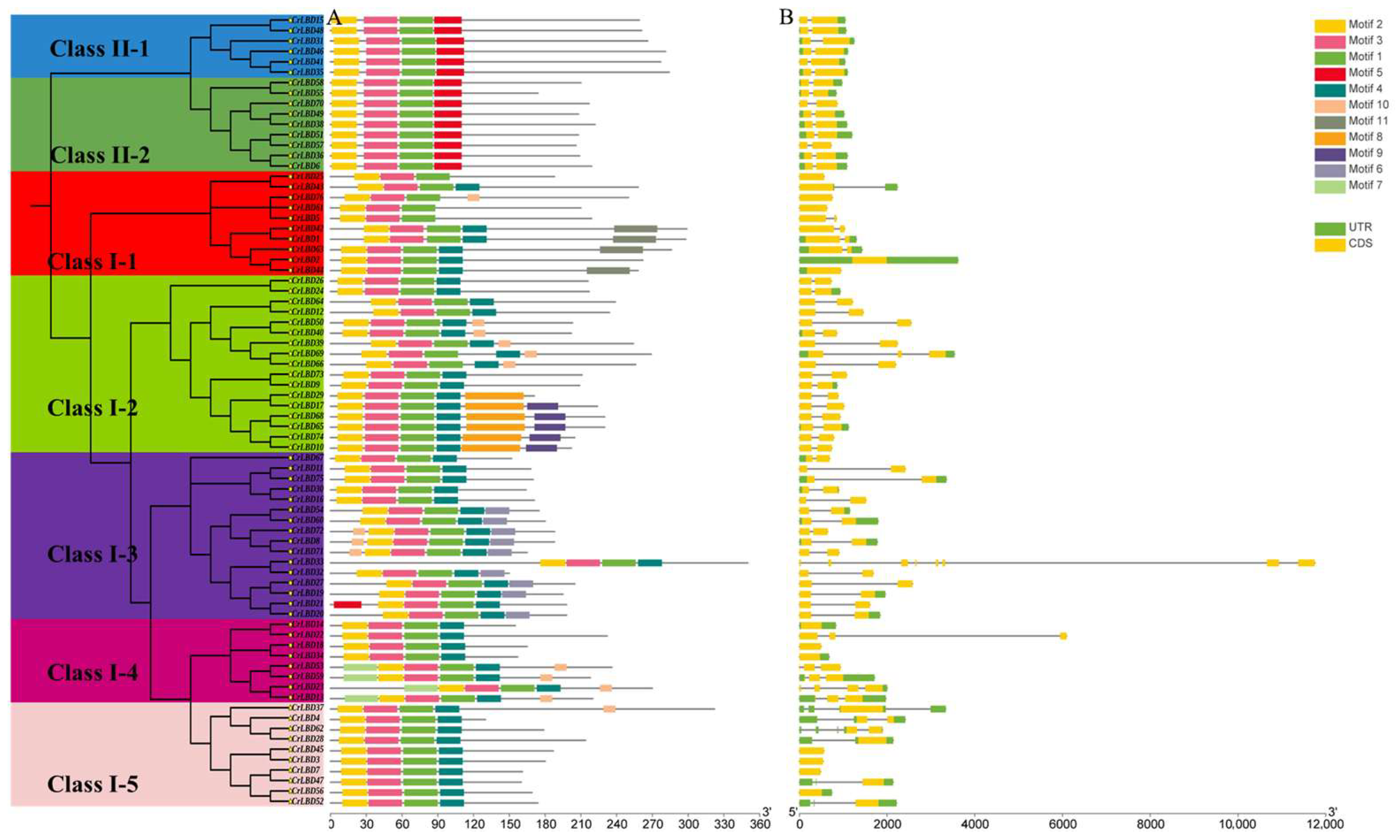
2.4. Analysis of CrLBDs Structure, Conserved Motifs and Cis Acting Elements
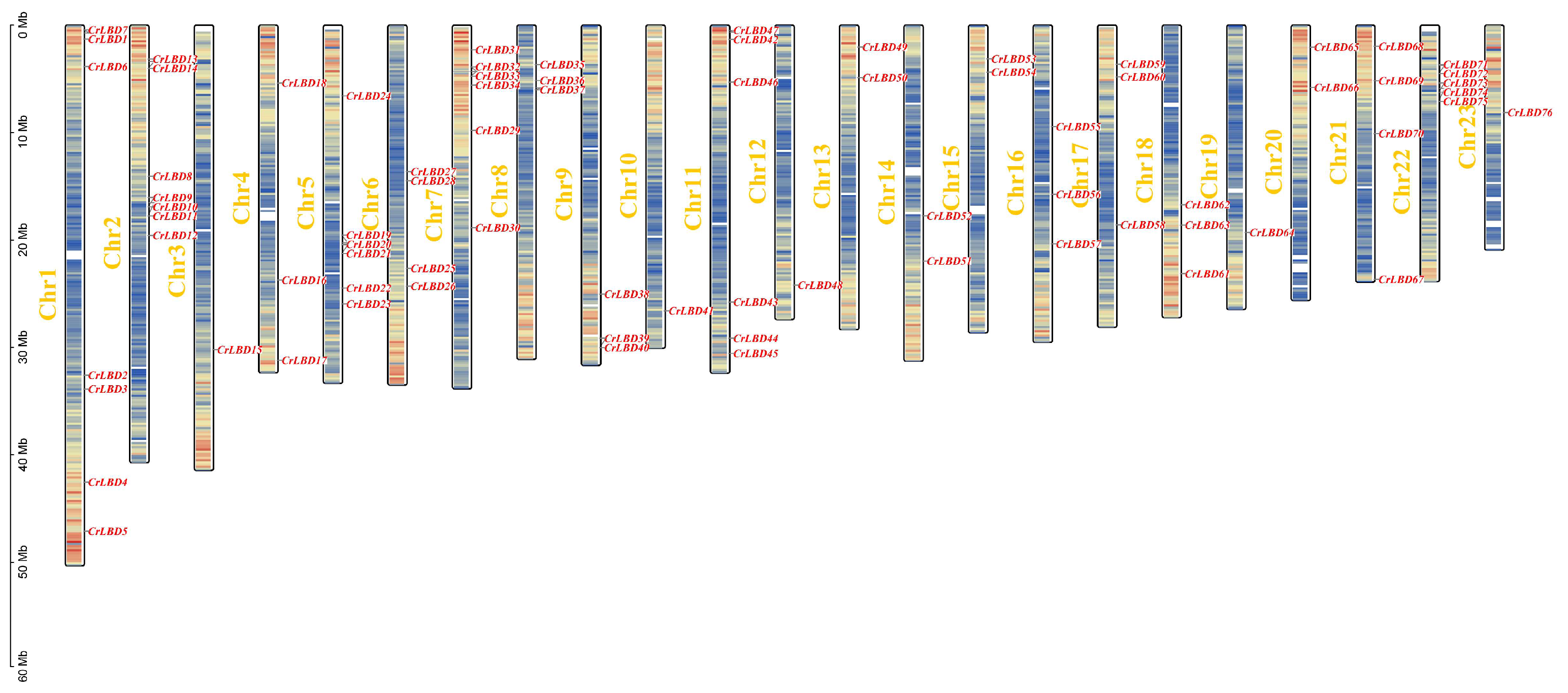
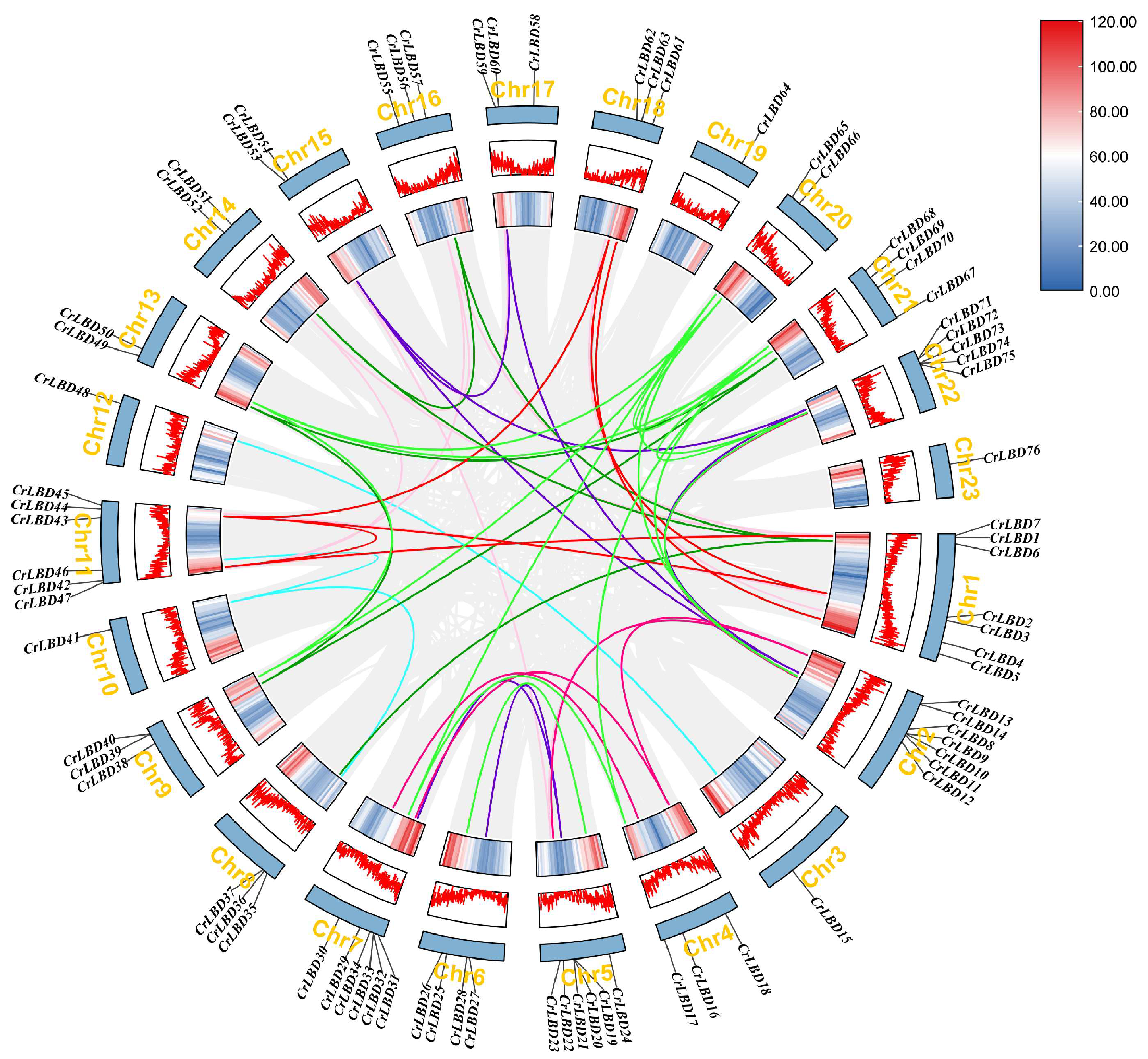
2.5. Collinearity and Chromosome Location Analysis
2.6. Analysis About CrLBD Genes Expression of qRT-PCR During Flower Development
2.7. Gene Expression Profiling and Co-Expression Network Analysis to Investigate CrLBD Genes Regulation
3. Results
3.1. Genome-Wide Identification and Phylogenetic Analysis of the LBD Gene Family in C. retusus
3.2. Analysis of Chromosomal Location, Collinearity and Replication of LBD Gene Family in C. retusus
3.3. Analysis of Gene Structure, Conserved Motifs and Conserved Domains of CrLBD Gene Family in C. retusus
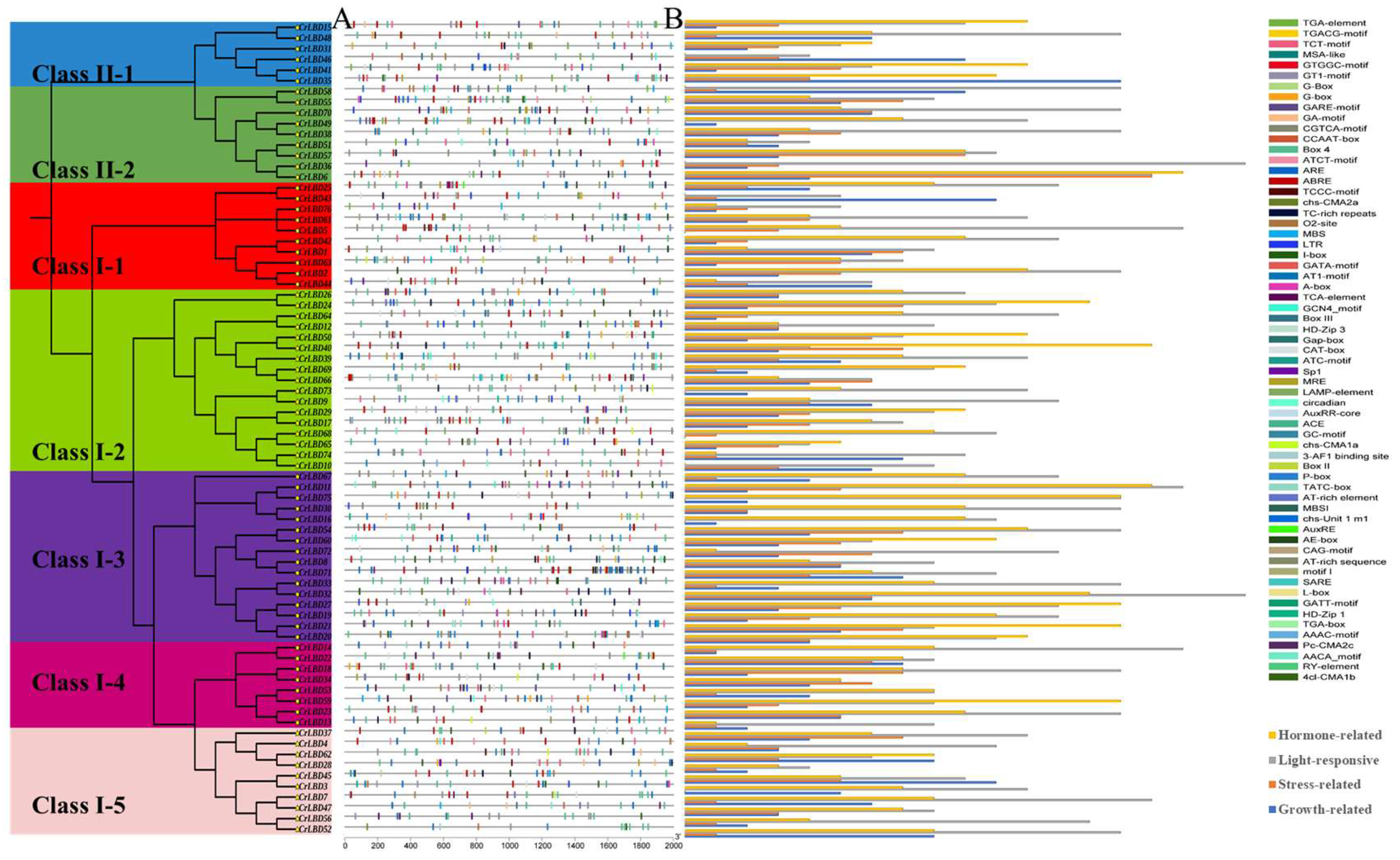
3.4. Identification and Enrichment Analysis of Cis-Acting Elements in the CrLBD Promoter of C. retusus
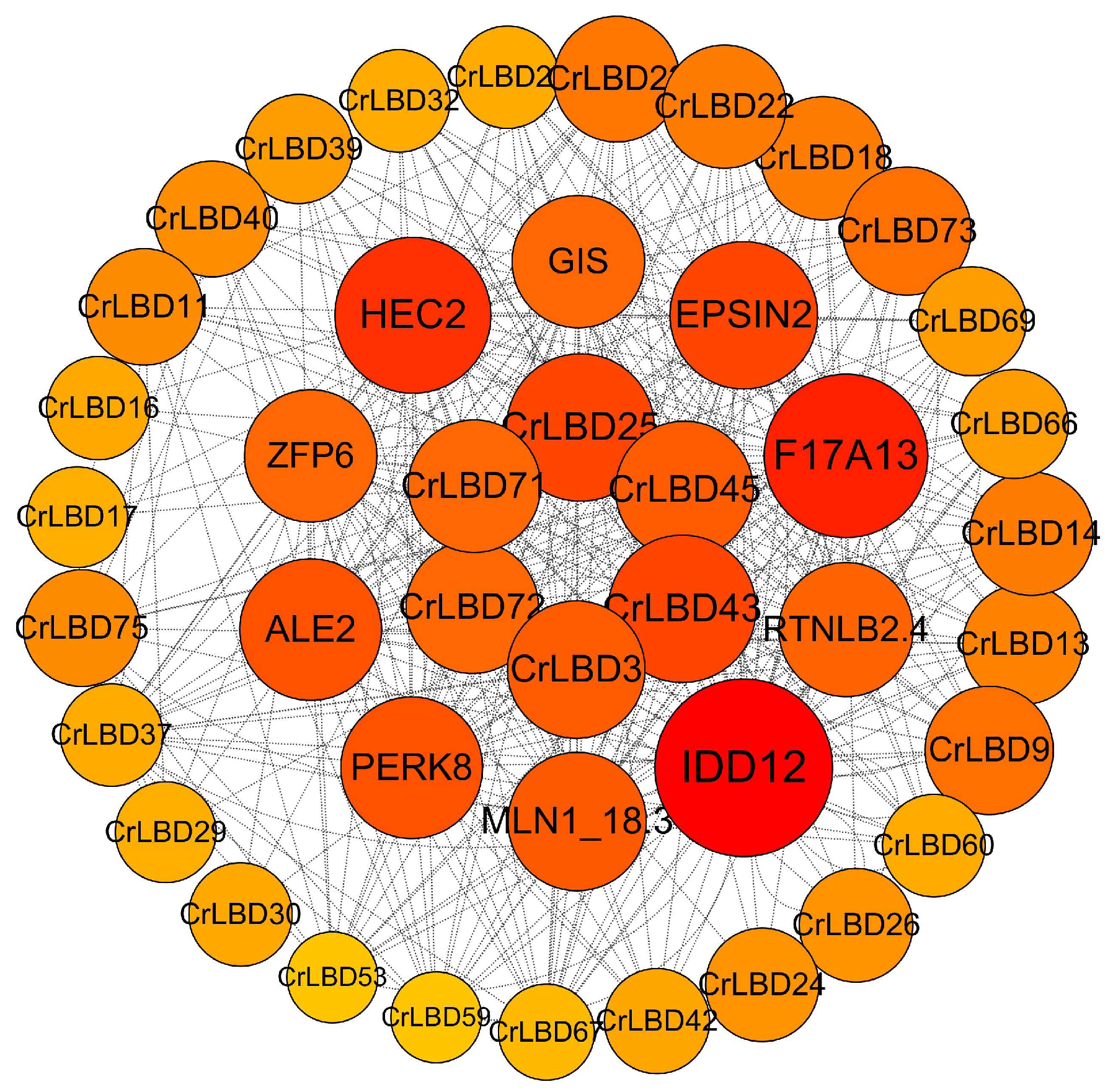
3.5. The Regulatory Network of the CrLBD Gene Family in C. retusus
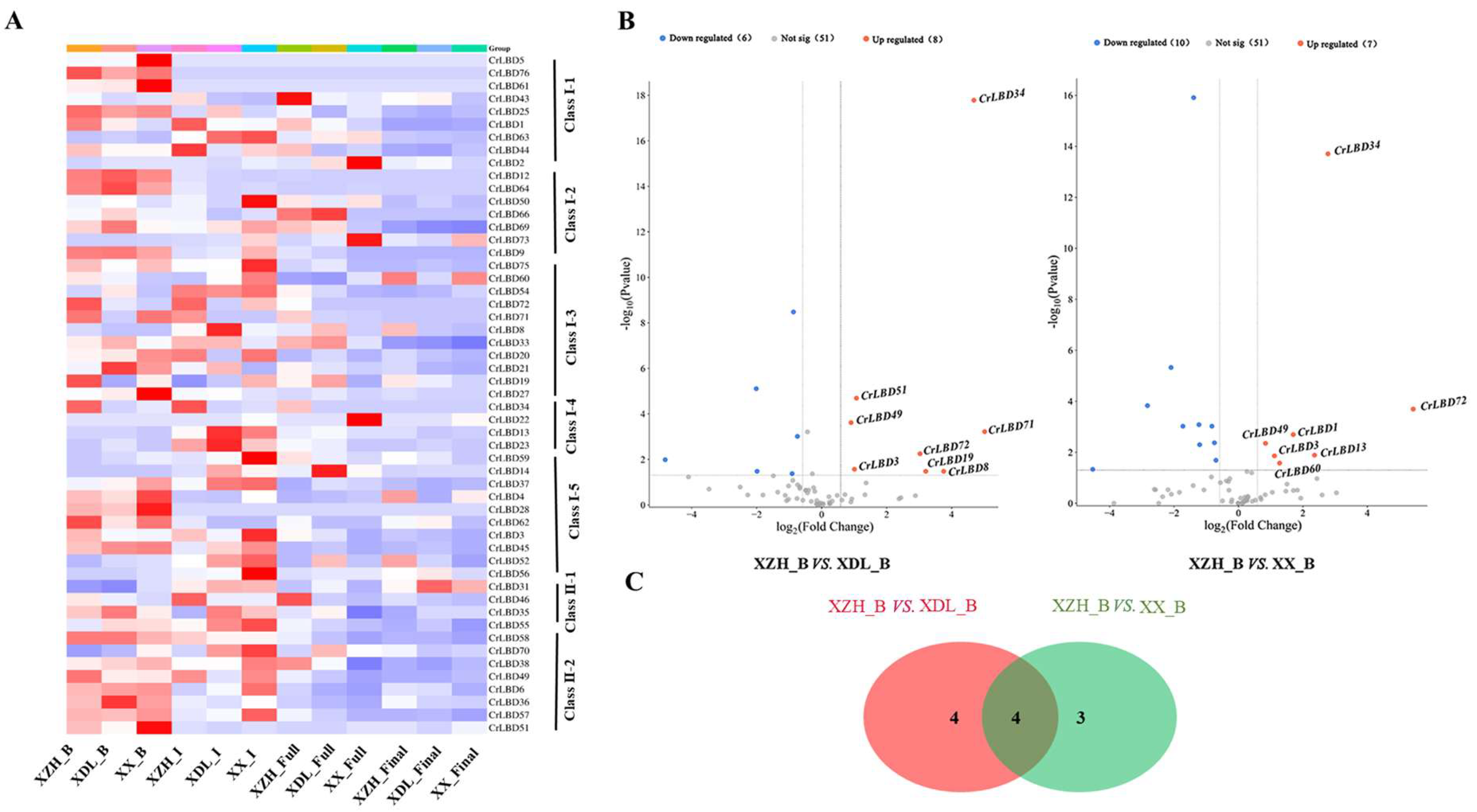
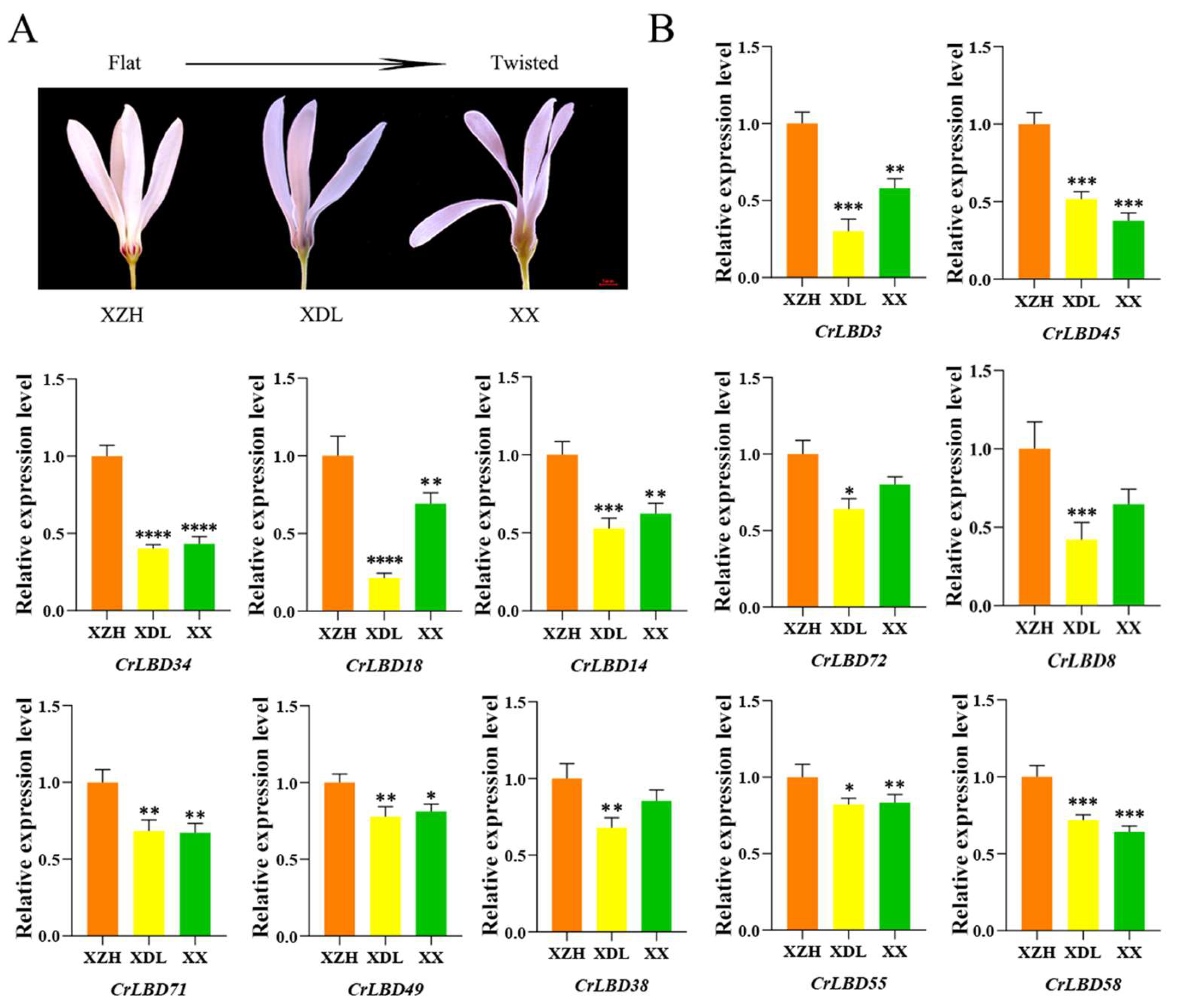
3.6. Analysis of Differential Expression Patterns of LBD Genes Related to Flower Morphological Diversity of C. retusus
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bretzel, F.; Vannucchi, F.; Romano, D.; Malorgio, F.; Benvenuti, S.; Pezzarossa, B. Wildflowers: From conserving biodiversity to urban greening—A review. Urban For. Urban Green. 2016, 20, 428–436. [Google Scholar] [CrossRef]
- Ren, Y.; Ge, Y.; Ma, D.; Song, X.; Shi, Y.; Pan, K.; Chang, J. Enhancing plant diversity and mitigating BVOC emissions of urban green spaces through the introduction of ornamental tree species. Urban For. Urban Green. 2017, 27, 305–313. [Google Scholar] [CrossRef]
- De-kui, Z.; Qi-bai, X.; Ri-ming, H.A.O. Study on distribution and utilization of plants in genus osmanthus. J. Southwest For. Univ. 2004, 24, 23–26. [Google Scholar] [CrossRef]
- Atchudan, R.; Edison, T.N.J.I.; Chakradhar, D.; Perumal, S.; Shim, J.J.; Lee, Y.R. Facile green synthesis of nitrogen-doped carbon dots using Chionanthus retusus fruit extract and investigation of their suitability for metal ion sensing and biological applications. Sens. Actuators B Chem. 2017, 246, 497–509. [Google Scholar] [CrossRef]
- He, Y.; Liu, L.; Yang, S.; Dong, M.; Yuan, W.; Shang, F. Characterization of the complete chloroplast genome of Chinese fringetree (Chionanthus retusus). Conserv. Genet. Resour. 2017, 9, 431–434. [Google Scholar] [CrossRef]
- Yang, Y.; He, S. A Review on the Application of Chionanthus retusus in Greening Urban Gardens. Kunming Univ. 2022, 44, 110–115. [Google Scholar]
- Song, J.; Oak, M.; Hong, S. Morphological traits in an androdioecious species, Chionanthus retusus (Oleaceae). Flora 2016, 223, 129–137. [Google Scholar] [CrossRef]
- Song, J.; Hong, S. Identity and localization of floral scent components in an androdioecious species, Chionanthus retusus (Oleaceae). Asia-Pac. Biodivers. 2020, 13, 288–294. [Google Scholar] [CrossRef]
- Shuai, B.; Reynaga-Pena, C.; Springer, P. The Lateral Organ Boundaries Gene Defines a Novel, Plant-Specific Gene Family. Plant Physiol. 2002, 129, 747–761. [Google Scholar] [CrossRef]
- Lu, H.; Dai, Z.; Li, L.; Wang, J.; Miao, X.; Shi, Z. OsRAMOSA2 shapes panicle architecture through regulating pedicel length. Front. Plant Sci. 2017, 8, 1538. [Google Scholar] [CrossRef]
- Majer, C.; Hochholdinger, F. Defining the boundaries: Structure and function of LOB domain proteins. Trends Plant Sci. 2011, 16, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Li, A.; Zhang, Y.; Dai, Z. Progress of LBD gene family in higher plants. Mol. Plant Breed. 2006, 3, 301–308. [Google Scholar]
- Yang, Y.; Yu, X.; Wu, P. Comparison and evolution analysis of two rice subspecies LATERAL ORGAN BOUNDARIES domain gene family and their evolutionary characterization from Arabidopsis. Mol. Phylogenet. Evol. 2006, 39, 248–262. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, S.; Su, L.; Liu, X.; Hao, Y. A genome-wide analysis of the LBD (LATERAL ORGAN BOUNDARIES domain) gene family in Malus domestica with a functional characterization of MdLBD11. PLoS ONE 2013, 8, e57044. [Google Scholar] [CrossRef]
- Dang, H.; Yu, C.; Nan, S.; Li, Y.; Du, S.; Zhao, K.; Wang, S. Genome-wide identification and gene expression networks of LBD transcription factors in Populus trichocarpa. BMC Genom. 2024, 25, 920. [Google Scholar] [CrossRef]
- Goh, T.; Toyokura, K.; Yamaguchi, N.; Okamoto, Y.; Uehara, T.; Kaneko, S.; Takebayashi, Y.; Kasahara, H.; Ikeyama, Y.; Okushima, Y.; et al. Lateral root initiation requires the sequential induction of transcription factors LBD16 and PUCHI in Arabidopsis thaliana. New Phytol. 2019, 224, 749–760. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, P.; Di, P.; Wang, Y. Genome-wide analysis of LBD genes in the medicinal plant Panax ginseng reveals the roles and molecular mechanisms of PgLBD18 and PgLBD49 in regulating lateral root development. Ind. Crops Prod. 2025, 232, 121232. [Google Scholar] [CrossRef]
- Semiarti, E.; Ueno, Y.; Tsukaya, H.; Iwakawa, H.; Machida, C.; Machida, Y. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana regulates formation of a symmetric lamina, establishment of venation and repression of meristem-related homeobox genes in leaves. Development 2001, 128, 1771–1783. [Google Scholar] [CrossRef]
- Wang, L.; Wang, W.; Xu, D.; Chang, Z.; Cui, X. Overexpression of Arabidopsis ASL25/LBD28 gene affects leaf morphogenesis. Acta Bot. Boreali Occident. 2010, 30, 888–893. [Google Scholar]
- Kim, M.J.; Kim, M.; Kim, J. Combinatorial interactions between LBD10 and LBD27 are essential for male gametophyte development in Arabidopsis. Plant Signal. Behav. 2015, 10, e1044193. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Z.; Ma, B.; Hou, Q.; Wan, X. Phylogeny and functions of LOB domain proteins in plants. Mol. Sci. 2020, 21, 2278. [Google Scholar] [CrossRef]
- Koppolu, R.; Anwar, N.; Sakuma, S.; Tagiri, A.; Lundqvist, U.; Pourkheirandish, M.; Rutten, T.; Seiler, C.; Himmelbach, A.; Ariyadasa, R.; et al. Six-rowed spike4 (Vrs4) controls spikelet determinacy and row-type in barley. Proc. Natl. Acad. Sci. USA 2013, 110, 13198–13203. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Liu, C.; Liu, C.; Zhao, Y.; Xu, R. Genomewide analysis of the lateral organ boundaries domain gene family in Vitis vinifera. J. Genet. 2016, 95, 515–526. [Google Scholar] [CrossRef] [PubMed]
- Goh, T.; Joi, S.; Mimura, T.; Fukaki, H. The establishment of asymmetry in Arabidopsis lateral root founder cells is regulated by LBD16/ASL18 and related LBD/ASL proteins. Development 2012, 139, 883–893. [Google Scholar] [CrossRef] [PubMed]
- Cho, C.; Jeon, E.; Pandey, S.; Ha, S.; Kim, J. LBD13 positively regulates lateral root formation in Arabidopsis. Planta 2019, 249, 1251–1258. [Google Scholar] [CrossRef]
- Jeon, E.; Young Kang, N.; Cho, C.; Joon Seo, P.; Chung Suh, M.; Kim, J. LBD14/ASL17 positively regulates lateral root formation and is involved in ABA response for root architecture in Arabidopsis. Plant Cell Physiol. 2017, 58, 2190–2201. [Google Scholar] [CrossRef]
- Naito, T.; Yamashino, T.; Kiba, T.; Koizumi, N.; Kojima, M.; Sakakibara, H.; Mizuno, T. A link between cytokinin and ASL9 (ASYMMETRIC LEAVES 2 LIKE 9) that belongs to the AS2/LOB (LATERAL ORGAN BOUNDARIES) family genes in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 2007, 71, 1269–1278. [Google Scholar] [CrossRef]
- Li, C.; Zou, X.; Zhang, C.; Shao, Q.; Liu, J.; Liu, B.; Li, H.; Zhao, T. OsLBD3-7 Overexpression Induced Adaxially Rolled Leaves in Rice. PLoS ONE 2016, 11, e0156413. [Google Scholar] [CrossRef]
- Li, C.; Zhu, S.; Zhang, H.; Chen, L.; Cai, M.; Wang, J.; Chai, J.; Wu, F.; Cheng, Z.; Guo, X.; et al. OsLBD37 and OsLBD38, two class II type LBD proteins, are involved in the regulation of heading date by controlling the expression of Ehd1 in rice. Biochem. Biophys. Res. Commun. 2017, 486, 720–725. [Google Scholar] [CrossRef]
- Wang, Y.; Song, J.; Wang, Z. ASYMMETRIC LEAVES2-LIKE38, one member of AS2/LOB gene family, involves in regulating ab-adaxial patterning in Arabidopsis lateral organs. Acta Physiol. Plant. 2015, 37, 185. [Google Scholar] [CrossRef]
- Causier, B.; Ashworth, M.; Guo, W.; Davies, B. The TOPLESS Interactome: A Framework for Gene Repression in Arabidopsis. Plant Physiol. 2012, 158, 423–438. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Wang, Z.; He, X.; Zhou, H.; Xu, Y.; Hong, L. PRESSED FLOWER works downstream of ASYMMETRIC LEAVES 2 to affect sepal flatness in Arabidopsis. Exp. Bot. 2025, 76, 4101–4113. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wu, L.; Li, J.; Cao, Q.; Gong, P.; Wang, L.; He, C. Heterometric expression of an LBD gene via LBD-TCP assembly regulates floral organ size and fruit weight in Physalis. Hortic. Res. 2025, 12, uhaf211. [Google Scholar] [CrossRef] [PubMed]
- Bortiri, E.; Chuck, G.; Vollbrecht, E.; Rocheford, T.; Martienssen, R.; Hake, S. ramosa2 Encodes a LATERAL ORGAN BOUNDARY Domain Protein That Determines the Fate of Stem Cells in Branch Meristems of Maize. Plant Cell 2006, 18, 574–585. [Google Scholar] [CrossRef]
- Wang, J.; Xu, D.; Sang, Y.L.; Sun, M.; Liu, C.; Niu, M.; Li, J. A telomere-to-telomere gap-free reference genome of Chionanthus retusus provides insights into the molecular mechanism underlying petal shape changes. Hortic. Res. 2024, 11, uhae249. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.; Frank, M.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bryant, S. CD-Search: Protein domain annotations on the fly. Nucleic Acids Res. 2004, 32, W327–W331. [Google Scholar] [CrossRef]
- Bailey, T.; Boden, M.; Buske, F.; Frith, M.; Grant, C.; Clementi, L.; Ren, J.; Li, W.; Noble, W. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Ma, B.; Wu, J.; Shi, L.; Yang, Y.; Wang, B.; Zheng, Y.; Su, S.; Yao, Y.; Xue, W.; Porth, I.; et al. Lilac (Syringa oblata) genome provides insights into its evolution and molecular mechanism of petal color change. Commun. Biol. 2022, 5, 686. [Google Scholar] [CrossRef] [PubMed]
- Rao, G.; Zhang, J.; Liu, X.; Lin, C.; Xin, H.; Xue, L.; Wang, C. De novo assembly of a new Olea europaea genome accession using nanopore sequencing. Hortic. Res. 2021, 8, 64. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Finn, R.; Clements, J.; Eddy, S. HMMER web server: Interactive sequence similarity searching. Nucleic Acids Res. 2011, 39, W29–W37. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Fan, S.; Zhang, D.; Xing, L.; Qi, S.; Du, L.; Wu, H.; Han, M. Phylogenetic analysis of IDD gene family and characterization of its expression in response to flower induction in Malus. Mol. Genet. Genom. 2017, 292, 755–771. [Google Scholar] [CrossRef]
- Ballester, P.; Martínez-Godoy, M.; Ezquerro, M.; Navarrete-Gómez, M.; Trigueros, M.; Rodríguez-Concepción, M.; Ferrándiz, C. A transcriptional complex of NGATHA and bHLH transcription factors directs stigma development in Arabidopsis. Plant Cell 2021, 33, 3645–3657. [Google Scholar] [CrossRef]
- Yang, C.; Sun, L.; Yasin, M.; Zulqarnain, H.; Ali, B.; Liu, Y.; Gan, Y. GIS mediates GA signaling to directly target the expression of SPL15 to regulate trichome development in Arabidopsis thaliana. Plant Physiol. Biochem. 2025, 225, 109975. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, R.; Cheng, Y.; Lei, P.; Song, W.; Zheng, W.; Nie, X. Genome-wide identification, evolution, and expression analysis of LBD transcription factor family in bread wheat (Triticum aestivum L.). Front. Plant Sci. 2021, 12, 721253. [Google Scholar] [CrossRef]
- Chalfun-Junior, A.; Franken, J.; Mes, J.; Marsch-Martinez, N.; Pereira, A.; Angenent, G. ASYMMETRIC LEAVES2-LIKE1 gene, a member of the AS2/LOB family, controls proximal–distal patterning in Arabidopsis petals. Plant Mol. Biol. 2005, 57, 559–575. [Google Scholar] [CrossRef]
- Gendron, J.; Liu, J.; Fan, M.; Bai, M.; Wenkel, S.; Springer, P.; Barton, M.; Wang, Z. Brassinosteroids regulate organ boundary formation in the shoot apical meristem of Arabidopsis. Proc. Natl. Acad. Sci. USA 2012, 109, 21152–21157. [Google Scholar] [CrossRef]
- Sun, Y.; Fan, X.; Cao, D.; Tang, W.; He, K.; Zhu, J.; He, J.; Bai, M.; Zhu, S.; Oh, E.; et al. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev. Cell 2010, 19, 765–777. [Google Scholar] [CrossRef]
- Bell, E.; Lin, W.; Husbands, A.; Yu, L.; Jaganatha, V.; Jablonska, B.; Mangeon, A.; Neff, M.; Girke, T.; Springer, P. Arabidopsis LATERAL ORGAN BOUNDARIES negatively regulates brassinosteroid accumulation to limit growth in organ boundaries. Proc. Natl. Acad. Sci. USA 2012, 109, 21146–21151. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Guo, L.; Wang, H.; Wu, Y.; Zhao, S.; Song, W.; Li, J.; Wang, J. Genome-Wide Identification and Expression Pattern Analysis of CrLBD Family Reveal Their Involvement in Floral Development in Chionanthus retusus. Horticulturae 2025, 11, 1429. https://doi.org/10.3390/horticulturae11121429
Wang M, Guo L, Wang H, Wu Y, Zhao S, Song W, Li J, Wang J. Genome-Wide Identification and Expression Pattern Analysis of CrLBD Family Reveal Their Involvement in Floral Development in Chionanthus retusus. Horticulturae. 2025; 11(12):1429. https://doi.org/10.3390/horticulturae11121429
Chicago/Turabian StyleWang, Mengmeng, Liyang Guo, Haiyan Wang, Yuzhu Wu, Shicong Zhao, Wenjing Song, Jihong Li, and Jinnan Wang. 2025. "Genome-Wide Identification and Expression Pattern Analysis of CrLBD Family Reveal Their Involvement in Floral Development in Chionanthus retusus" Horticulturae 11, no. 12: 1429. https://doi.org/10.3390/horticulturae11121429
APA StyleWang, M., Guo, L., Wang, H., Wu, Y., Zhao, S., Song, W., Li, J., & Wang, J. (2025). Genome-Wide Identification and Expression Pattern Analysis of CrLBD Family Reveal Their Involvement in Floral Development in Chionanthus retusus. Horticulturae, 11(12), 1429. https://doi.org/10.3390/horticulturae11121429



