Genome-Wide Identification and Characterization of Tomato Acyl-CoA Oxidase Family Genes ACX
Abstract
1. Introduction
2. Materials and Methods
2.1. The Genome-Wide Identification of ACX Gene Family Members in Solanum lycopersicum
2.2. Physicochemical Property Analysis of the SlACX Gene Family
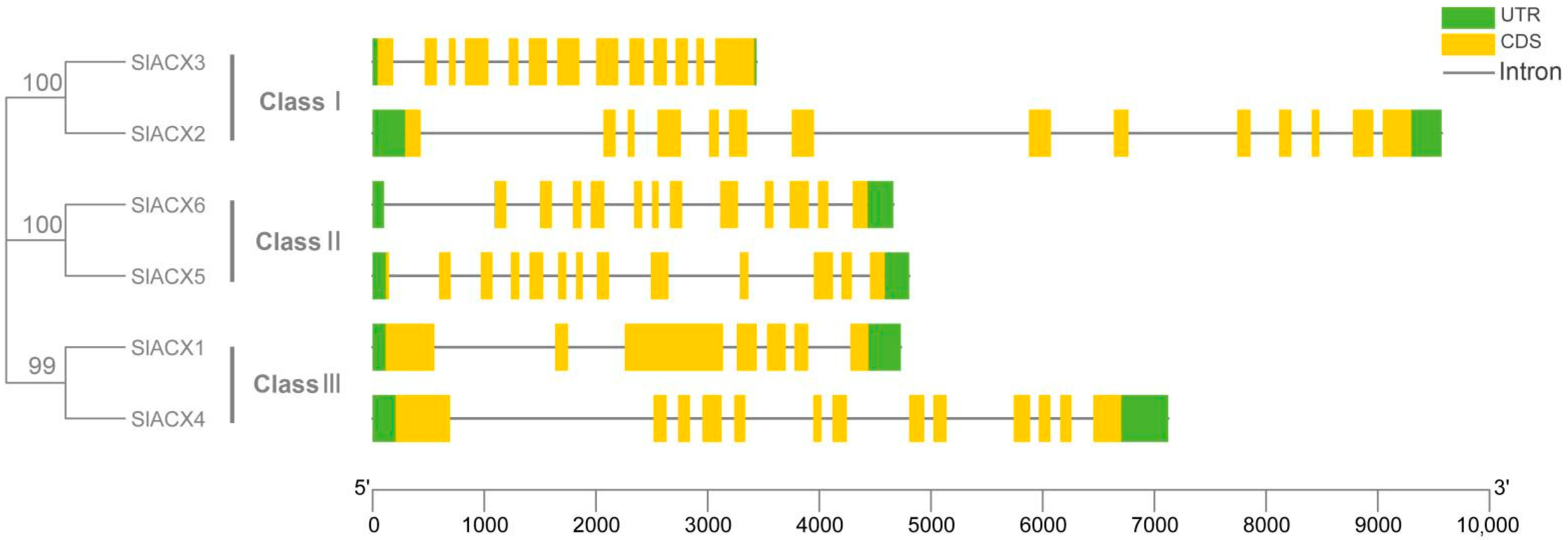
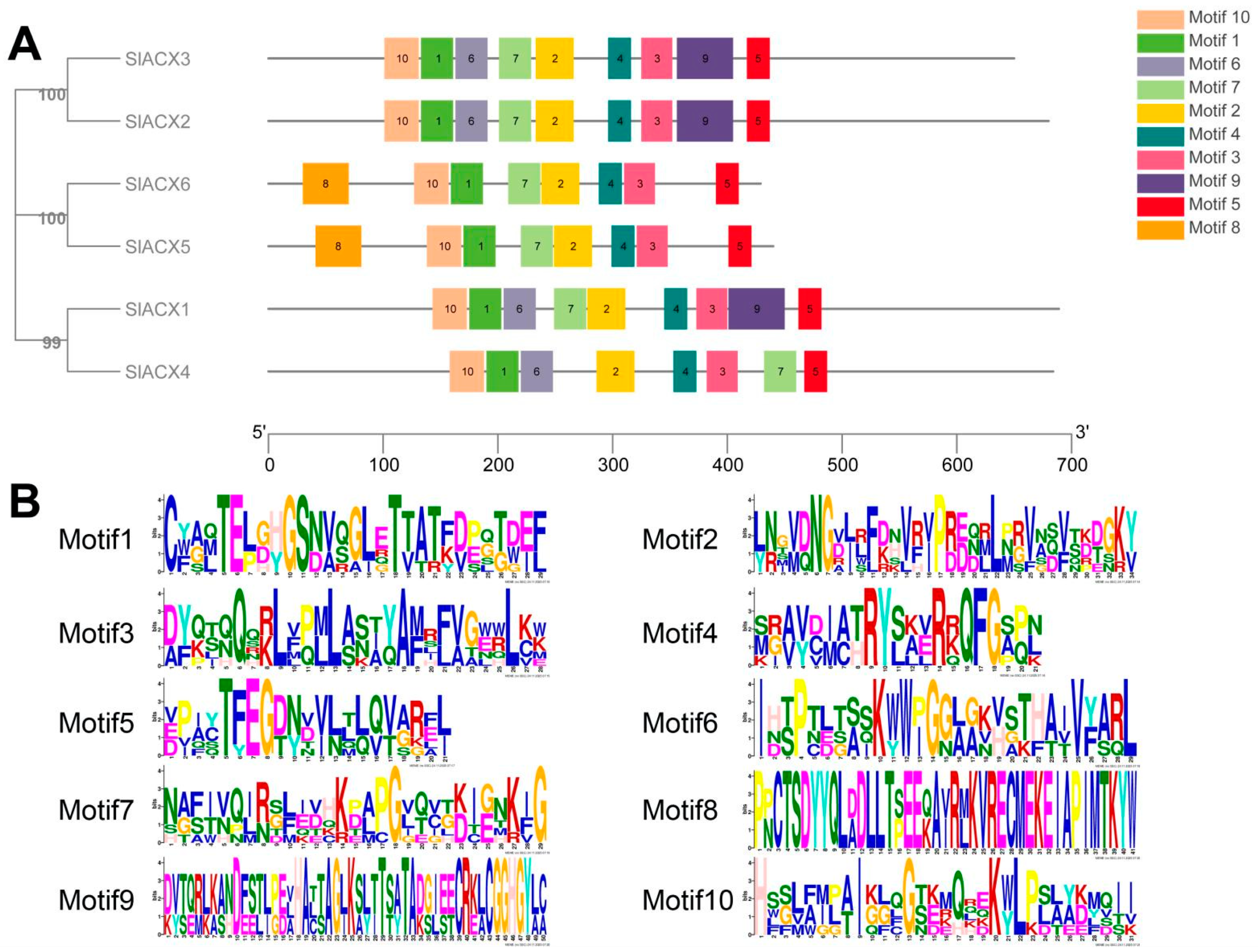
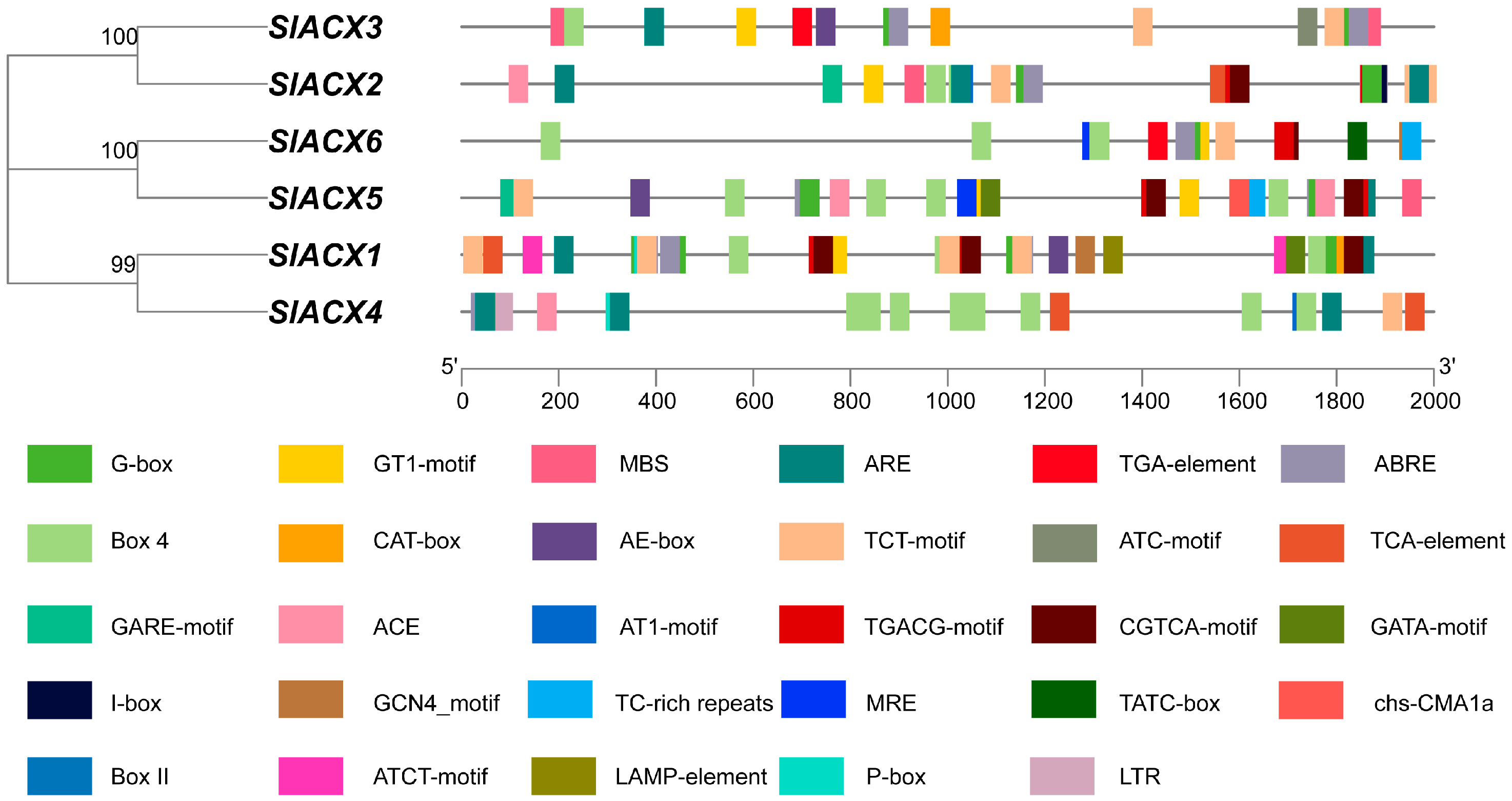
2.3. SlACX Family Gene Structure and Motifs Analysis
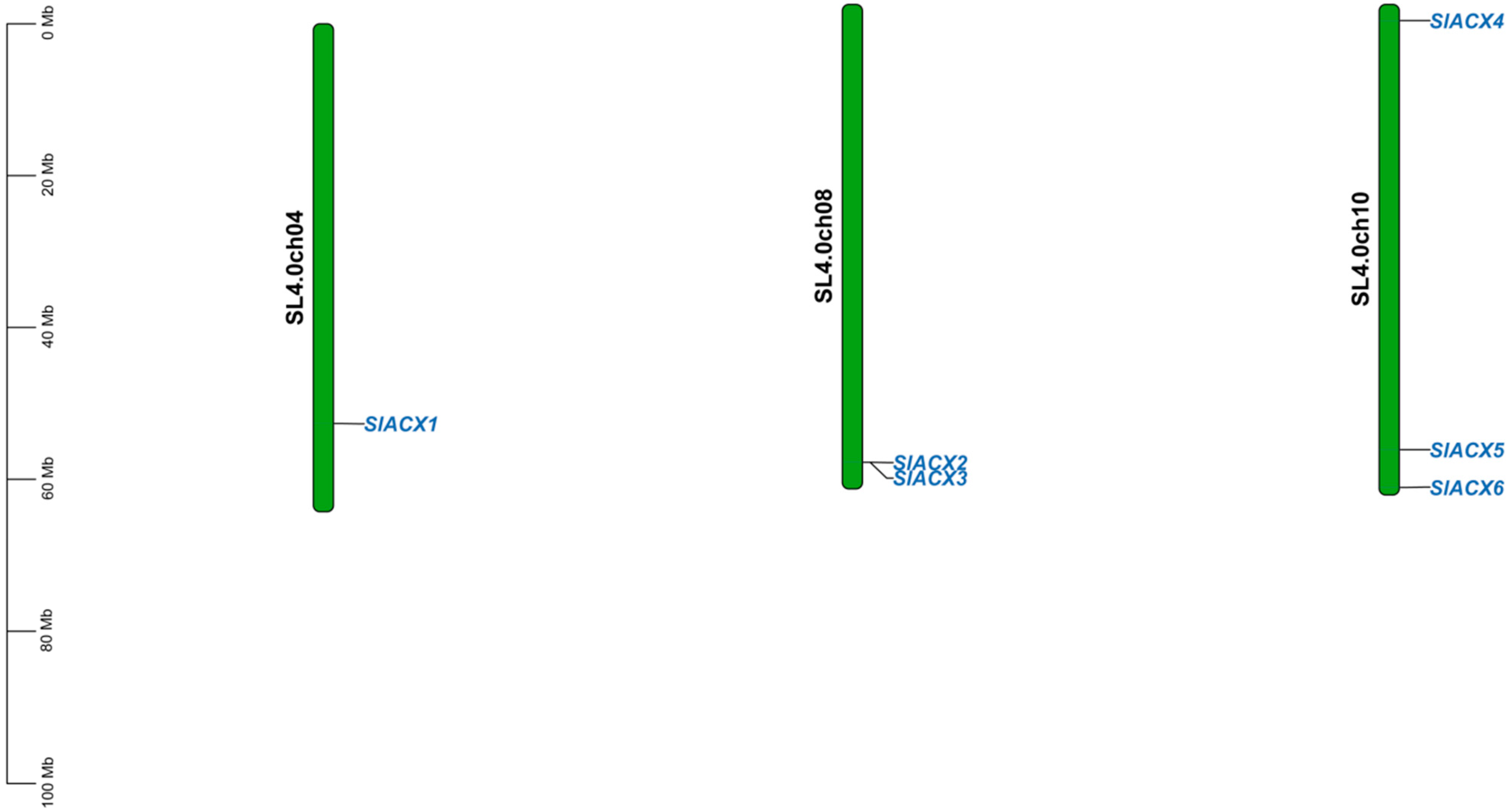
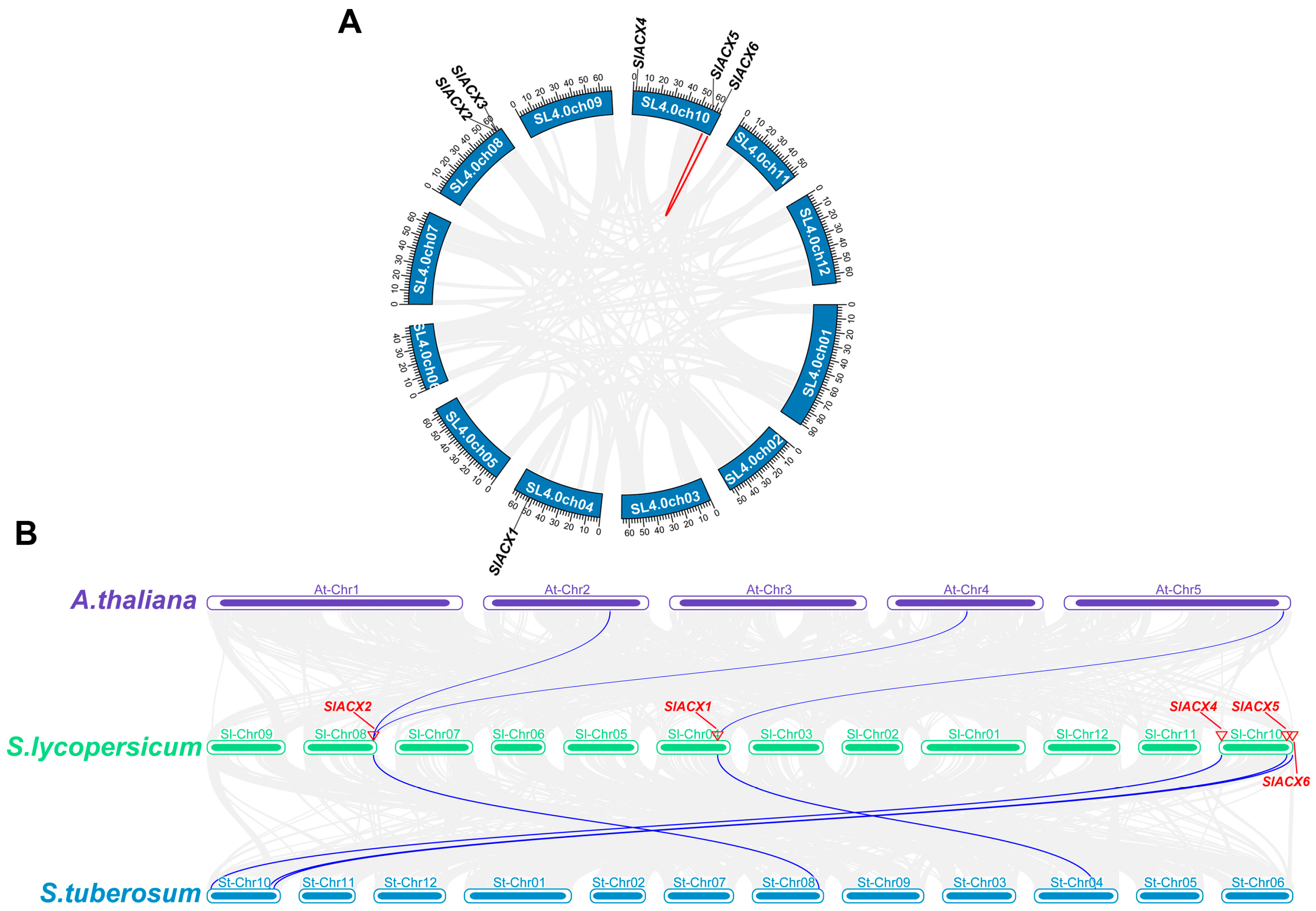
2.4. Chromosomal Localization Analysis of the SlACX Gene Family
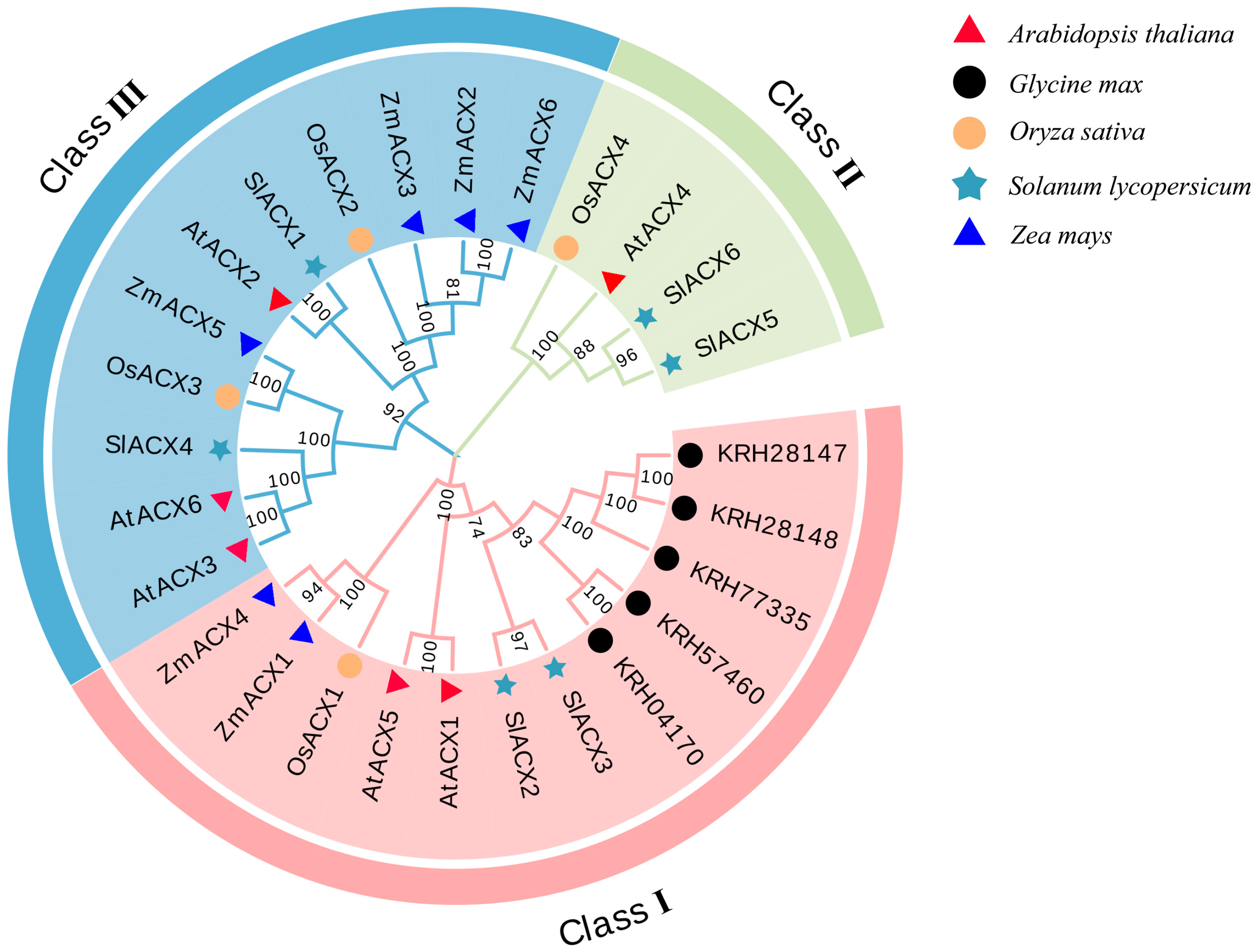
2.5. Phylogenetic Tree and Collinearity Analysis of the SlACX Gene Family
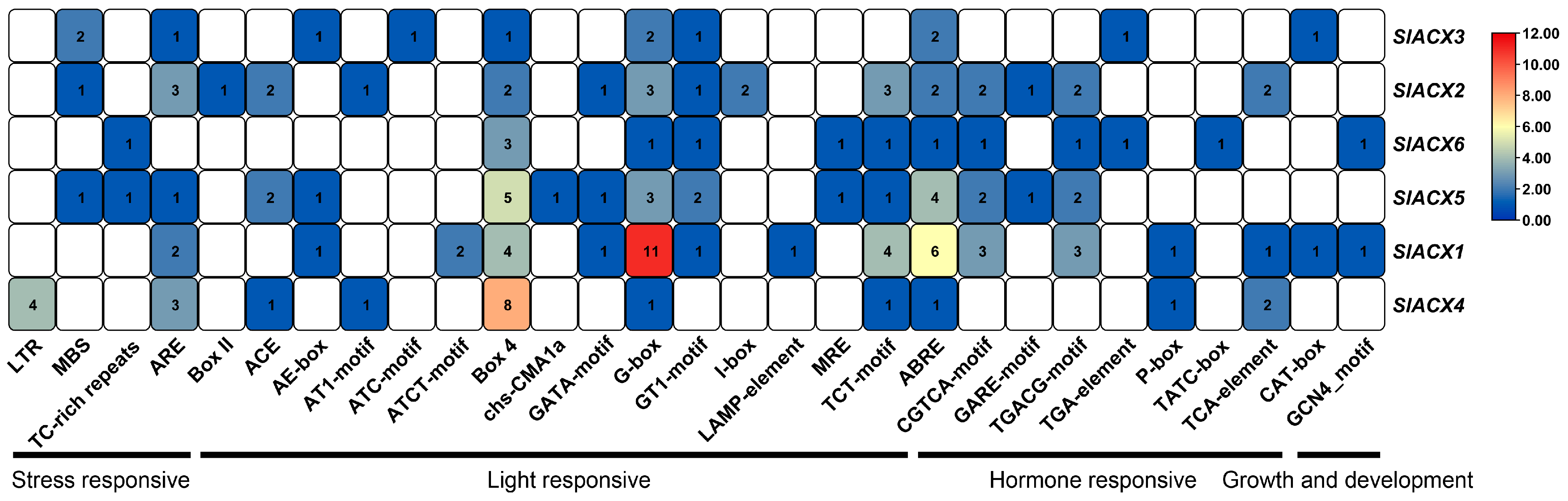
2.6. Analysis of Cis-Acting Elements of the SlACX Gene Family
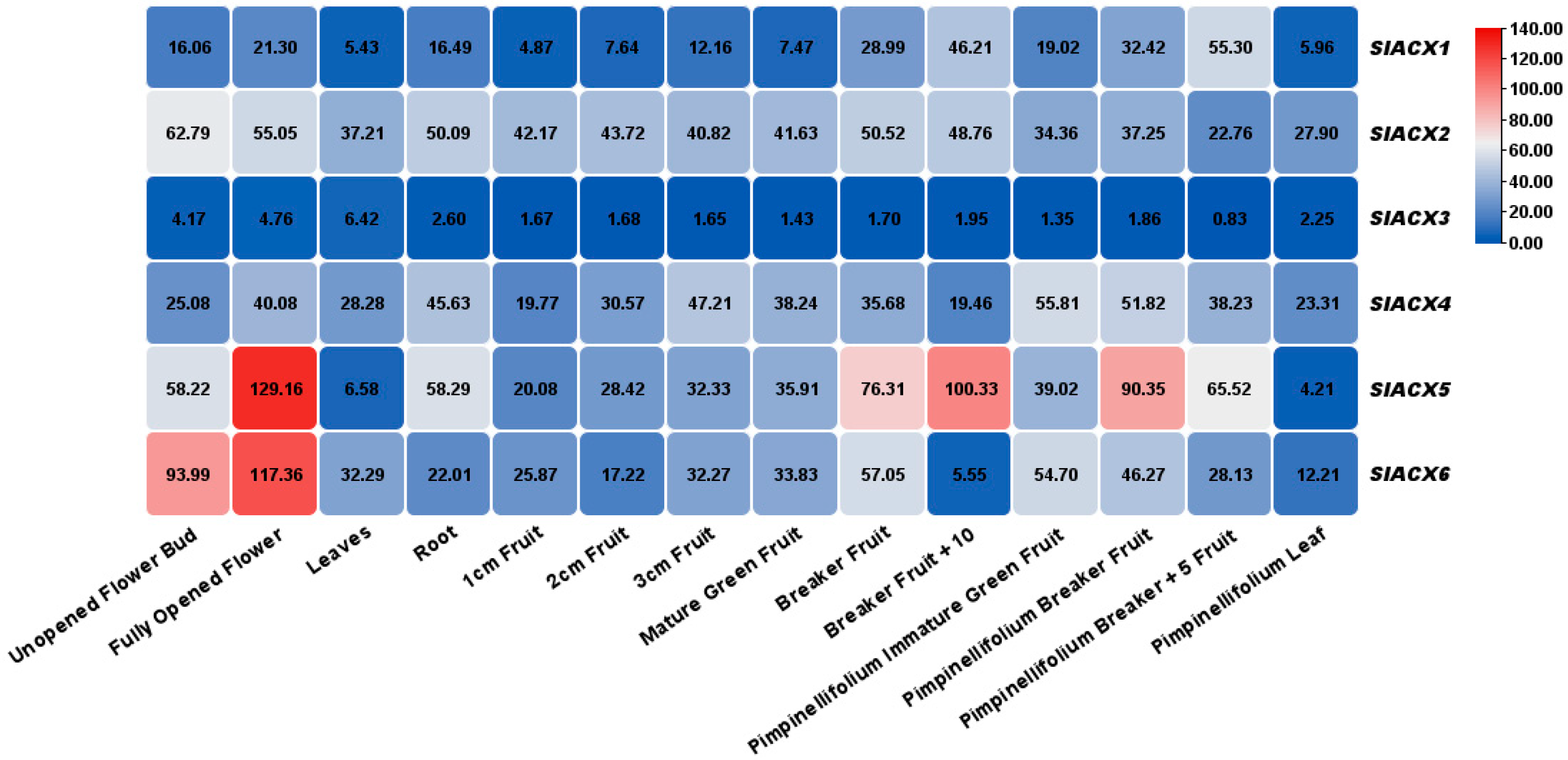
2.7. Analysis of Tissue Specificity and Expression Under Different Stress Treatments
2.8. RNA Extraction and Real-Time Fluorescence Quantitative Analysis
2.9. Statistical Analysis
3. Results
3.1. Genome-Wide Identification of SlACX Genes in Solanum lycopersicum
3.2. Analysis of Conserved Motifs and Gene Structure of the SlACX Family Genes
3.3. Chromosomal Location of Tomato SlACX Genes
3.4. Conserved Motif Analysis of Tomato SlACX Proteins
3.5. Phylogenetic Analysis of SlACX Family Members
3.6. Analysis of Gene Duplication and Collinearity of SlACX Genes
3.7. Cis-Acting Element Analysis of Tomato SlACX Genes
3.8. Expression Analysis of SlACX Genes in Different Organs
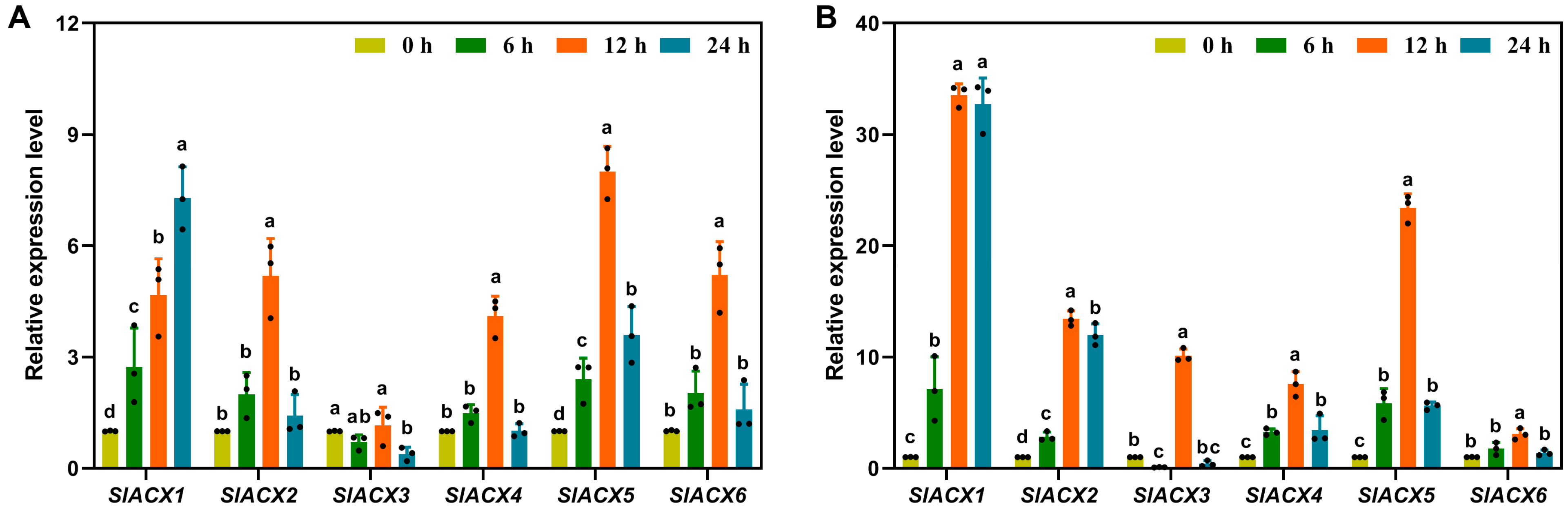
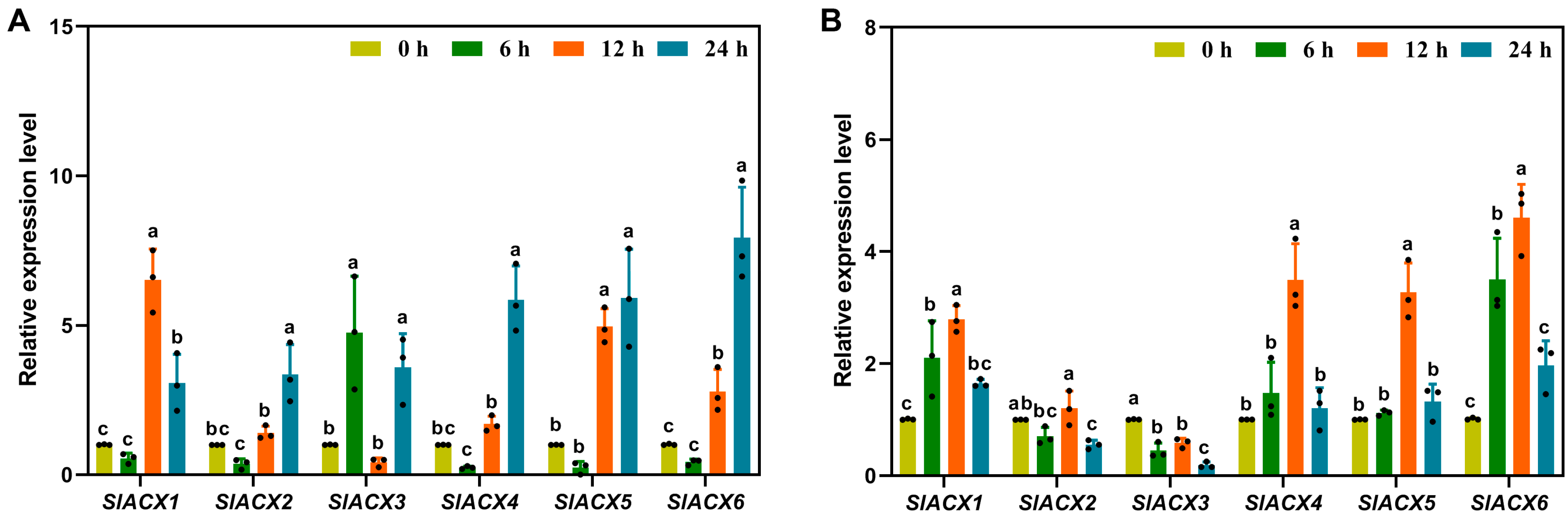
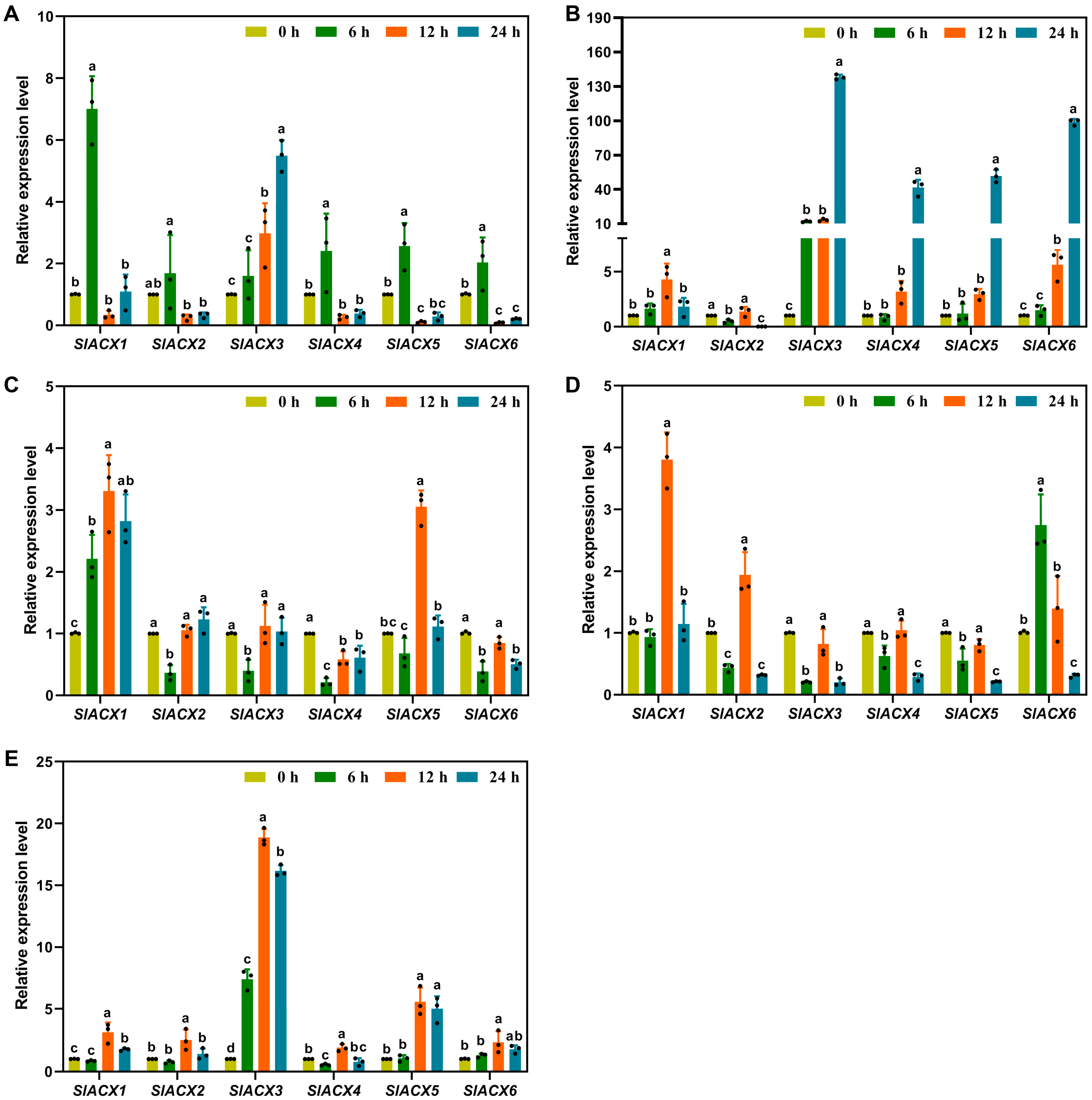
3.9. Expression Analysis of Tomato SlACX Genes Under Hormonal and Abiotic Stress
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Li, Y.; Zhang, F.; Chen, B.; Du, W.; Fang, S.; Wang, G.; Hu, J.; Li, L.; Hu, Z.; Jia, Z.; et al. Identification of MFS gene family in tomato and functional characterization of SlZIF1/SlMFS4 under salt stress. Plant Physiol. Biochem. PPB 2025, 229, 110554. [Google Scholar] [CrossRef]
- He, R.; Ma, W.; Zhou, F.; Cao, H.; Kang, Z.; Dong, J.; Xing, J. Molecular characterization of acyl-CoA oxidase (ACX) family genes in maize reveals their role in disease resistance. Genes 2025, 16, 486. [Google Scholar] [CrossRef]
- Xin, Z.; Chen, S.; Ge, L.; Li, X.; Sun, X. The involvement of a herbivore-induced acyl-CoA oxidase gene, CsACX1, in the synthesis of jasmonic acid and its expression in flower opening in tea plant (Camellia sinensis). Plant Physiol. Biochem. 2019, 135, 132–140. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Han, T.; Yuan, H.; Yu, Z.; Zhang, L.; Zhang, B.; Zhai, S.; Zheng, S.; Lu, Y. CATALASE2 functions for seedling postgerminative growth by scavenging H2O2 and stimulating ACX2/3 activity in Arabidopsis. Plant Cell Environ. 2017, 40, 2720–2728. [Google Scholar] [CrossRef] [PubMed]
- Xi, W.; Zhang, B.; Liang, L.; Shen, J.; Wei, W.; Xu, C.; Allan, A.; Ferguson, I.; Chen, K. Postharvest temperature influences volatile lactone production via regulation of acyl-CoA oxidases in peach fruit. Plant Cell Environ. 2012, 35, 534–545. [Google Scholar] [CrossRef] [PubMed]
- Jia, D.; Bin, L.; Yan, H. Molecular cloning and characterization of three novel genes related to fatty acid degradation and their responses to abiotic stresses in Gossypium hirsutum L. J. Integr. Agric. 2012, 12, 582–588. [Google Scholar]
- Rafeiza, B.; Raquel, A.; Zolman, B.K. Peroxisomal acyl-CoA oxidase 4 activity differs between Arabidopsis accessions. Plant Mol. Biol. 2012, 78, 45–58. [Google Scholar]
- Yang, H.; Bai, X.; Muhammad, T.; Jia, C.; Baike, W.; Yu, Q.; Wang, J. GHMP gene family: Identification, evolutionary and expression analysis under various exogenous hormones and abiotic stress in tomato. BMC Plant Biol. 2025, 25, 850. [Google Scholar] [CrossRef]
- Mo, F.; Xue, X.; Wang, J.; Mozhen, C.; Liu, S.; Zhao, L.; Chen, X. Genome-wide analysis of KCS genes in tomato and functional characterization of SlKCS8 and SlKCS10 in drought tolerance. Plant Physiol. Biochem. 2025, 222, 109783. [Google Scholar] [CrossRef]
- Can, H.; Dogan, I.; Uras, E.M. Genome-wide screening of mitogen-activated protein kinase (MAPK) gene family and expression profile under heavy metal stress in Solanum lycopersicum. Biotechnol. Lett. 2025, 47, 27. [Google Scholar] [CrossRef]
- Cui, X.; Gu, J.; Liu, P.; Zhen, R.; Zhang, Y.; Wang, F.; Qi, M.; Liu, Y.; Li, T. Genome-wide identification and characterization of the thioredoxin (TRX) gene family in tomato (Solanum lycopersicum) and a functional analysis of SlTRX2 under salt stress. Plant Physiol. Biochem. 2025, 220, 109478. [Google Scholar] [CrossRef]
- Sun, H.; Yang, J.; Fan, B.; Wang, Y.; Chen, G.; Cheng, G. Genome-wide analysis of BURP domain-containing gene family in Solanum lycopersicum and functional analysis of SlRD1 under drought and salt stresses. Int. J. Mol. Sci. 2024, 25, 12539. [Google Scholar] [CrossRef]
- Xu, J.; Cui, J.; He, Q.; Liu, Y.; Lu, X.; Qi, J.; Xiong, J.; Yu, W.; Li, C. Genome-wide identification of HIPP and mechanism of SlHIPP4/7/9/21/26/32 mediated phytohormones response to Cd, osmotic, and salt stresses in tomato. Plant Physiol. Biochem. PPB 2024, 217, 109220. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Zhang, J.; Li, J. Genome-wide identification and functional analysis of the trehalose-6-phosphate phosphatase (TPP) gene family in tomato (Solanum lycopersicum L.) and the role of SlTPP3 under NaCl stress. Sci. Hortic. 2025, 345, 114161. [Google Scholar] [CrossRef]
- Almeida, J.; Fons, P.L.; Drapal, M.; Kit, L.; Eugenia, M.; Paul, D. The FIBRILLIN multigene family in tomato, their roles in plastoglobuli structure and metabolism. Plant J. Cell Mol. Biol. 2025, 123, e70447. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Dai, L.; Yang, Y.; Zhu, Y. Identification and expression pattern analysis of the PP2C gene family response to salt stress in tomato. Russ. J. Plant Physiol. 2025, 72, 122. [Google Scholar] [CrossRef]
- Cao, H.; Wang, D.; Li, X.; Zhang, Y.; Su, D.; Wang, L.; Xu, K.; Li, Z. Genome-wide identification and expression profiling of SlGeBP gene family in response to hormone and abiotic stresses in Solanum lycopersicum L. Int. J. Mol. Sci. 2025, 26, 6008. [Google Scholar] [CrossRef]
- Arif, M.; Abbas, H.; Mahmood, N.; Muhammad, U.; Muhammad, A.; Shahbaz, A.; Yao, X.; Xu, R.; Li, L. Genome-Wide analysis of the NRAMP gene family in Arabidopsis thaliana: Identification, expression and response to multiple heavy metal stresses and phytohormones. BMC Plant Biol. 2025, 25, 1305. [Google Scholar] [CrossRef]
- Zhang, H.; Yu, Z.; Yao, X.; Chen, J.; Chen, X.; Zhou, H.; Lou, Y.; Ming, F.; Jin, Y. Genome-wide identification and characterization of small auxin-up RNA (SAUR) gene family in plants: Evolution and expression profiles during normal growth and stress response. BMC Plant Biol. 2021, 21, 4. [Google Scholar] [CrossRef]
- Cheng, S.; Hao, L.; Sun, J.; Ren, H.; Song, H. Genome-wide analysis of the SnRK gene family in Caragana Korshinskii and their expression profiling under drought and nitrogen deposition. BMC Genom. 2025, 26, 838. [Google Scholar] [CrossRef]
- Fu, G.; Yang, Y.; Mahmood, T.; Liu, X.; Xie, Z.; Zhao, Z.; Dong, Y.; Tian, Y.; Jehanzeb, F.; Iram, S.; et al. Genome-Wide Characterization of the ABI3 Gene Family in Cotton. Genes 2025, 16, 854. [Google Scholar] [CrossRef]
- Martina, J.; Thomas, D. Phylogenetic analysis of the expansion of the MATH-BTB gene family in the grasses. Plant Signal. Behav. 2014, 9, e28242. [Google Scholar]
- Yue, H.; Yang, Y.; Cha, G. Genomic colinearity and transcriptional regulatory networks of BES1 gene family in horticultural Plants particularly kiwifruit and peach. Horticulturae 2023, 9, 971. [Google Scholar] [CrossRef]
- Lv, M.; Lan, X.; Zhang, D.; Juye, Z.; Zhi, L.; Wei, Q.; Cui, Y. Evolution and diversification of adenine phosphoribosyltransferase (APT) gene family revealed their roles on flowering time in Brassica napus L. Ind. Crops Prod. 2025, 234, 121627. [Google Scholar] [CrossRef]
- Bhar, S.; Prajapati, V.D.; Gonzalez, S.M.; Yoon, C.-S.; Kevin, M.; Laura, S.; Kari, B.B.; Rebecca, A.B. Comparative metabolomics identifies the roles of acyl-CoA oxidases in the biosynthesis of ascarosides and a complex family of secreted n-Acylethanolamines. ACS Chem. Biol. 2025, 20, 1298–1308. [Google Scholar] [CrossRef]
- Zhang, L.; Li, H.; Gao, L.; Qi, Y.; Fu, W.; Li, X.; Zhou, X.; Gao, Q.; Gao, Z.; Jia, H. Acyl-CoA oxidase 1 is involved in γ-decalactone release from peach (Prunus persica) fruit. Plant Cell Rep. 2017, 36, 829–842. [Google Scholar] [CrossRef]
- Zhou, Z.; Naoufal, L.; Dounya, K.; Cao, H.; Zhang, K.; Dong, J.; Xing, J. Genome-wide identification and analysis of soybean acyl-ACP thioesterase gene family reveals the role of GmFAT to improve fatty acid composition in soybean seed. Theor. Appl. Genet. 2021, 134, 3611–3623. [Google Scholar] [CrossRef]
- Rylott, E.; Rogers, C.; Gilday, A.; Edgell, T.; Larson, T.; Graham, I. Arabidopsis mutants in short- and medium-chain acyl-CoA oxidase activities accumulate acyl-CoAs and reveal that fatty acid β-Oxidation is essential for embryo development. J. Biol. Chem. 2003, 278, 21370–21377. [Google Scholar] [CrossRef]
- Zhu, X.; Hou, Z.; Xu, X.; Xiong, Z.; Chen, Y.; Yang, L.; Liu, Z.; Fang, Z. TMT-based comparative proteomics reveals the role of acyl-CoA oxidase 4 in enhancing the drought stress tolerance in common buckwheat (Fagopyrum esculentum). Int. J. Biol. Macromol. 2022, 215, 262–271. [Google Scholar] [CrossRef]
- Pedersen, L.; Henriksen, A. Acyl-CoA Oxidase 1 from Arabidopsis thaliana. Structure of a key enzyme in plant lipid metabolism. J. Mol. Biol. 2004, 345, 487–500. [Google Scholar] [CrossRef]
- Kim, C.M.; Kim, H.T.; Park, H.J.; Lee, C.; Cho, S. Expression of rice acyl-CoA oxidase isoenzymes in response to wounding. J. Plant Physiol. 2006, 164, 665–668. [Google Scholar] [CrossRef]
| Gene Name | Sequence (F) | Sequence (R) |
|---|---|---|
| SlACX1 | AGAACACTTACGCAACCCTAACTTC | TATTCCAAGCACCAAATCCTCCAAG |
| SlACX2 | CTCAAGAGGAAGCCATATGTTAAGG | CTGCTGTTTATCTGTGCCCTGTC |
| SlACX3 | CAACCACTGCCACTGCTGATG | CTGAAGTAGTAGCACGACGTTATCTC |
| SlACX4 | AAAAGGAGTAAGAAACCGGG | CAGGTCTCGTTCCCGTAGGC |
| SlACX5 | CAACACCAGCGTCCGTCTTTC | TTTCCATGCACTCTCTCACTTTCAG |
| SlACX6 | TGTCACAGGTACTTGAAGGAGAGG | ATAGCCGCCAACCAACAAGAATC |
| Gene | Gene ID | Gene Locus | ORF (bp) | Amino Acid | Molecular Weight | pI | ACX Domain Locations |
|---|---|---|---|---|---|---|---|
| SlACX1 | Solyc04g054890.3.1.ITAG4.0 | Chr04 | 2067 | 688 | 77,398.58 | 8.74 | 176–292; 323–484; 529–686 |
| SlACX2 | Solyc08g078390.4.1.ITAG4.0 | Chr08 | 1995 | 678 | 75,624.54 | 8.35 | 17–132; 134–247; 479–656 |
| SlACX3 | Solyc08g078400.3.1.ITAG4.0 | Chr08 | 1950 | 649 | 72,643.34 | 6.31 | 17–132; 134–246; 479–597 |
| SlACX4 | Solyc10g008110.4.1.ITAG4.0 | Chr10 | 2052 | 683 | 76,734.67 | 8.15 | 191–300; 331–489; 529–669 |
| SlACX5 | Solyc10g076600.2.1.ITAG4.0 | Chr10 | 1320 | 439 | 48,316.04 | 8.22 | 56–167; 171–263; 277–423 |
| SlACX6 | Solyc10g085200.2.1.ITAG4.0 | Chr10 | 1287 | 428 | 47,117.45 | 7.55 | 46–156; 160–252; 266–412 |
| Protein | Alpha Helix (%) | Beta Turn (%) | Random Coil (%) |
|---|---|---|---|
| SlACX1 | 55.09 | 5.81 | 28.05 |
| SlACX2 | 59.59 | 4.72 | 27.14 |
| SlACX3 | 60.09 | 4.31 | 27.27 |
| SlACX4 | 53.94 | 5.20 | 27.24 |
| SlACX5 | 46.01 | 5.24 | 34.85 |
| SlACX6 | 49.77 | 7.01 | 29.91 |
| Gene | Peroxisome | Endoplasmic Reticulum | Plasma Membrane | Chloroplast | Nucleus | Vacuole | Mitochondria | Cytoplasm |
|---|---|---|---|---|---|---|---|---|
| SlACX1 | — | 3 | 1 | 4 | 1.5 | 2 | 1 | 1 |
| SlACX2 | — | — | — | 1 | 2 | 1 | — | 10 |
| SlACX3 | 7 | 3 | 2 | 1 | — | 1 | — | — |
| SlACX4 | — | — | 1 | 5 | 4 | — | 3 | — |
| SlACX5 | 1 | 5 | 5 | 1 | — | 2 | — | — |
| SlACX6 | 12 | — | — | — | — | — | — | 2 |
| Motif | Width (aa) | Motif Sequence |
|---|---|---|
| Motif 1 | 29 | CYALTELGHGSNVQGLETTATFDPGTDEF |
| Motif 2 | 34 | LNGVDNGVJLFDNVRIPRDDLLPRVADVSKDGKY |
| Motif 3 | 28 | DYQTQQQKLVPLLASTYAFRFVGWRLKK |
| Motif 4 | 21 | SGAVDIATRYSAVRKQFGAPN |
| Motif 5 | 21 | EPIYTFEGDNDVLLLQVARFL |
| Motif 6 | 29 | IHSPTLTASKWWPGGLGKVSTHAIVYARL |
| Motif 7 | 29 | NAFIVQJRSLEDHKPAPGVQVTDIGNKIG |
| Motif 8 | 41 | PNCTSDYYQLDDLLTPEEKAIRLKVRECMEKEIAPIMTKYW |
| Motif 9 | 50 | DVTQRLKANDFSTLPEVHACTAGLKSLTTSATADGIEECRKLCGGHGYLC |
| Motif 10 | 31 | HSGLFIPAIKLQGSEMQKEKWLPSAYDMQII |
| Element | Sequence | Description |
|---|---|---|
| LTR | CCGAAA | cis-acting element involved in low-temperature responsiveness |
| MBS | CAACTG | MYB binding site involved in drought inducibility |
| TC-rich repeats | ATTCTCTAAC/GTTTTCTTAC | cis-acting element involved in defense and stress responsiveness |
| ARE | AAACCA | cis-acting regulatory element essential for the anaerobic induction |
| Box II | CCACGTGGC | part of a light-responsive element |
| ACE | CTAACGTATT/GACACGTATG | cis-acting element involved in light responsiveness |
| AE-box | AGAAACTT/AGAAACAA | part of a module for light response |
| AT1-motif | AATTATTTTTTATT | part of a light-responsive module |
| ATC-motif | AGTAATCT | part of a conserved DNA module involved in light responsiveness |
| ATCT-motif | AATCTAATCC | part of a conserved DNA module involved in light responsiveness |
| Box 4 | ATTAAT | part of a conserved DNA module involved in light responsiveness |
| chs-CMA1a | TTACTTAA | part of a light-responsive element |
| GATA-motif | GATAGGG/AAGGATAAGG/AAGATAAGATT | part of a light-responsive element |
| G-box | CCACGTAA/TACGTG/TAACACGTAG/GCCACGTGGA/CACGTC/TCCACATGGCA/CACGTG/CACGTT | cis-acting regulatory element involved in light responsiveness |
| GT1-motif | GGTTAA/GGTTAAT | light-responsive element |
| I-box | AAGATAAGGCT/AGATAAGG | part of a light-responsive element |
| LAMP-element | CTTTATCA | part of a light-responsive element |
| MRE | AACCTAA | MYB binding site involved in light responsiveness |
| TCT-motif | TCTTAC | part of a light-responsive element |
| ABRE | ACGTG/CACGTG/TACGTGTC | cis-acting element involved in the abscisic acid responsiveness |
| CGTCA-motif | CGTCA | cis-acting regulatory element involved in the MeJA responsiveness |
| GARE-motif | TCTGTTG | gibberellin-responsive element |
| TGACG-motif | TGACG | cis-acting regulatory element involved in the MeJA responsiveness |
| TGA-element | AACGAC | auxin-responsive element |
| P-box | CCTTTTG | gibberellin-responsive element |
| TATC-box | TATCCCA | cis-acting element involved in gibberellin responsiveness |
| TCA-element | CCATCTTTTT | cis-acting element involved in salicylic acid responsiveness |
| CAT-box | GCCACT | cis-acting regulatory element related to meristem expression |
| GCN4_motif | TGAGTCA | cis-regulatory element involved in endosperm expression |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.; Liu, Z.; Gao, Y.; Li, Q.; Wang, Q.; An, C. Genome-Wide Identification and Characterization of Tomato Acyl-CoA Oxidase Family Genes ACX. Horticulturae 2025, 11, 1426. https://doi.org/10.3390/horticulturae11121426
Wang C, Liu Z, Gao Y, Li Q, Wang Q, An C. Genome-Wide Identification and Characterization of Tomato Acyl-CoA Oxidase Family Genes ACX. Horticulturae. 2025; 11(12):1426. https://doi.org/10.3390/horticulturae11121426
Chicago/Turabian StyleWang, Chunlei, Zesheng Liu, Yanlong Gao, Qianbing Li, Qi Wang, and Caiting An. 2025. "Genome-Wide Identification and Characterization of Tomato Acyl-CoA Oxidase Family Genes ACX" Horticulturae 11, no. 12: 1426. https://doi.org/10.3390/horticulturae11121426
APA StyleWang, C., Liu, Z., Gao, Y., Li, Q., Wang, Q., & An, C. (2025). Genome-Wide Identification and Characterization of Tomato Acyl-CoA Oxidase Family Genes ACX. Horticulturae, 11(12), 1426. https://doi.org/10.3390/horticulturae11121426






