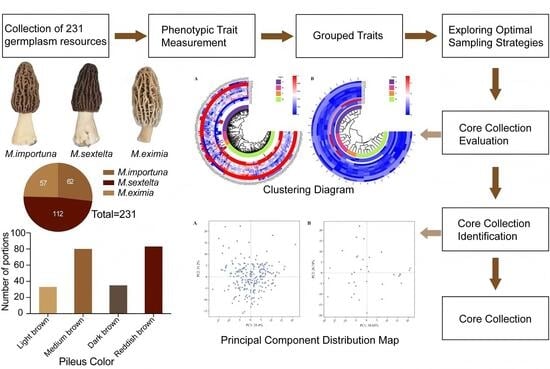
Construction of a Core Collection for Morchella Based on Phenotypic Traits from China
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. Field Trial
2.3. Data Organization
2.4. Screening of Sampling Strategies
2.5. Evaluation of Core Collection
2.6. Core Collection Validation
3. Results
3.1. Establishment of Core Collection
3.1.1. Analysis of Genetic Diversity in Morchella Germplasm Resources
3.1.2. Determination of Grouping and Sampling Methods
3.1.3. Sampling Strategy Determination
3.2. Core Collection Evaluation
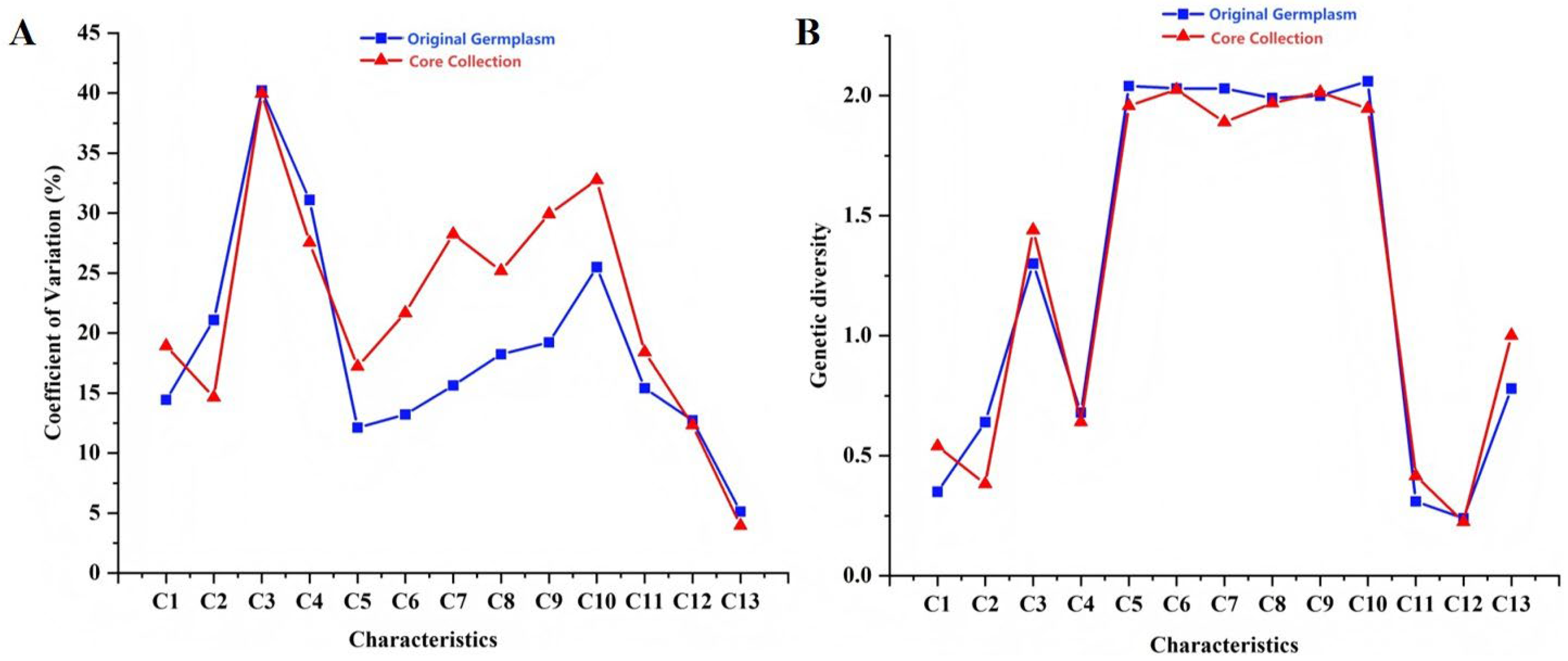
3.2.1. Comparison of Eigenvalues Between Original Germplasm and Core Collection
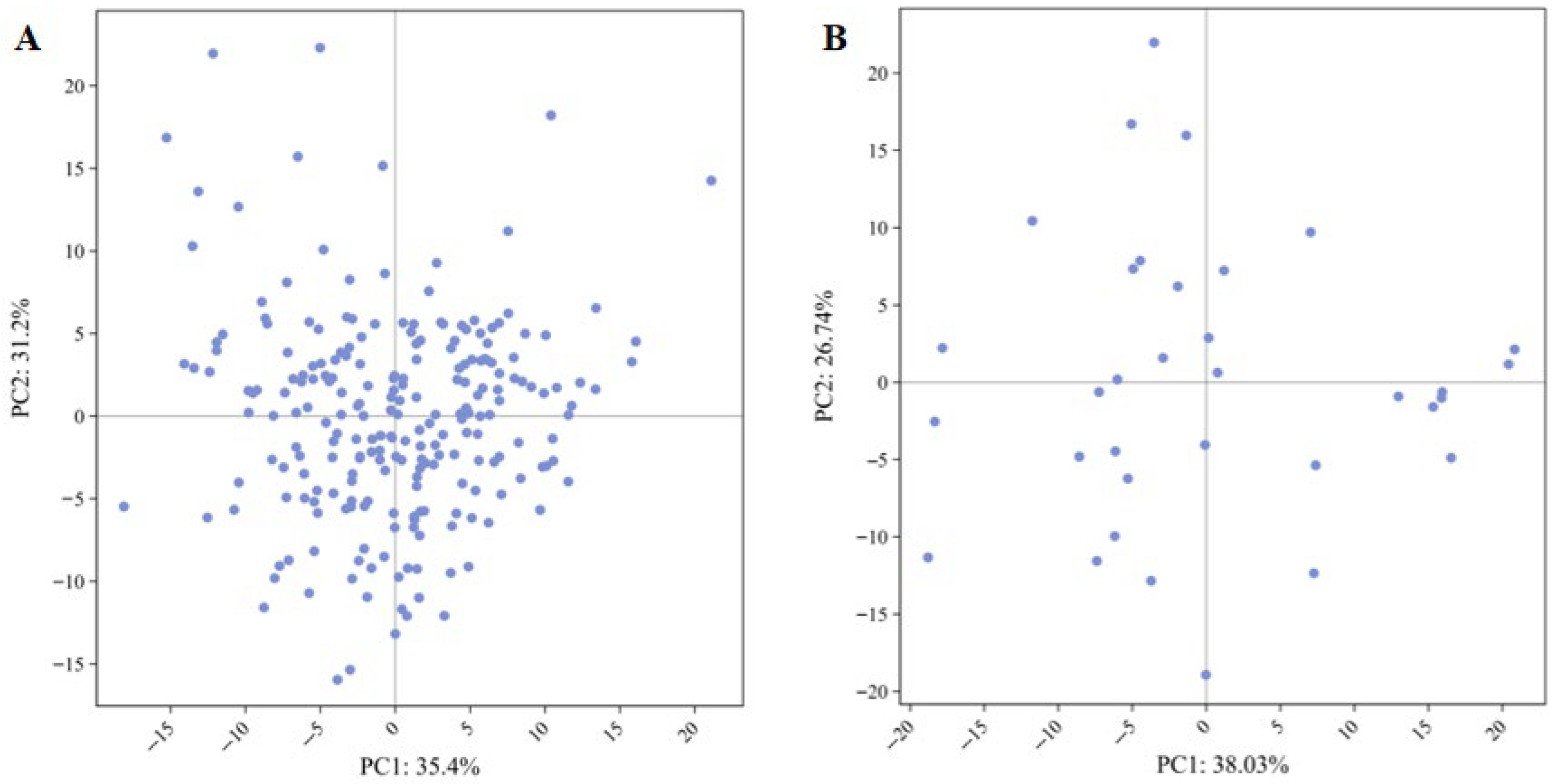
3.2.2. Cluster Analysis of Original Germplasm and Core Collection
3.3. Identification of Core Collection
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
| No. | Frequency Distribution (%) | Genetic Diversity (H’) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | ||
| C1 | 3.90 | 91.34 | 4.76 | 0.35 | |||||||
| C2 | 4.33 | 77.92 | 17.75 | 0.64 | |||||||
| C3 | 14.29 | 34.63 | 15.15 | 35.93 | 1.30 | ||||||
| C4 | 41.56 | 58.44 | 0.68 | ||||||||
| C5 | 2.60 | 3.03 | 10.39 | 13.42 | 19.48 | 19.05 | 20.35 | 4.76 | 4.76 | 2.16 | 2.04 |
| C6 | 1.30 | 3.46 | 10.39 | 16.45 | 18.18 | 24.68 | 10.39 | 8.66 | 3.03 | 3.46 | 2.03 |
| C7 | 1.73 | 3.03 | 7.79 | 18.18 | 23.38 | 19.48 | 12.12 | 6.06 | 5.19 | 3.03 | 2.03 |
| C8 | 4.33 | 10.39 | 15.15 | 22.51 | 22.51 | 12.55 | 5.19 | 3.90 | 3.46 | 1.99 | |
| C9 | 0.87 | 3.03 | 8.23 | 19.05 | 24.68 | 16.88 | 12.99 | 7.36 | 3.46 | 3.46 | 2.00 |
| C10 | 0.43 | 2.60 | 15.58 | 16.45 | 16.02 | 17.32 | 15.15 | 7.36 | 5.63 | 3.46 | 2.06 |
| C11 | 9.52 | 90.48 | 0.31 | ||||||||
| C12 | 6.49 | 93.51 | 0.24 | ||||||||
| C13 | 6.93 | 69.26 | 23.81 | 0.78 | |||||||
| Clustering Distance | Clustering Method | Overall Sampling Proportion | Core Collection | Percentage of Mean Difference (MD/%) | Percentage of Variance Difference (VD/%) | Coincidence Rate of Range (CR/%) | Change Rate of Coefficient of Variation (VR/%) | Variance of Phenotypic Frequency (VPF) | Phenotypic Retention Rate (RPR/%) |
|---|---|---|---|---|---|---|---|---|---|
| Euclidean Distance (D1) | Average Linkage (C1) | 10% | D1C1-10 | 0.00 | 33.34 | 87.30 | 135.78 | 0.07 | 52.00 |
| Single Linkage (C2) | D1C2-10 | 0.00 | 20.92 | 93.64 | 141.71 | 0.06 | 56.00 | ||
| Complete Linkage (C3) | D1C3-10 | 0.00 | 32.96 | 91.84 | 135.03 | 0.10 | 56.00 | ||
| Ward’s Method (C4) | D1C4-10 | 0.00 | 37.92 | 89.30 | 123.29 | 0.10 | 52.00 | ||
| Average Linkage (C5) | 15% | D1C5-15 | 0.00 | 36.96 | 94.01 | 125.85 | 0.08 | 78.00 | |
| Single Linkage (C6) | D1C6-15 | 0.00 | 25.05 | 95.13 | 136.94 | 0.06 | 81.00 | ||
| Complete Linkage (C7) | D1C7-15 | 0.00 | 44.55 | 97.77 | 139.34 | 0.09 | 75.00 | ||
| Ward’s Method (C8) | D1C8-15 | 0.00 | 50.25 | 94.74 | 129.07 | 0.10 | 72.00 | ||
| Average Linkage (C9) | 20% | D1C9-20 | 0.00 | 35.19 | 95.63 | 127.18 | 0.08 | 101.00 | |
| Single Linkage (C10) | D1C10-20 | 0.00 | 34.37 | 95.18 | 132.71 | 0.08 | 105.00 | ||
| Complete Linkage (C11) | D1C11-20 | 0.00 | 43.97 | 88.42 | 119.36 | 0.08 | 122.00 | ||
| Ward’s Method (C12) | D1C12-20 | 0.00 | 44.04 | 89.14 | 111.60 | 0.10 | 98.00 | ||
| Average Linkage (C13) | 25% | D1C13-25 | 0.00 | 40.12 | 90.76 | 117.92 | 0.08 | 122.00 | |
| Single Linkage (C14) | D1C14-25 | 0.00 | 29.09 | 95.18 | 130.64 | 0.07 | 125.00 | ||
| Complete Linkage (C15) | D1C15-25 | 0.00 | 45.88 | 97.08 | 122.73 | 0.09 | 115.00 | ||
| Ward’s Method (C16) | D1C16-25 | 0.00 | 43.83 | 96.29 | 119.88 | 0.09 | 117.00 | ||
| Average Linkage (C17) | 30% | D1C17-30 | 0.00 | 47.73 | 95.63 | 122.60 | 0.08 | 141.00 | |
| Single Linkage (C18) | D1C18-30 | 0.00 | 30.66 | 95.18 | 125.24 | 0.08 | 144.00 | ||
| Complete Linkage (C19) | D1C19-30 | 0.00 | 49.62 | 97.08 | 119.68 | 0.09 | 133.00 | ||
| Ward’s Method (C20) | D1C20-30 | 0.00 | 41.28 | 96.29 | 117.89 | 0.09 | 137.00 | ||
| Mahalanobis Distance (D2) | Average Linkage (C21) | 10% | D2C21-10 | 0.00 | 31.39 | 91.29 | 132.25 | 0.09 | 55.00 |
| Single Linkage (C22) | D2C22-10 | 0.00 | 19.95 | 92.72 | 137.35 | 0.07 | 54.00 | ||
| Complete Linkage (C23) | D2C23-10 | 0.00 | 14.57 | 91.05 | 138.83 | 0.06 | 58.00 | ||
| Ward’s Method (C24) | D2C24-10 | 0.00 | 35.48 | 92.05 | 129.32 | 0.07 | 57.00 | ||
| Average Linkage (C25) | 15% | D2C25-15 | 0.00 | 33.77 | 95.31 | 129.62 | 0.08 | 78.00 | |
| Single Linkage (C26) | D2C26-15 | 0.00 | 35.37 | 95.47 | 136.25 | 0.07 | 80.00 | ||
| Complete Linkage (C27) | D2C27-15 | 0.00 | 21.98 | 93.51 | 131.23 | 0.07 | 75.00 | ||
| Ward’s Method (C28) | D2C28-15 | 0.00 | 34.65 | 94.17 | 131.02 | 0.07 | 78.00 | ||
| Average Linkage (C29) | 20% | D2C29-20 | 0.00 | 32.08 | 95.49 | 128.52 | 0.07 | 101.00 | |
| Single Linkage (C30) | D2C30-20 | 0.00 | 32.15 | 95.52 | 131.94 | 0.07 | 104.00 | ||
| Complete Linkage (C31) | D2C31-20 | 0.00 | 22.41 | 93.51 | 126.56 | 0.07 | 98.00 | ||
| Ward’s Method (C32) | D2C32-20 | 0.00 | 37.35 | 94.53 | 125.37 | 0.08 | 99.00 | ||
| Average Linkage (C33) | 25% | D2C33-25 | 0.00 | 32.79 | 95.52 | 126.40 | 0.08 | 121.00 | |
| Single Linkage (C34) | D2C34-25 | 0.00 | 29.08 | 95.52 | 128.05 | 0.07 | 121.00 | ||
| Complete Linkage (C35) | D2C35-25 | 0.00 | 30.71 | 95.13 | 124.68 | 0.08 | 120.00 | ||
| Ward’s Method (C36) | D2C36-25 | 0.00 | 39.90 | 95.52 | 126.68 | 0.08 | 121.00 | ||
| Average Linkage (C37) | 30% | D2C37-30 | 0.00 | 34.57 | 95.52 | 121.82 | 0.08 | 137.00 | |
| Single Linkage (C38) | D2C38-30 | 0.00 | 33.91 | 95.52 | 125.14 | 0.07 | 143.00 | ||
| Complete Linkage (C39) | D2C39-30 | 0.00 | 35.24 | 95.18 | 121.77 | 0.08 | 140.00 | ||
| Ward’s Method (C40) | D2C40-30 | 0.00 | 46.35 | 95.52 | 121.38 | 0.08 | 137.00 |
| Germplasm | C1 | C2 | C3 | C4 | C5 | C6 | C7 | C8 | C9 | C10 | C11 | C12 | C13 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| M7 | 1 | 2 | 1 | 1 | 42.09 | 30.30 | 1.39 | 10.23 | 49.58 | 17.32 | 2 | 2 | 117 |
| M242 | 3 | 2 | 1 | 1 | 62.30 | 16.95 | 0.94 | 5.09 | 13.88 | 8.19 | 2 | 2 | 117 |
| M172 | 1 | 2 | 1 | 1 | 61.04 | 18.97 | 3.22 | 5.11 | 16.53 | 9.14 | 2 | 2 | 98 |
| M162 | 2 | 2 | 1 | 2 | 61.04 | 22.32 | 1.12 | 5.28 | 18.38 | 9.48 | 1 | 2 | 117 |
| M184 | 2 | 2 | 1 | 1 | 60.17 | 22.65 | 1.26 | 5.51 | 18.65 | 9.65 | 2 | 2 | 107 |
| M190 | 2 | 2 | 1 | 1 | 59.10 | 22.98 | 1.26 | 5.68 | 19.49 | 9.68 | 2 | 2 | 98 |
| M176 | 2 | 2 | 1 | 1 | 57.89 | 23.51 | 1.30 | 5.70 | 20.56 | 10.00 | 1 | 2 | 107 |
| M167 | 2 | 2 | 2 | 1 | 46.96 | 30.05 | 1.56 | 6.40 | 23.01 | 10.61 | 2 | 2 | 117 |
| M215 | 1 | 2 | 2 | 1 | 45.50 | 42.20 | 1.08 | 7.67 | 20.59 | 19.23 | 1 | 2 | 107 |
| M4 | 2 | 2 | 2 | 1 | 55.66 | 33.06 | 1.68 | 10.55 | 34.50 | 18.11 | 1 | 2 | 117 |
| M82 | 2 | 2 | 2 | 2 | 46.27 | 24.09 | 1.92 | 5.11 | 36.85 | 20.96 | 1 | 2 | 117 |
| M51 | 2 | 2 | 2 | 1 | 60.17 | 30.26 | 1.99 | 8.65 | 42.18 | 17.94 | 2 | 2 | 96 |
| M202 | 2 | 2 | 2 | 1 | 62.30 | 22.32 | 2.79 | 7.55 | 22.77 | 10.55 | 2 | 2 | 96 |
| M208 | 2 | 2 | 2 | 1 | 61.04 | 29.23 | 2.09 | 6.05 | 32.92 | 10.86 | 2 | 2 | 103 |
| M23 | 2 | 2 | 2 | 1 | 39.21 | 26.88 | 1.46 | 5.09 | 24.37 | 17.99 | 2 | 2 | 98 |
| M58 | 2 | 1 | 2 | 2 | 57.89 | 32.94 | 1.76 | 8.58 | 21.26 | 15.47 | 1 | 2 | 103 |
| M219 | 2 | 2 | 3 | 2 | 34.63 | 16.95 | 2.04 | 7.42 | 23.39 | 9.65 | 2 | 2 | 103 |
| M225 | 2 | 2 | 3 | 2 | 45.23 | 35.87 | 1.26 | 9.39 | 24.09 | 23.90 | 2 | 2 | 105 |
| M209 | 2 | 2 | 3 | 2 | 47.63 | 30.89 | 1.54 | 7.81 | 26.46 | 9.14 | 2 | 2 | 105 |
| M241 | 2 | 2 | 3 | 2 | 30.85 | 24.58 | 1.26 | 5.85 | 13.88 | 8.19 | 2 | 2 | 117 |
| M238 | 1 | 2 | 3 | 2 | 44.09 | 28.06 | 1.57 | 6.25 | 19.49 | 16.32 | 2 | 2 | 98 |
| M90 | 2 | 2 | 3 | 2 | 48.88 | 28.87 | 1.69 | 6.77 | 34.30 | 20.47 | 2 | 2 | 103 |
| M117 | 2 | 2 | 3 | 2 | 50.93 | 36.79 | 1.38 | 8.09 | 37.20 | 18.05 | 2 | 2 | 103 |
| M220 | 2 | 1 | 3 | 2 | 52.52 | 41.14 | 1.28 | 9.98 | 28.54 | 22.49 | 2 | 1 | 107 |
| M84 | 2 | 2 | 4 | 2 | 37.84 | 29.20 | 1.30 | 7.22 | 39.80 | 22.15 | 2 | 2 | 117 |
| M222 | 2 | 2 | 4 | 2 | 34.57 | 36.94 | 0.94 | 7.42 | 18.38 | 22.35 | 2 | 1 | 107 |
| M230 | 2 | 2 | 4 | 2 | 48.85 | 43.47 | 1.12 | 14.03 | 22.30 | 21.93 | 2 | 2 | 103 |
| M218 | 2 | 2 | 4 | 2 | 35.41 | 35.51 | 1.00 | 8.67 | 30.73 | 25.35 | 2 | 1 | 107 |
| M221 | 3 | 3 | 4 | 2 | 32.21 | 22.98 | 1.40 | 5.75 | 21.08 | 14.38 | 2 | 2 | 98 |
| M121 | 2 | 2 | 4 | 2 | 37.78 | 18.97 | 1.99 | 5.51 | 33.05 | 10.87 | 2 | 2 | 103 |
| M102 | 2 | 2 | 4 | 2 | 46.15 | 30.29 | 1.52 | 8.35 | 23.32 | 21.34 | 2 | 2 | 103 |
| M140 | 2 | 2 | 4 | 2 | 50.46 | 30.93 | 1.63 | 5.70 | 30.39 | 12.53 | 2 | 2 | 103 |
| M100 | 2 | 3 | 4 | 2 | 41.41 | 23.81 | 1.74 | 6.18 | 28.09 | 10.85 | 2 | 2 | 103 |
| M232 | 2 | 2 | 4 | 2 | 46.39 | 23.51 | 1.97 | 5.28 | 16.53 | 11.27 | 2 | 2 | 103 |
References
- Du, X.F.; Zhao, Q.; Yang, Z.L. Diversity, evolutionary history and cultivation of morels: A review. Mycosystema 2014, 33, 183–197. [Google Scholar]
- Tietel, Z.; Masaphy, S. True morels (Morchella)-nutritional and phytochemical composition, health benefits and flavor: A review. Crit. Rev. Food Sci. Nutr. 2018, 58, 1888–1901. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.T.; Lu, Z.P.; Gao, X.Y.; Liu, M.L.; Sa, W.; Liang, J.; Wang, L.; Yin, W.; Shang, Q.H.; Li, Z.H. Maximum Entropy Modeling the Distribution Area of Morchella Dill. ex Pers. Species in China under Changing Climate. Biology 2022, 11, 1027. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.F.; Zhang, Z.; Shang, Y.H.; Zheng, Y. Extraction, structure and antioxidant activity of the polysaccharides from morels (Morchella spp.): A review. Int. J. Biol. Macromol. 2024, 264, 130656. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Tang, J.; Peng, W.H.; Gang, B.C.; Huang, Z.Q.; Wang, Y. A New Morchella Cultivar‘Chuan Yangdujun 6’. Acta Hortic. Sin. 2017, 44, 2431–2432. [Google Scholar]
- Tang, J.; Wang, Y.; Xu, Y.Y.; Tan, H.; He, X.L.; Yu, Y.; Chen, Y.; Peng, W.H. Research Progress on Key Issues of Morel Industry Development. J. Fungal Res. 2021, 19, 217–231. [Google Scholar]
- Frankel, O.H. Genetic Perspectives of Germplasm Conservation. In Genetic Manipulation: Impact on Man and Society; Cambridge University Press: Cambridge, UK, 1984; pp. 161–170. [Google Scholar]
- Brown, A.H. Core collections: A practical approach to genetic resources management. Genome 1989, 31, 818–824. [Google Scholar] [CrossRef]
- Lei, M.L.; Liu, X.; Zhang, J.H.; Wang, Y.N.; Huang, R.; Du, H.L. The Genetic Representation of Local Wheat Varieties in Shanxi Province was Studied by Agronomic Characters. Mol. Plant Breed. 2020, 18, 6853–6872. [Google Scholar]
- Song, J.M.; Arif, M.; Zi, Y.; Sze, S.H.; Zhang, M.P.; Zhang, H.B. Molecular and genetic dissection of the UsDA rice mini-core collection using high-density SNP markers. Plant Sci. 2021, 308, 110910. [Google Scholar] [CrossRef]
- Wang, Z.Q.; Yan, T.X.; Yan, X.W.; Liang, J.C.; Le, M.W.; Sun, J. Diversity analysis of sesame germplasm resources and core germplasm construction. J. Plant Genet. Resour. 2025, 26, 455–470. [Google Scholar]
- Guo, Y.C.; Zhang, L.L.; Chen, S.Y.; Qi, J.M.; Fang, P.P.; Tao, A.F.; Zhang, L.M.; Zhang, L.W. DNA molecular identity card construction of core germplasm using jute. Acta Agron. Sin. 2021, 47, 80–93. [Google Scholar] [CrossRef]
- Zhao, X.R.; Chen, X.T.; Xue, W.; Wang, L.; Cai, X.R.; Lin, B.S.; Liu, X.J.; Cui, J.H. Construction of Core Potato Germplasm Resources in North Hebei based on Phenotypic Traits. J. Nucl. Agric. Sci. 2024, 38, 805–818. [Google Scholar]
- Zhang, T.; Wang, X.Y.; Guo, Q.W.; Wei, J.; Fang, P.P.; Xie, L.F.; Zhao, D.F.; Li, C.S. Construction of Core Germplasm Collection Based on Phenotypic Characters in Luffa spp. Chin. J. Trop. Crops 2025, 46, 106–114. [Google Scholar]
- Liu, J.; Liao, K.; Cao, Q.; Sun, Q.; Liu, H.; Jia, Y. Construction of core germplasm resources of Xinjiang wild apricot using phenotypic traits. J. Fruit Sci. 2015, 32, 787–796. [Google Scholar]
- Yin, M.Y.; Wu, B.; Wuyun, T.N.; Pang, Y.J.; Zhang, X.L. Construction of core germplasm of Pinus sylvestris var. mongolica based on phenotypic traits and SSR markers. J. Cent. South Univ. For. Technol. 2025, 459, 13. [Google Scholar]
- Li, J.W.; Su, J.S.; Zhang, F.; Fang, W.M.; Guan, Z.Y.; Chen, S.M.; Chen, F.D. Construction of Core Collection of Traditional Chrysanthemum morifolium Based on Phenotypic Traits. Sci. Agric. Sin. 2021, 54, 3514–3526. [Google Scholar]
- Wang, L.; Wang, J.M.; Wang, W.; Wang, L.; Wang, L.J.; Yan, X.C.; Tan, M.L. Core sunflower germplasm was constructed based on phenotypic diversity. Chin. J. Oil Crops Sci. 2021, 43, 1052–1060. [Google Scholar]
- Chen, C.; Ding, C.J.; Huang, Q.J.; Li, Z.H.; Zhang, J.; Liu, N.; Li, B.; Su, X.H. Construction of phenotypic core collection of Populus deltoides. For. Res. 2021, 34, 1–11. [Google Scholar]
- Zhai, L.N.; Tang, Q.J.; Zhou, B.J.; Zhou, S.Z.; Wang, H.J.; Yun, Y.; Han, Y.S.; Wang, Q.Y.; Yan, X.W.; Xing, F.N. Analysis of genetiediversity and construetion of core gemplasm of common wildrice in Hainan. J. Plant Genet. Resour. 2024, 25, 1624–1636. [Google Scholar]
- Zhao, L. Analysis of Genetic Diversity and Establishment of Core Collection of Rice Germplasm in Ningxia and Xinjiang. Master’s Thesis, Ningxia University, Yinchuan, China, 2018. [Google Scholar]
- Gao, L.H.; Bao, D.P.; Xu, Z.; Li, Y.; Lu, H.; Tan, Y.S.; Shang, X.D.; Chen, H.Y.; Wang, R.J.; Wu, Y.Y. Construction of core collection and DNA fingerprinting of Flammulina filiformis based on genetic diversity analysis. Mycosystema 2021, 40, 3214–3230. [Google Scholar]
- Oh, Y.L.; Choi, I.G.; Kong, W.S.; Jang, K.Y.; Oh, M.Y.; Im, J.H. Evaluating genetic diversity of Agaricus bisporus accessions through phylogenetic analysis using single-nucleotide polymorphism(SNP) markers. Mycobiology 2020, 49, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.K.; Cai, Z.X.; Guo, Z.J.; Lu, Y.P.; Chen, M.Y.; Liao, J.H. A Preliminary Report on Resequencing 18 Representative Strains of Agaricus bisporus. Fujian J. Agric. Sci. 2019, 34, 1167–1172. [Google Scholar]
- Chen, Y.; Tang, J.; Peng, W.H.; Gan, B.C.; Huang, Z.Q.; Wang, Y.; Jiang, L.; Min, J.; Liu, L.X. High-efficiency cultivation model and technology of Morchella in Sichuan. Edible Med. Mushrooms 2016, 24, 151–154. [Google Scholar]
- NY/T 4221—2022; Guidelines for the Conduct of Tests for Distinctness, Uniformity and Stability. Chinese Agriculture Press: Beijing, China, 2022.
- Liu, Z.T.; Qiao, S.Y.; Wu, Y.H.; Zhao, S.Y. Genetics, 3rd ed.; Higher Education Press: Beijing, China, 2015; p. 194. [Google Scholar]
- Hu, J.; Zhu, J.; Xu, H.M. Methods of constructing core collections by stepwise clustering with three sampling strategies based on the genotypic values of crops. Theor. Appl. Genet. 2000, 101, 264–268. [Google Scholar] [CrossRef]
- Liu, Z.C.; Zhang, C.Y.; Zhang, Y.M.; Zhang, X.Y.; Wu, C.J.; Wang, H.B.; Shi, J.; Chen, X.S. Study on method of constructing core collection of Malus sieversii based on quantitative traits. Sci. Agric. Sin. 2010, 43, 358–370. [Google Scholar]
- Lyu, X.; Liu, G.X.; Li, Y.X.; Ji, Y.; Li, Y.; Li, Y.Y.; Cheng, Y.Z.; Wu, Z.D.; Zhang, X.W. Current Status and Prospects of Plant Core Germplasm Research. J. Plant Genet. Resour. 2025, 26, 1693–1707. [Google Scholar]
- El Caid, M.B.; Lachheb, M.; Lagram, K.; Wang, X.; Serghini, M.A. Ecotypic variation and environmental influence on saffron (Crocus sativus L.) vegetative growth: A multivariate performance analysis. J. Appl. Res. Med. Aromat. Plants 2024, 43, 100601. [Google Scholar] [CrossRef]
- Qi, Y.W.; Fan, L.N.; Luo, Q.W.; Wang, Q.N.; Chen, Y.S.; Huang, Z.X.; Liu, R.; Liu, S.M.; Deng, H.H.; Li, Q.W. Establishment of Saccharum spontaneum L. core collections. Acta Agron. Sin. 2013, 39, 649–656. [Google Scholar] [CrossRef]
- Dai, P.H.; Sun, J.L.; He, S.P.; Wang, L.R.; Jia, Y.H.; Pan, Z.E.; Pang, B.Y.; Du, X.M.; Wang, M. Comprehensive evaluation and genetic diversity analysis of phenotypic traits of core collection in upland cotton. Sci. Agric. Sin. 2016, 49, 3694–3708. [Google Scholar]
- Ai, Y.; Chen, L.; Lan, S.R.; Xie, T.X.; Chen, J.; Peng, D.H. Construction of core collection of Cymbidium ensifolium varieties based on morphological traits. Mol. Plant Breed. 2019, 17, 7924–7934. [Google Scholar]
- Lei, G.; Zhou, K.H.; Fang, R.; Wu, Y.; Chen, X.J. Studies on the constructing of pepper core collection based on phenotypic data. Acta Bot. Boreali-Occident. Sin. 2016, 36, 804–810. [Google Scholar]
- Li, H.F.; Chen, T.Y.; Huang, Y.M.; Wu, C.R.; Li, Y.Q.; Lu, S.Q.; Chen, X.T. Sampling strategies of sweet potato core collection based on morphological traits. J. Plant Genet. Resour. 2013, 14, 87–93. [Google Scholar]
- Duan, H.J.; Cao, S.; Zheng, H.Q.; Hu, D.H.; Lin, J.; Cui, B.B.; Lin, H.Z.; Hu, R.Y.; Wu, B.; Sun, Y.H.; et al. Genetic Characterization of Chinese fir from Six Provinces in Southern China and Construction of a Core Collection. Sci. Rep. 2017, 7, 138–148. [Google Scholar] [CrossRef] [PubMed]
- Miao, L.M.; Wang, S.Y.; Zou, M.H.; Li, J.B.; Kong, L.J.; Yu, X.L. Review of the studies on core collection for horticultural crops. J. Plant Genet. Resour. 2016, 17, 791–800. [Google Scholar]

| No. | Germplasm | Species | Source |
|---|---|---|---|
| 1 | M1–M48, M48–M60, M82, M215–M217 | M. importuna | Chengdu City, Sichuan Province |
| 2 | M62–M72, M86–M106, M110–M111, M115–M119, M125, M129–M148, M153–M160, M218–M219, M221–M222, M227, M230, M236–M237 | M. sextelata | Chengdu City, Sichuan Province |
| 3 | M73–M74, M220 | Shiyan City, Hubei Province | |
| 4 | M75, M112, M223–M224 | Mianyang City, Sichuan Province | |
| 5 | M76 | Zunyi City, Guizhou Province | |
| 6 | M77 | Dazhou City, Sichuan Province | |
| 7 | M78, M232–M233 | Hanzhong City, Shaanxi Province | |
| 8 | M79 | Taiyuan City, Shanxi Province | |
| 9 | M80–M81 | Lanzhou City, Gansu Province | |
| 10 | M82–M83, M114 | Nanyang City, Henan Province | |
| 11 | M84–M85 | Neijiang City, Sichuan Province | |
| 12 | M107–M109 | Jinan City, Shandong Province | |
| 13 | M113 | Guangzhou City, Guangdong Province | |
| 14 | M120 | Changsha City, Hunan Province | |
| 15 | M121–M123 | Suzhou City, Anhui Province | |
| 16 | M124, M229, M231, M240–M241 | Kunming City, Yunnan Province | |
| 17 | M126, M234–M235 | Bazhong City, Sichuan Province | |
| 18 | M127 | Shenyang City, Liaoning Province | |
| 19 | M225–M226 | Hohhot City, Inner Mongolia Autonomous Region | |
| 20 | M228 | Shijiazhuang City, Hebei Province | |
| 21 | M161, M166–M175, M177–M178, M180, M182–M185, M187–M197, M199–M202, M205, M207–M214, M238–M239 | M. eximia | Chengdu City, Sichuan Province |
| 22 | M162, M165 | Hanzhong City, Shanxi Province | |
| 23 | M163 | Yibin City, Sichuan Province | |
| 24 | M164 | Kangding City, Sichuan Province | |
| 25 | M176 | Urumqi City, Xinjiang Uygur Autonomous Region | |
| 26 | M179 | Mianyang City, Sichuan Province | |
| 27 | M181 | Shenyang City, Liaoning Province | |
| 28 | M186, M242 | Kunming City, Yunnan Province | |
| 29 | M198 | Bazhong City, Sichuan Province |
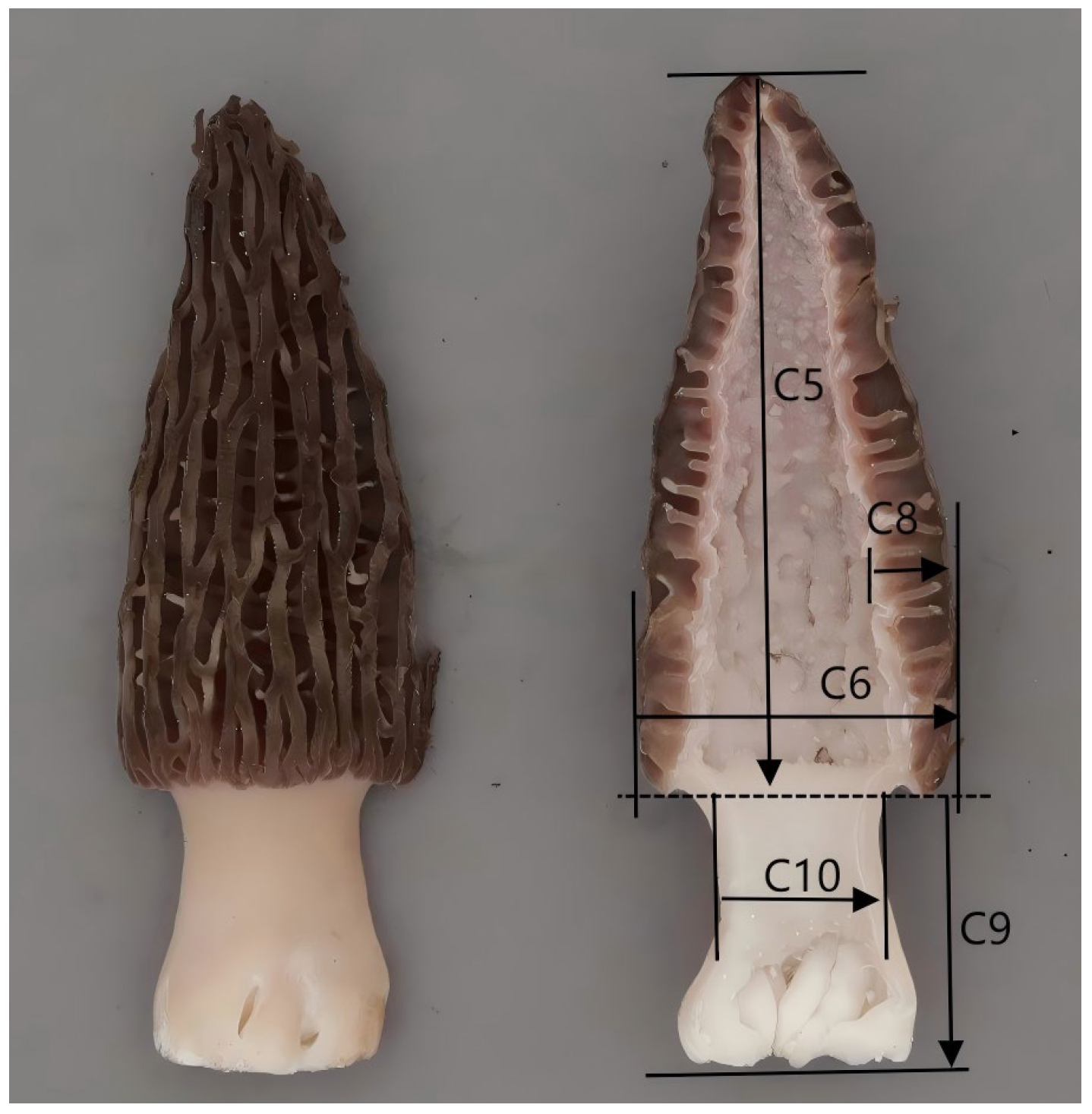
| No. | Characteristics | Characteristics Type | Method of Observation |
|---|---|---|---|
| C1 | Cap: Shape (Triangular = 1; Ovate = 2; Rectangular = 3) | QL | VG |
| C2 | Cap: Lateral Ridges Density (Sparse = 1; Moderate = 2; Dense = 3) | QL | VG |
| C3 | Cap: Color (Light Brown = 1; Medium Brown = 2; Dark Brown = 3; Reddish Brown = 4) | QL | VG |
| C4 | Cap: Base Shape (Concave = 1; Convex = 2) | QL | VG |
| C5 | Cap: Length | QN | MS |
| C6 | Cap: Width | QN | MS |
| C7 | Cap: Length/Width Ratio | QN | MS |
| C8 | Cap: Thickness | QN | MS |
| C9 | Stem: Length | QN | MS |
| C10 | Stem: Diameter | QN | MS |
| C11 | Stem: Longitudinal Section Shape (Rectangular = 1; Trapezoidal = 2) | QL | VG |
| C12 | Stem: Color (White = 1; Yellowish-white = 2) | QL | VG |
| C13 | Maturity Period (Early = 93; Mid = 103; Late = 113) | QL | MG |
| Characteristics | Min | Max | Average Value | Standard Deviation | Coefficient of Variation |
|---|---|---|---|---|---|
| C5 | 30.85 | 62.3 | 47.74 | 5.79 | 12.12 |
| C6 | 16.95 | 43.47 | 28.67 | 3.79 | 13.22 |
| C7 | 0.94 | 2.79 | 1.69 | 0.26 | 15.64 |
| C8 | 5.05 | 14.03 | 7.37 | 1.34 | 18.23 |
| C9 | 13.88 | 49.58 | 26.56 | 5.10 | 19.22 |
| C10 | 7.22 | 25.35 | 15.33 | 3.91 | 25.49 |
| Sampling Method | Data Grouping | Proportion of Each Group | Total Sample Intensity | Original Germplasm | |||||
|---|---|---|---|---|---|---|---|---|---|
| 10% | 15% | 20% | 25% | 30% | Number with in the Group | Intra-Group Proportion | |||
| Simple Proportion | I | 13.04 | 3 | 5 | 7 | 8 | 10 | 33 | 14.29 |
| II | 34.78 | 8 | 12 | 16 | 20 | 24 | 80 | 34.63 | |
| III | 17.39 | 4 | 5 | 7 | 9 | 11 | 35 | 15.15 | |
| IV | 34.78 | 8 | 12 | 17 | 21 | 25 | 83 | 35.93 | |
| Logarithmic Proportion | I | 22.06 | 5 | 7 | 10 | 13 | 15 | 33 | 14.29 |
| II | 27.64 | 6 | 9 | 13 | 16 | 19 | 80 | 34.63 | |
| III | 22.43 | 5 | 8 | 11 | 13 | 16 | 35 | 15.15 | |
| IV | 27.87 | 7 | 10 | 13 | 16 | 20 | 83 | 35.93 | |
| Square Root Proportion | I | 19.33 | 4 | 7 | 9 | 11 | 13 | 33 | 14.29 |
| II | 30.10 | 7 | 10 | 14 | 17 | 21 | 80 | 34.63 | |
| III | 19.91 | 5 | 7 | 9 | 12 | 14 | 35 | 15.15 | |
| IV | 30.66 | 7 | 10 | 15 | 18 | 22 | 83 | 35.93 | |
| Diversity Proportion | I | 21.34 | 5 | 7 | 10 | 12 | 15 | 33 | 14.29 |
| II | 28.19 | 6 | 9 | 13 | 16 | 19 | 80 | 34.63 | |
| III | 22.24 | 5 | 8 | 10 | 13 | 16 | 35 | 15.15 | |
| IV | 28.22 | 7 | 10 | 14 | 17 | 20 | 83 | 35.93 | |
| Characteristics | Coefficient of Variation | Genetic Diversity | Average Value | Standard Deviation | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Original Germplasm | Core Collection | Original Germplasm | Core Collection | Original Germplasm | Core Collection | p Value (t-Test) | Original Germplasm | Core Collection | p Value (F-Test) | |
| C1 | 14.44 | 21.43 | 0.35 | 0.58 | 2.01 | 1.94 | 0.24 | 0.29 | 0.42 | 0.24 |
| C2 | 21.10 | 17.15 | 0.64 | 0.44 | 2.13 | 2.00 | 0.10 | 0.45 | 0.34 | 0.10 |
| C3 | 40.21 | 42.47 | 1.30 | 1.38 | 2.73 | 2.62 | 0.59 | 1.10 | 1.11 | 0.59 |
| C4 | 31.10 | 30.04 | 0.68 | 0.67 | 1.58 | 1.62 | 0.71 | 0.49 | 0.49 | 0.71 |
| C5 | 12.12 | 19.71 | 2.04 | 1.84 | 47.74 | 48.37 | 0.59 | 5.79 | 9.53 | 0.59 |
| C6 | 13.22 | 24.18 | 2.03 | 1.90 | 28.67 | 28.46 | 0.79 | 3.79 | 6.88 | 0.79 |
| C7 | 15.64 | 30.75 | 2.03 | 1.78 | 1.69 | 1.57 | 0.19 | 0.26 | 0.48 | 0.04 * |
| C8 | 18.23 | 27.69 | 1.99 | 1.85 | 7.37 | 7.17 | 0.59 | 1.34 | 1.99 | 0.47 |
| C9 | 19.22 | 32.42 | 2.00 | 1.89 | 26.56 | 26.07 | 0.64 | 5.10 | 8.45 | 0.64 |
| C10 | 25.49 | 35.26 | 2.06 | 1.83 | 15.33 | 15.19 | 0.85 | 3.91 | 5.35 | 0.85 |
| C11 | 15.41 | 20.91 | 0.31 | 0.47 | 1.90 | 1.82 | 0.15 | 0.29 | 0.38 | 0.15 |
| C12 | 12.73 | 14.84 | 0.24 | 0.30 | 1.94 | 1.91 | 0.62 | 0.25 | 0.28 | 0.62 |
| C13 | 5.13 | 6.46 | 0.78 | 0.99 | 101.81 | 105.97 | 0.00 * | 5.23 | 6.85 | 0.00 * |
| Principal Component | Eigen Value | Contribution Rate (%) | Total Contribution Rate (%) | |||
|---|---|---|---|---|---|---|
| Original Germplasm | Core Collection | Original Germplasm | Core Collection | Original Germplasm | Core Collection | |
| 1 | 3.09 | 3.65 | 23.78 | 28.04 | 23.78 | 28.04 |
| 2 | 2.01 | 2.43 | 15.46 | 18.71 | 39.25 | 46.75 |
| 3 | 1.62 | 1.63 | 12.43 | 12.57 | 51.68 | 59.31 |
| 4 | 1.17 | 1.21 | 9.00 | 9.33 | 60.67 | 68.64 |
| Characteristics | Original Germplasm | Core Collection | ||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 1 | 2 | 3 | 4 | |
| C1 | −0.08 | −0.17 | −0.01 | 0.62 | −0.23 | 0.33 | −0.22 | −0.49 |
| C2 | −0.15 | −0.40 | 0.22 | −0.60 | −0.62 | 0.48 | −0.00 | 0.22 |
| C3 | 0.59 | −0.62 | 0.32 | 0.08 | 0.23 | 0.88 | 0.21 | −0.00 |
| C4 | 0.60 | −0.52 | 0.18 | 0.25 | 0.22 | 0.73 | −0.01 | −0.15 |
| C5 | −0.13 | 0.59 | 0.72 | 0.05 | −0.03 | −0.84 | 0.12 | −0.11 |
| C6 | 0.68 | 0.47 | −0.05 | −0.31 | 0.88 | 0.10 | −0.09 | 0.17 |
| C7 | −0.66 | 0.09 | 0.61 | 0.31 | −0.35 | −0.29 | 0.66 | 0.28 |
| C8 | 0.54 | 0.50 | 0.01 | 0.21 | 0.74 | 0.05 | −0.01 | 0.32 |
| C9 | 0.44 | 0.30 | 0.35 | −0.02 | 0.20 | 0.11 | −0.09 | 0.81 |
| C10 | 0.79 | 0.07 | 0.32 | −0.12 | 0.77 | 0.33 | −0.11 | 0.29 |
| C11 | 0.15 | −0.42 | 0.22 | −0.01 | −0.07 | 0.45 | 0.56 | −0.06 |
| C12 | −0.43 | 0.12 | −0.05 | −0.18 | −0.64 | −0.24 | 0.00 | 0.36 |
| C13 | 0.39 | 0.22 | −0.52 | 0.26 | −0.00 | −0.04 | −0.88 | 0.08 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, X.; Chen, Y.; Liu, L.; Tang, J.; Liu, S.; Xie, L.; Li, Y. Construction of a Core Collection for Morchella Based on Phenotypic Traits from China. Horticulturae 2025, 11, 1274. https://doi.org/10.3390/horticulturae11111274
Cao X, Chen Y, Liu L, Tang J, Liu S, Xie L, Li Y. Construction of a Core Collection for Morchella Based on Phenotypic Traits from China. Horticulturae. 2025; 11(11):1274. https://doi.org/10.3390/horticulturae11111274
Chicago/Turabian StyleCao, Xuelian, Ying Chen, Lixu Liu, Jie Tang, Shishi Liu, Liyuan Xie, and Yiping Li. 2025. "Construction of a Core Collection for Morchella Based on Phenotypic Traits from China" Horticulturae 11, no. 11: 1274. https://doi.org/10.3390/horticulturae11111274
APA StyleCao, X., Chen, Y., Liu, L., Tang, J., Liu, S., Xie, L., & Li, Y. (2025). Construction of a Core Collection for Morchella Based on Phenotypic Traits from China. Horticulturae, 11(11), 1274. https://doi.org/10.3390/horticulturae11111274