Pathogen Survey in Agrocybe chaxingu and Characterization of the Dominant Pathogen Fuligo gyrosa
Abstract
1. Introduction
2. Materials and Methods
2.1. A. chaxingu Strain and Culture Conditions
2.2. Disease Survey and Statistical Analysis in A. chaxingu
2.3. Isolation and Purification of Potential Pathogens from A. chaxingu
2.4. Morphological Observation of Potential Pathogenic Microorganisms
2.5. Molecular Identification of Potential Pathogens
2.6. Construction of Phylogenetic Trees
2.7. Pathogenicity Assessment and Infection Process Observation of Predominant Potential Pathogens
3. Results
3.1. Major Diseases and Putative Pathogens of A. chaxingu
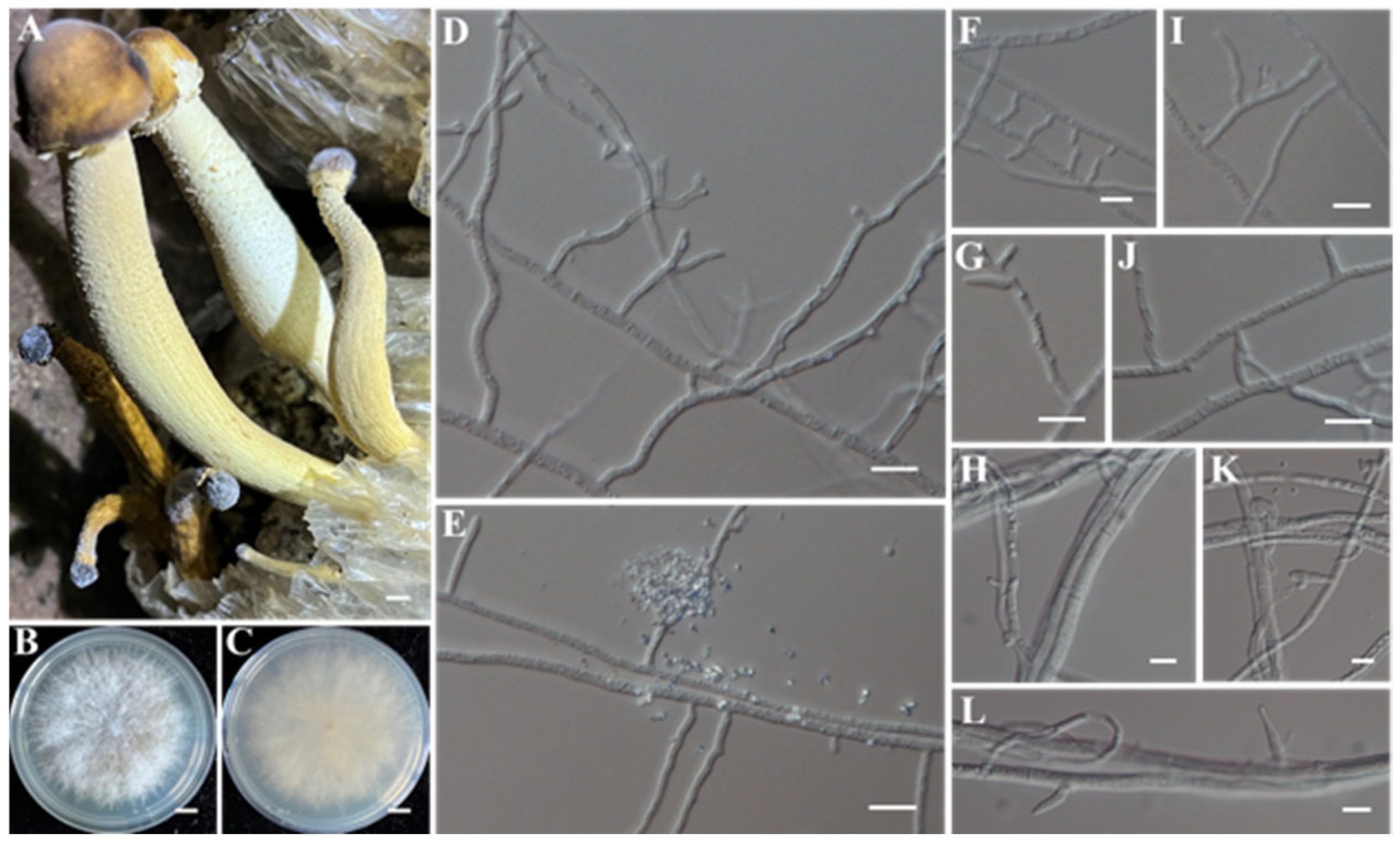
- (1)
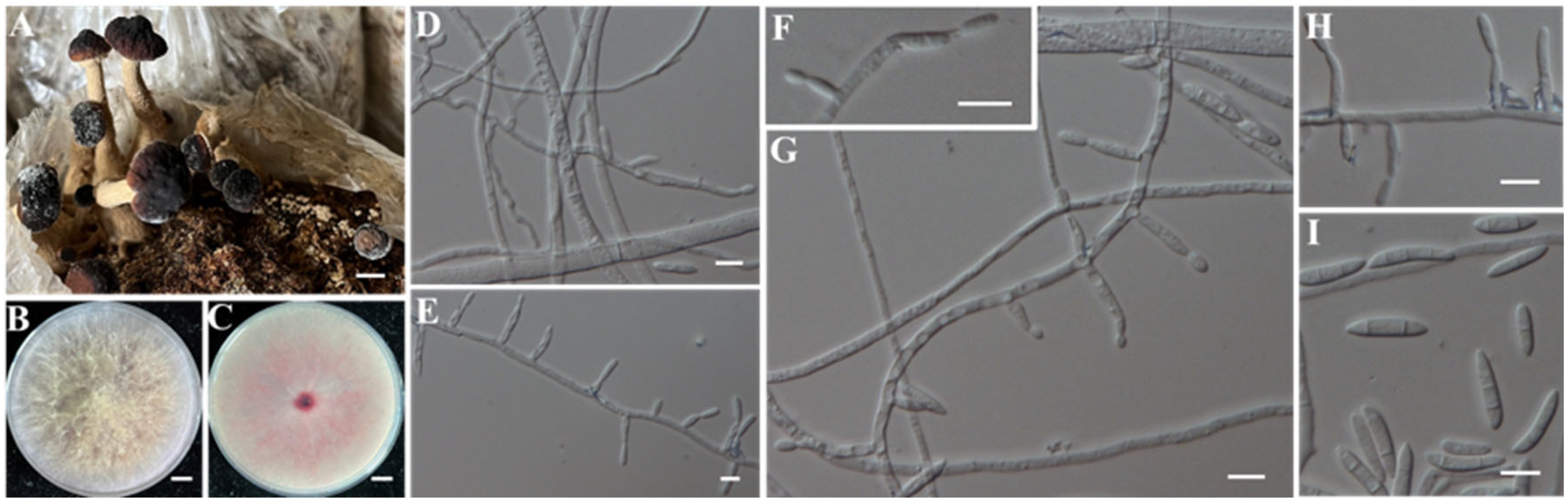
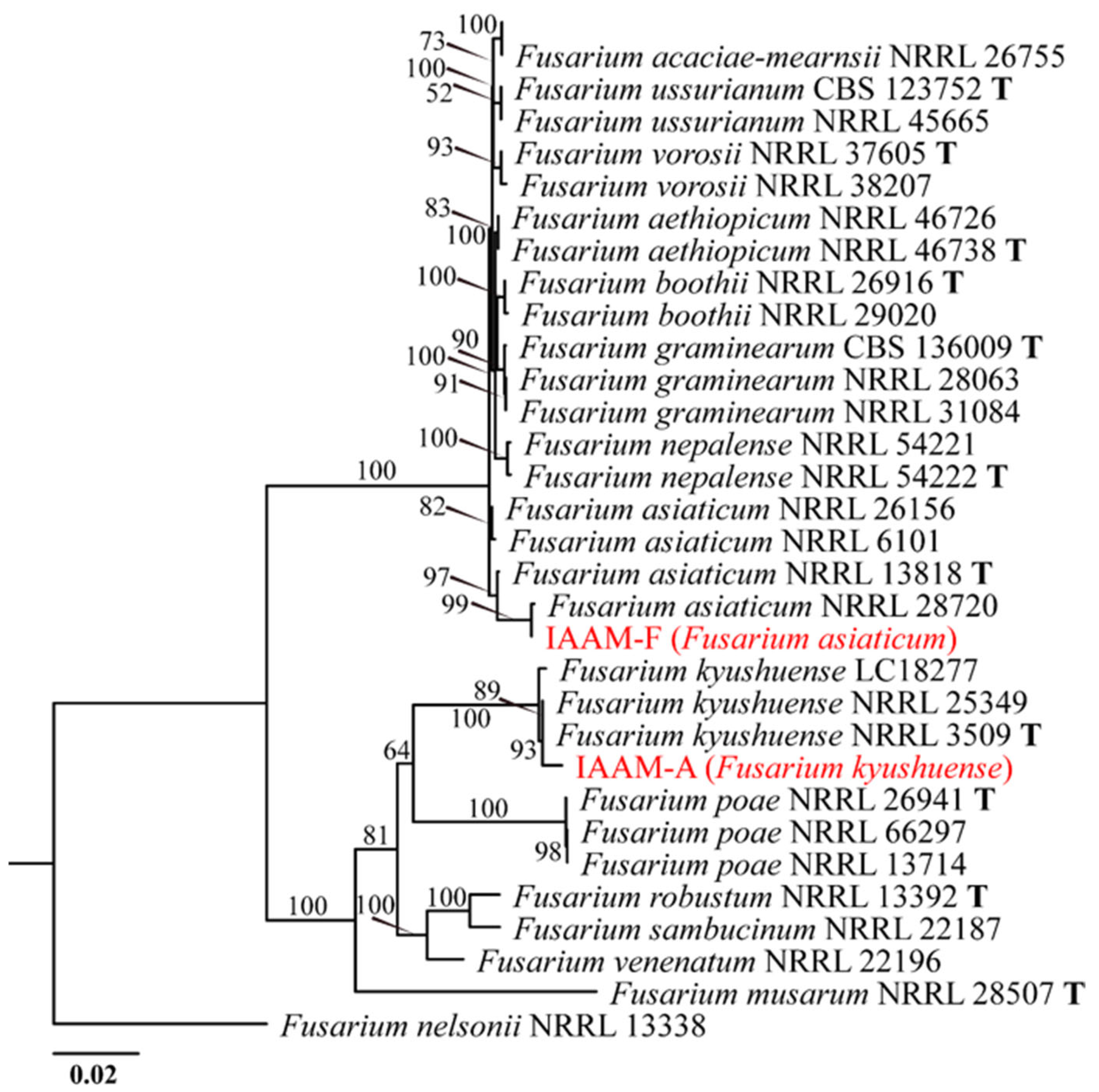
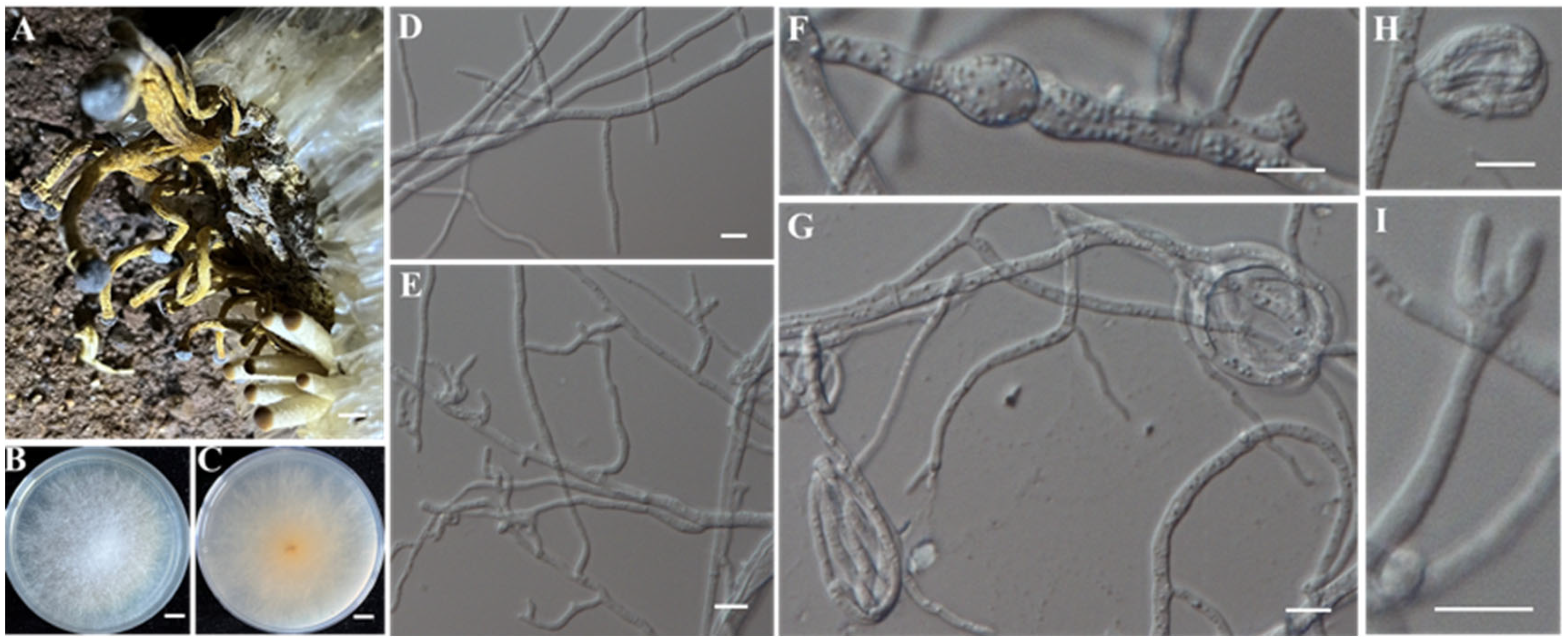
- Identification of Putative Pathogenic Fungi IAAM-A and IAAM-F
- (2)
- Identification of Putative Pathogenic Fungi IAAM-B, IAAM-C, and IAAM-O
- (3)
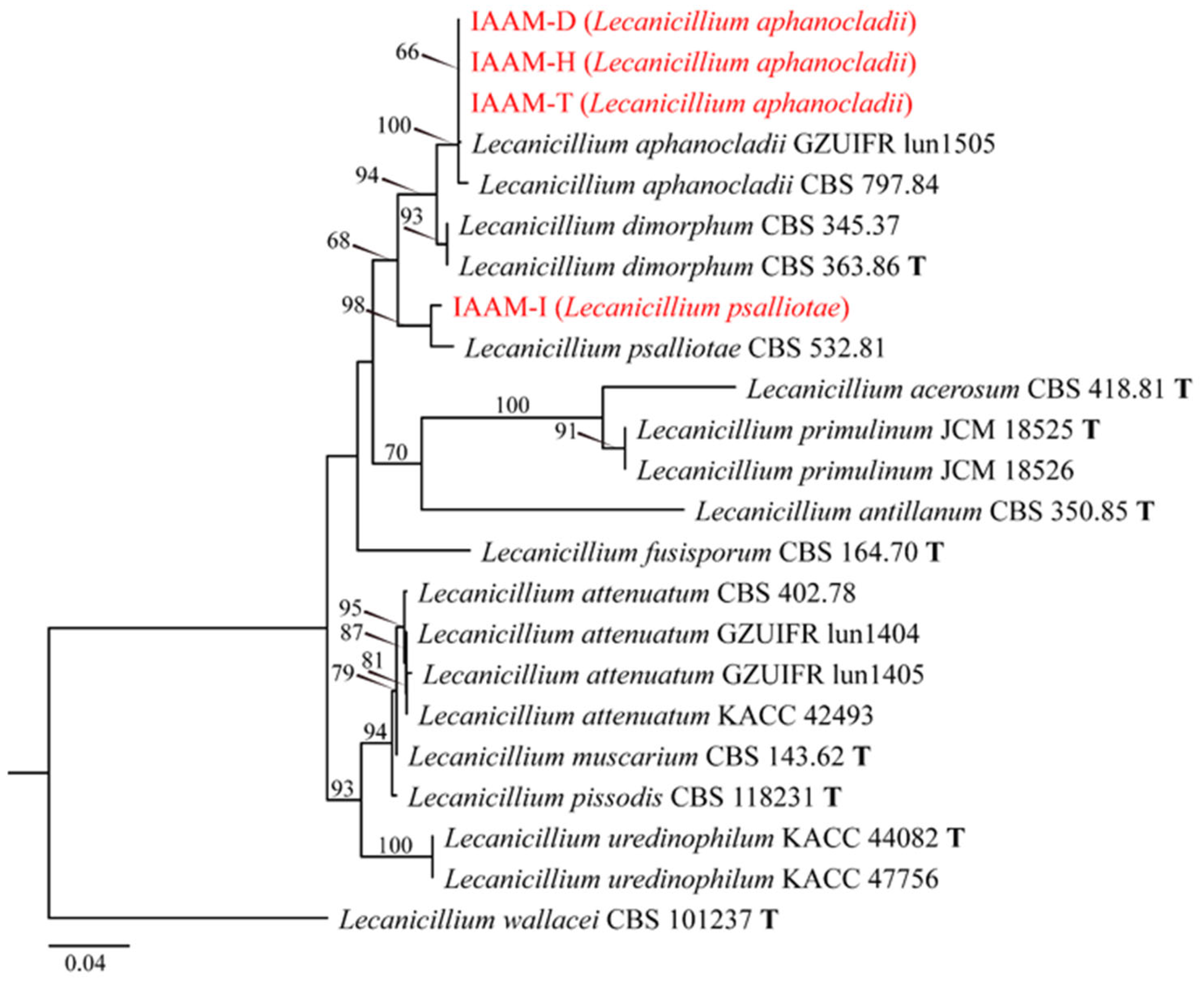
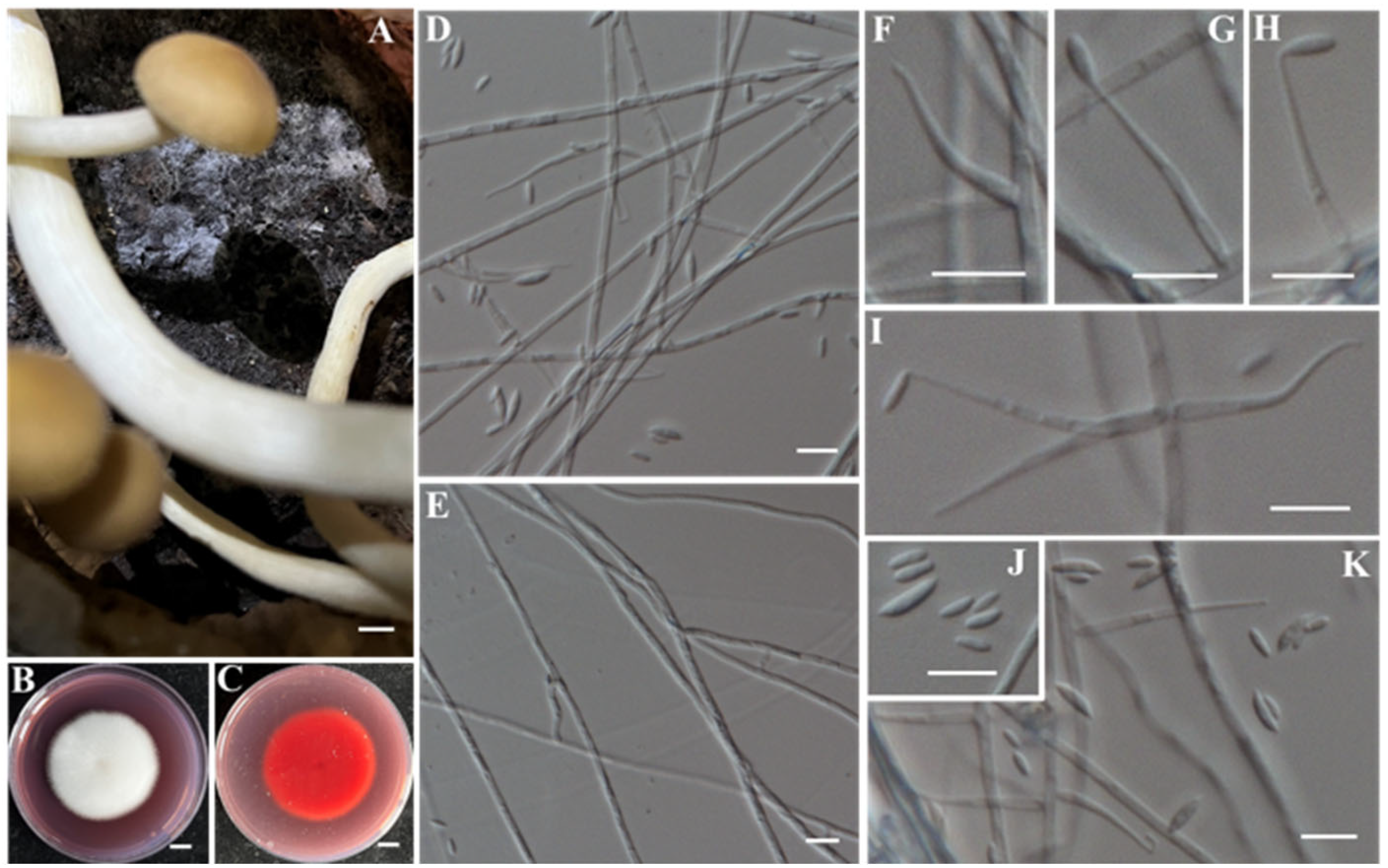
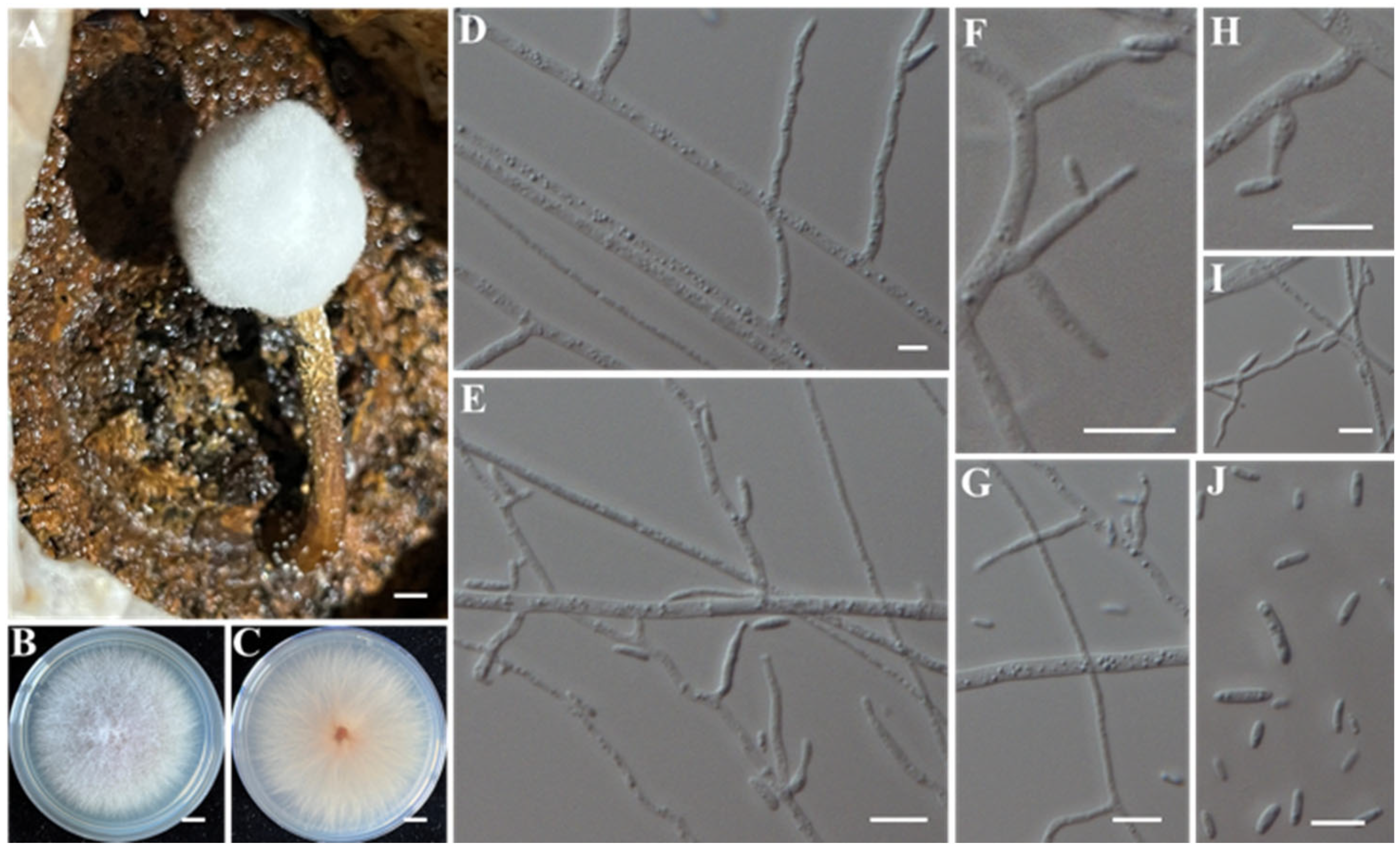
- Identification of Putative Pathogenic Fungi IAAM-D, IAAM-H, IAAM-I, and IAAM-T
- (4)
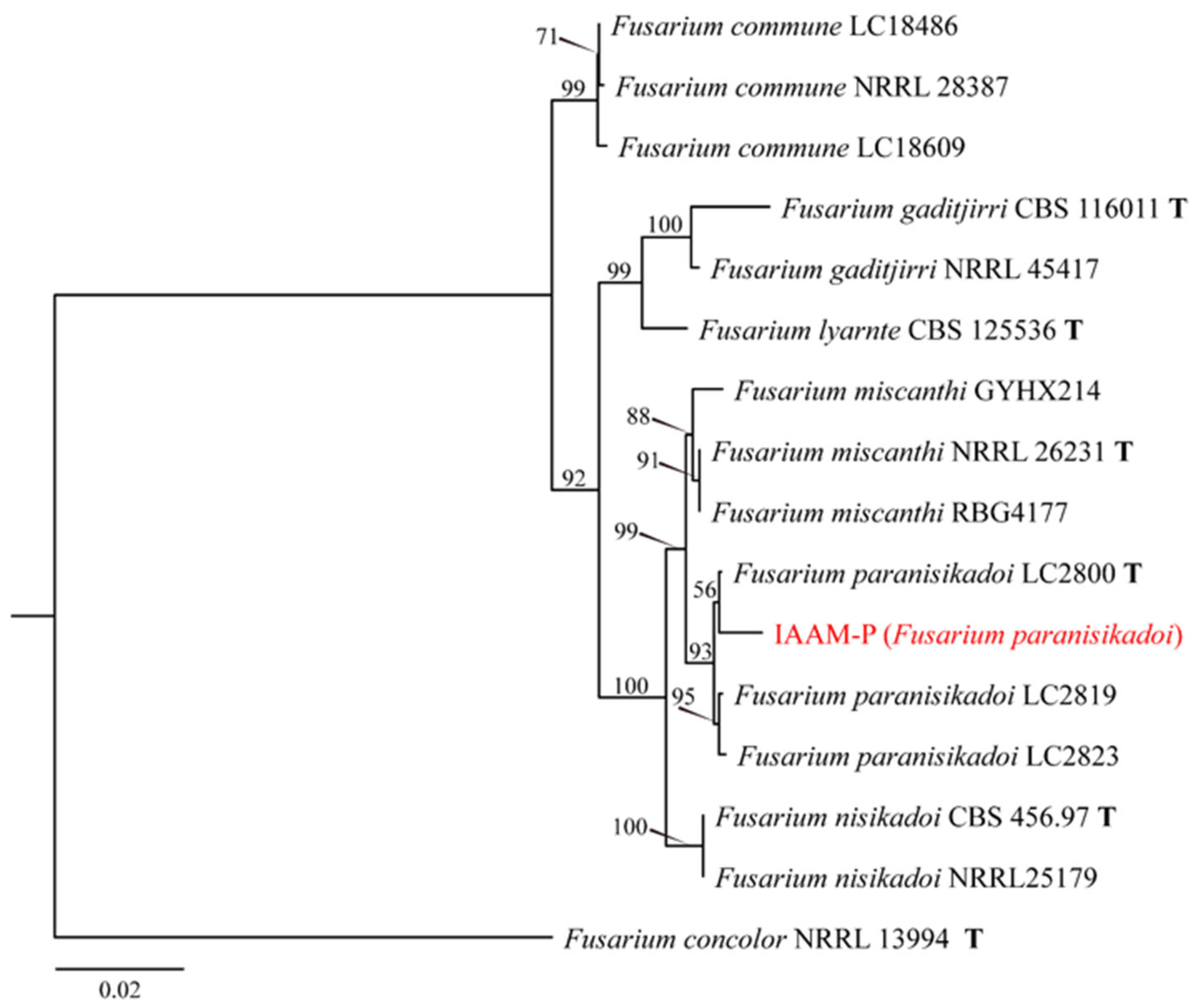
- Identification of Putative Pathogenic Fungus IAAM-P
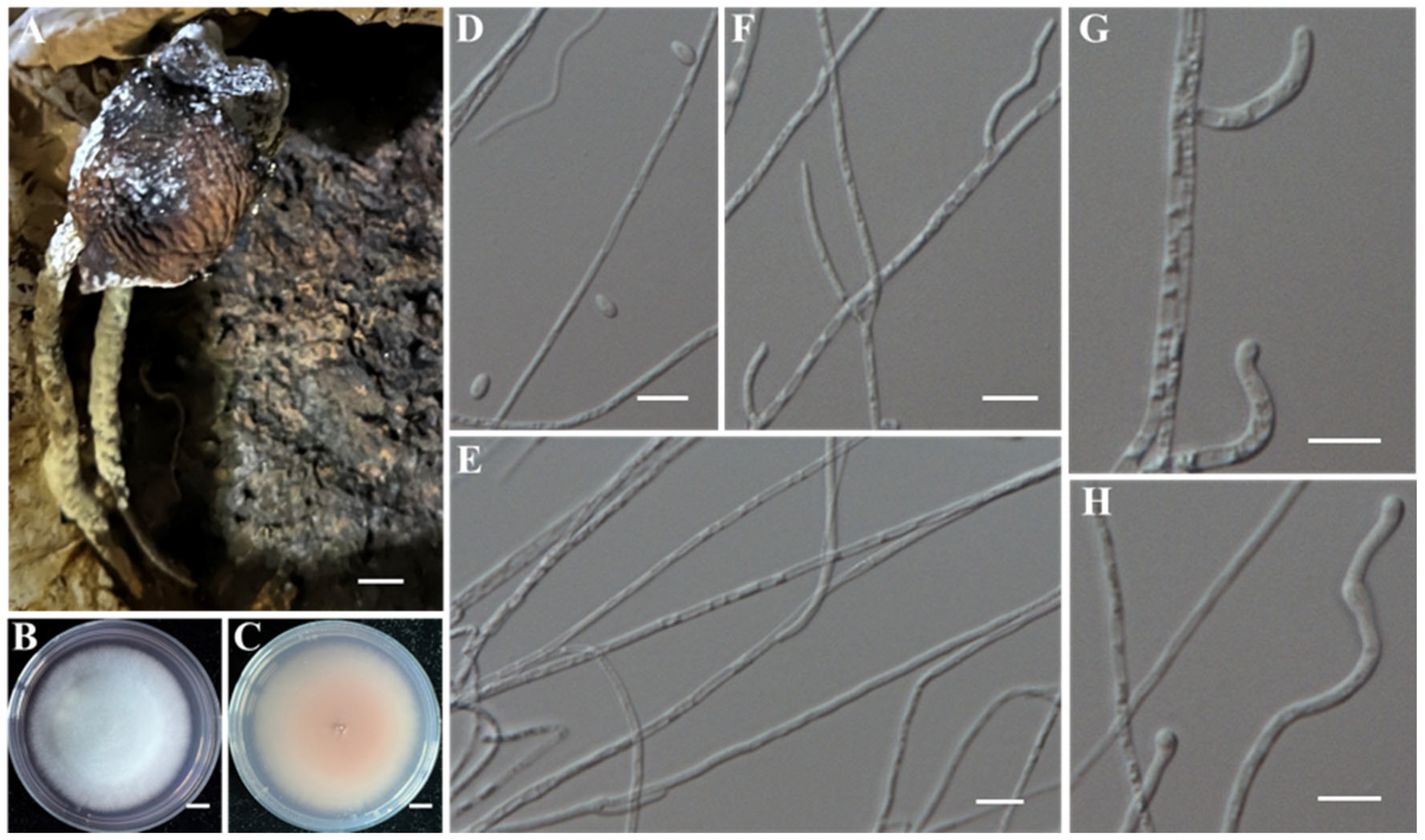
- (5)
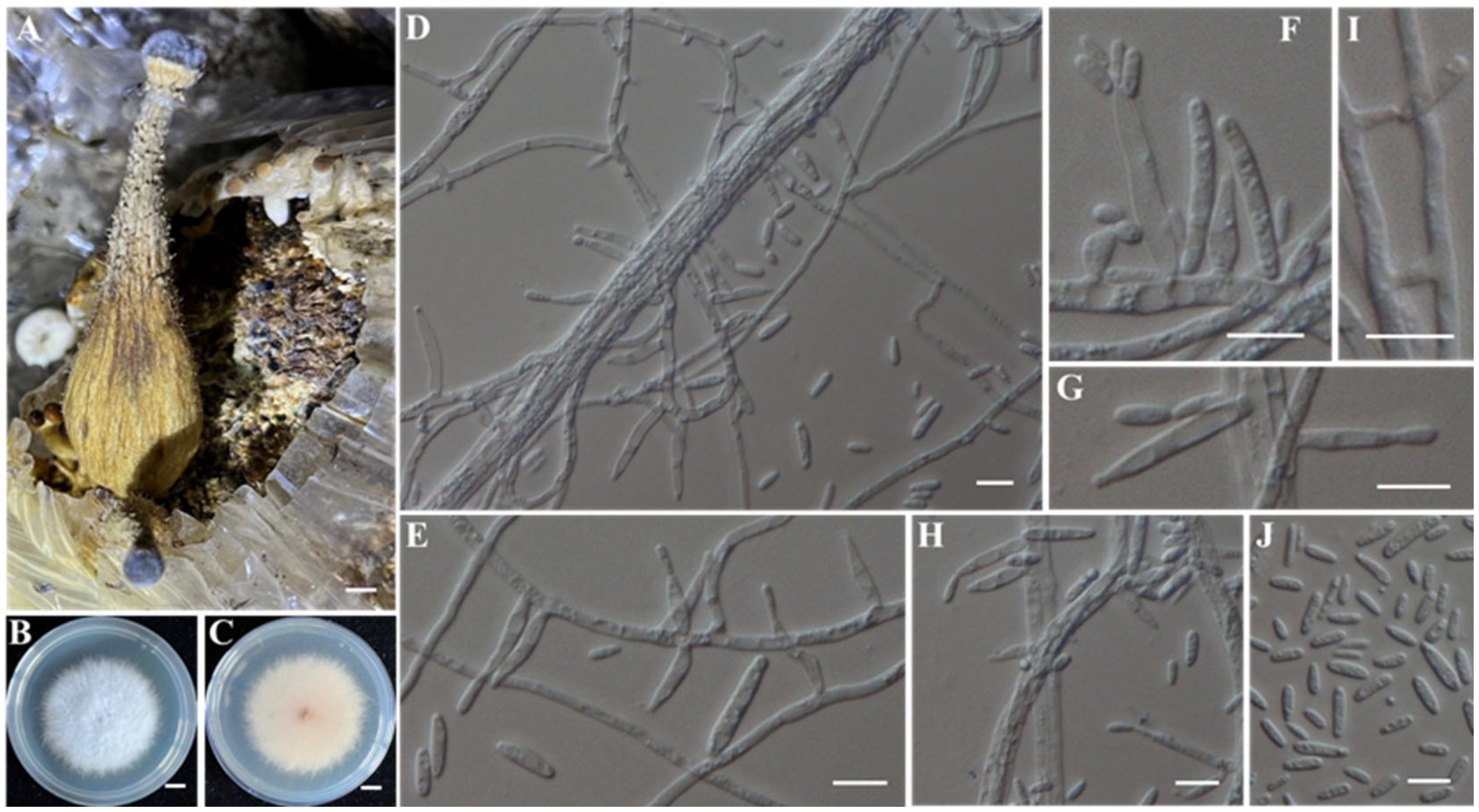
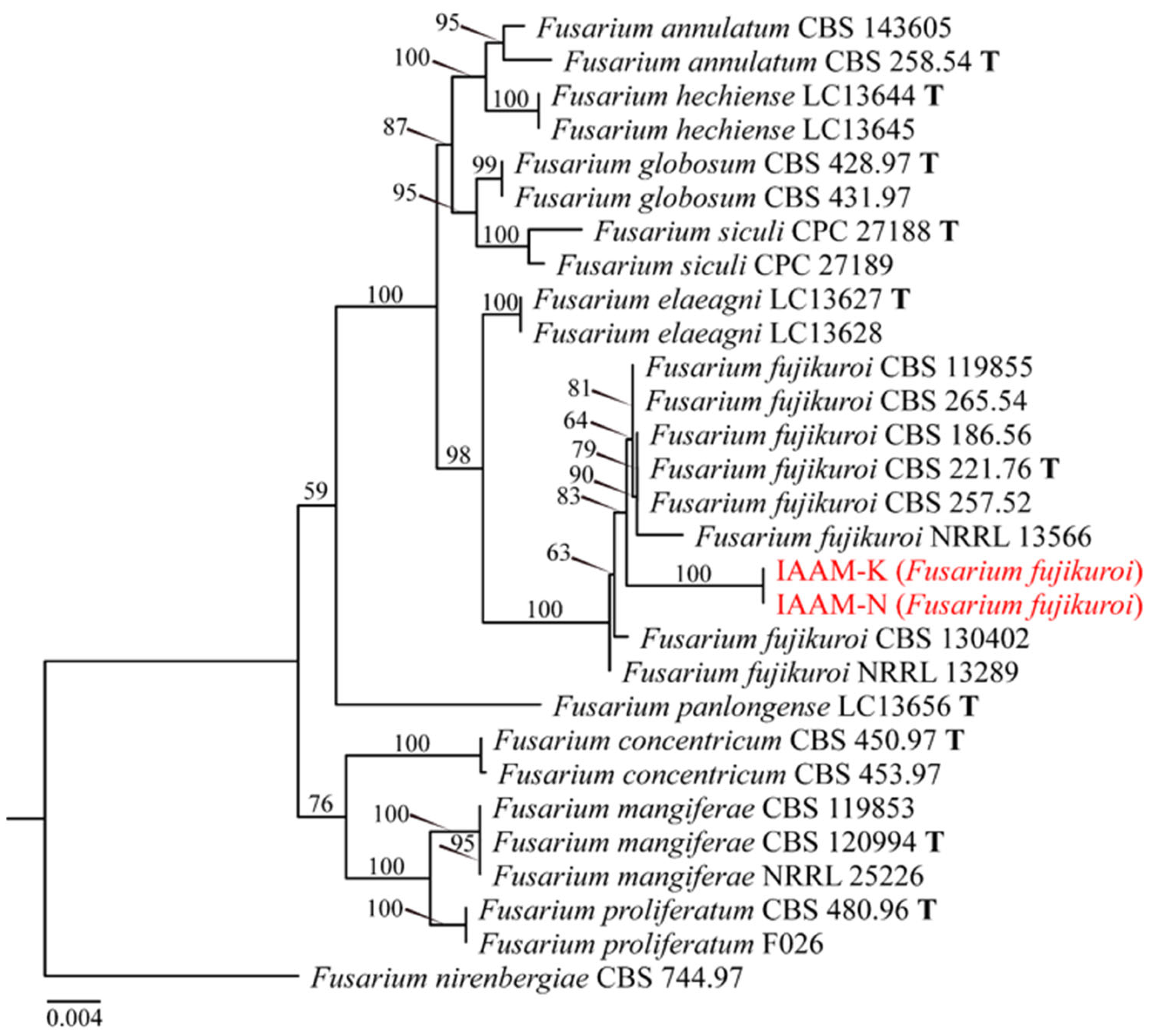
- Identification of Putative Pathogenic Fungi IAAM-K and IAAM-N
- (6)
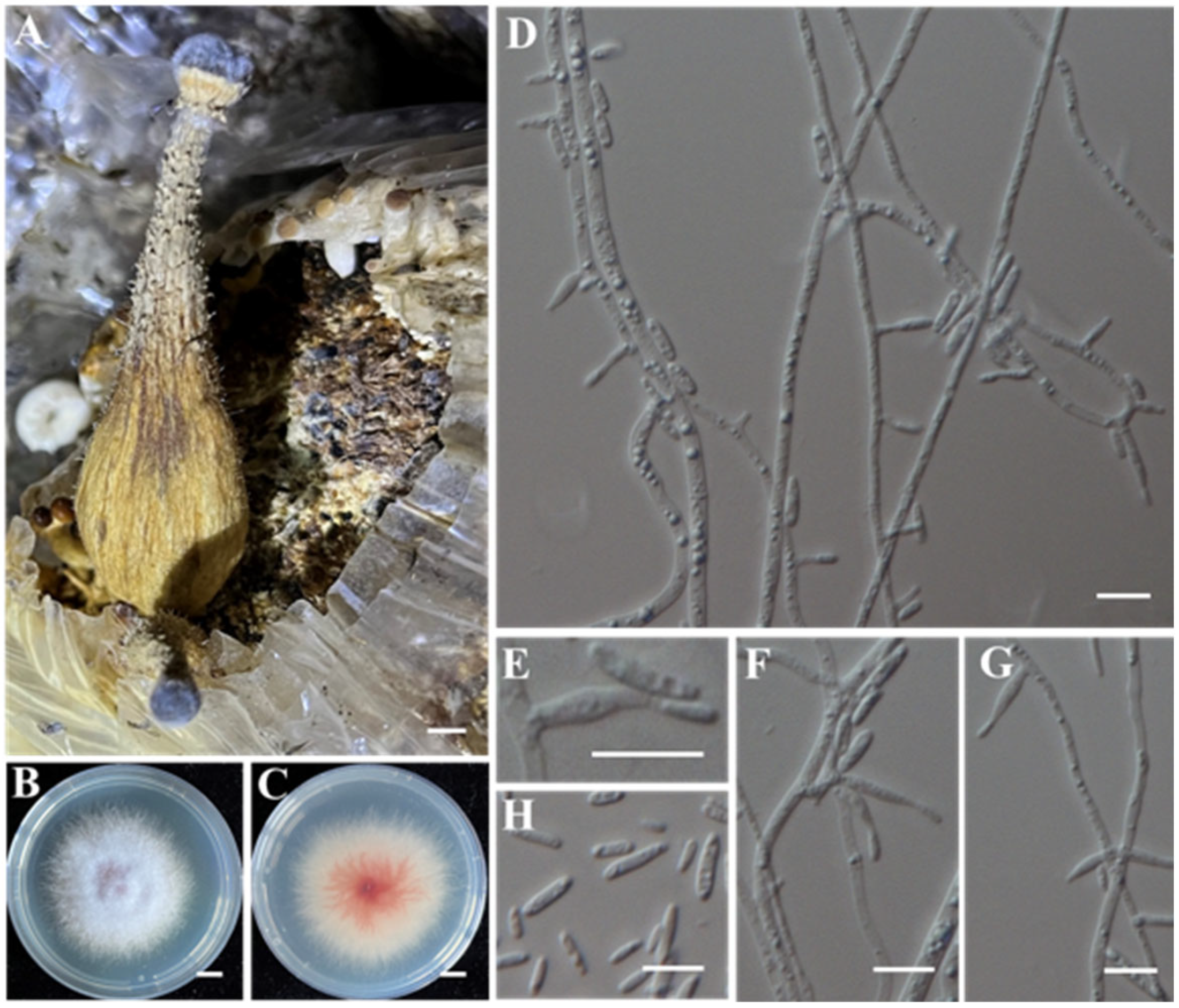
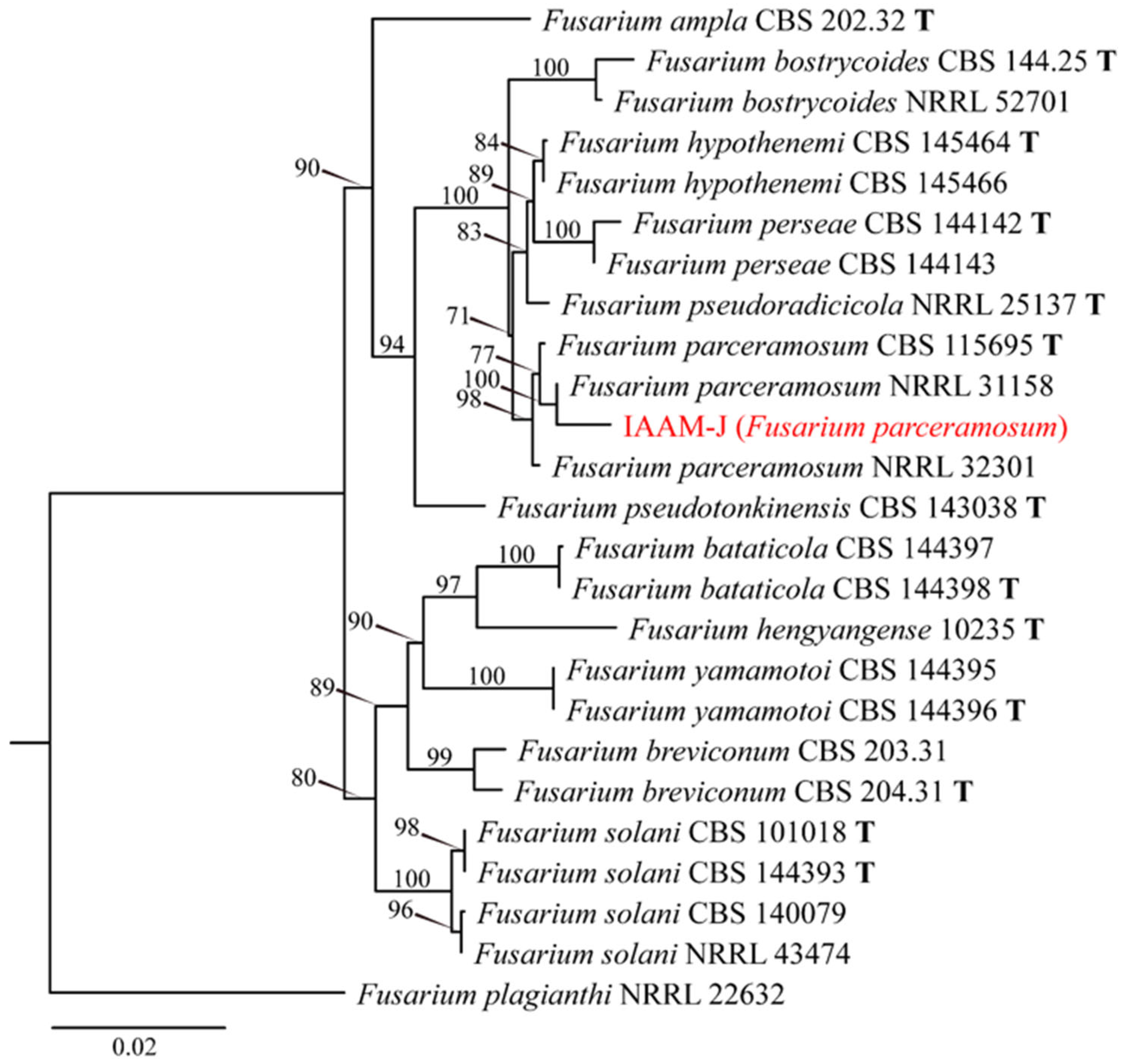
- Identification of Putative Pathogenic Fungus IAAM-J
- (7)
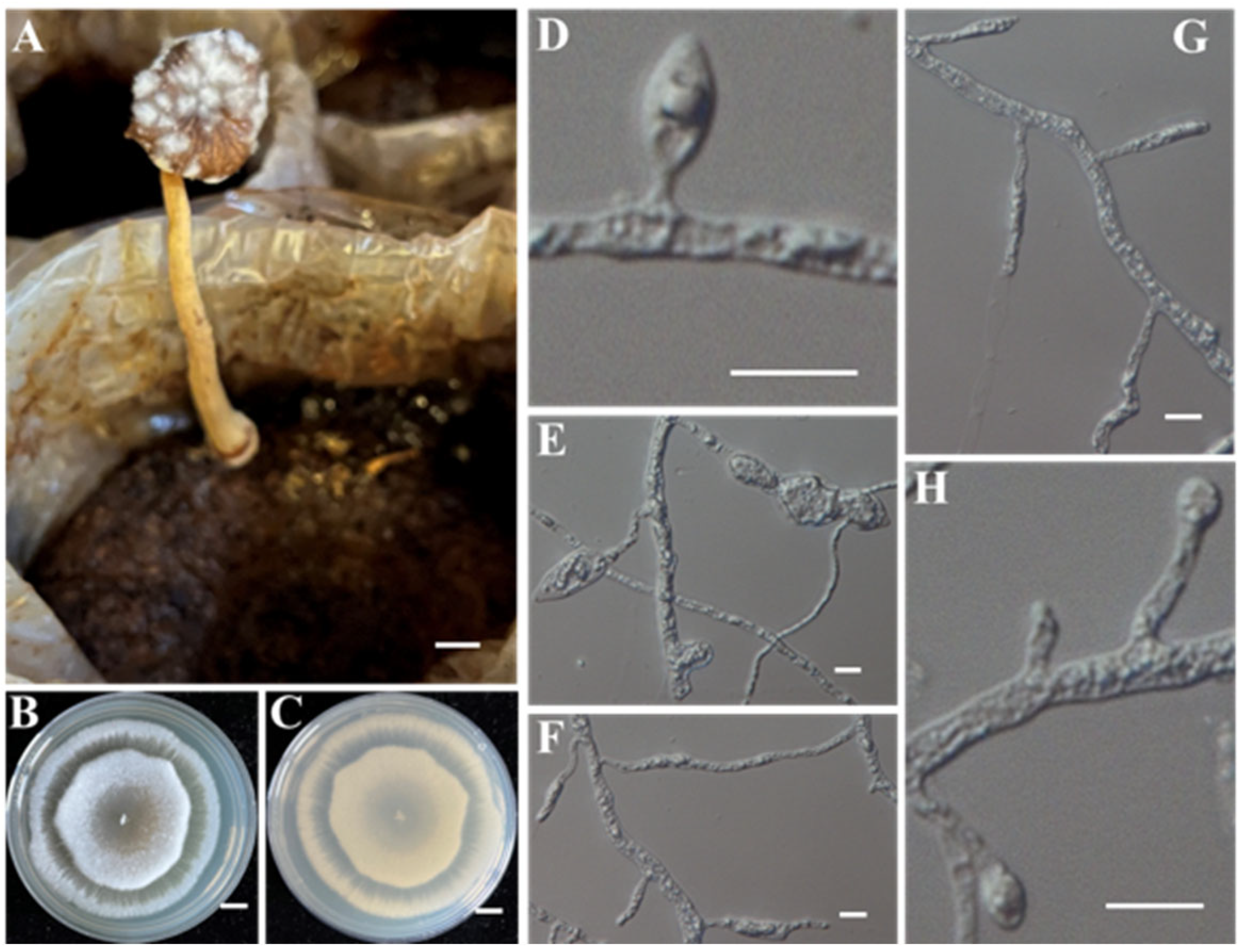
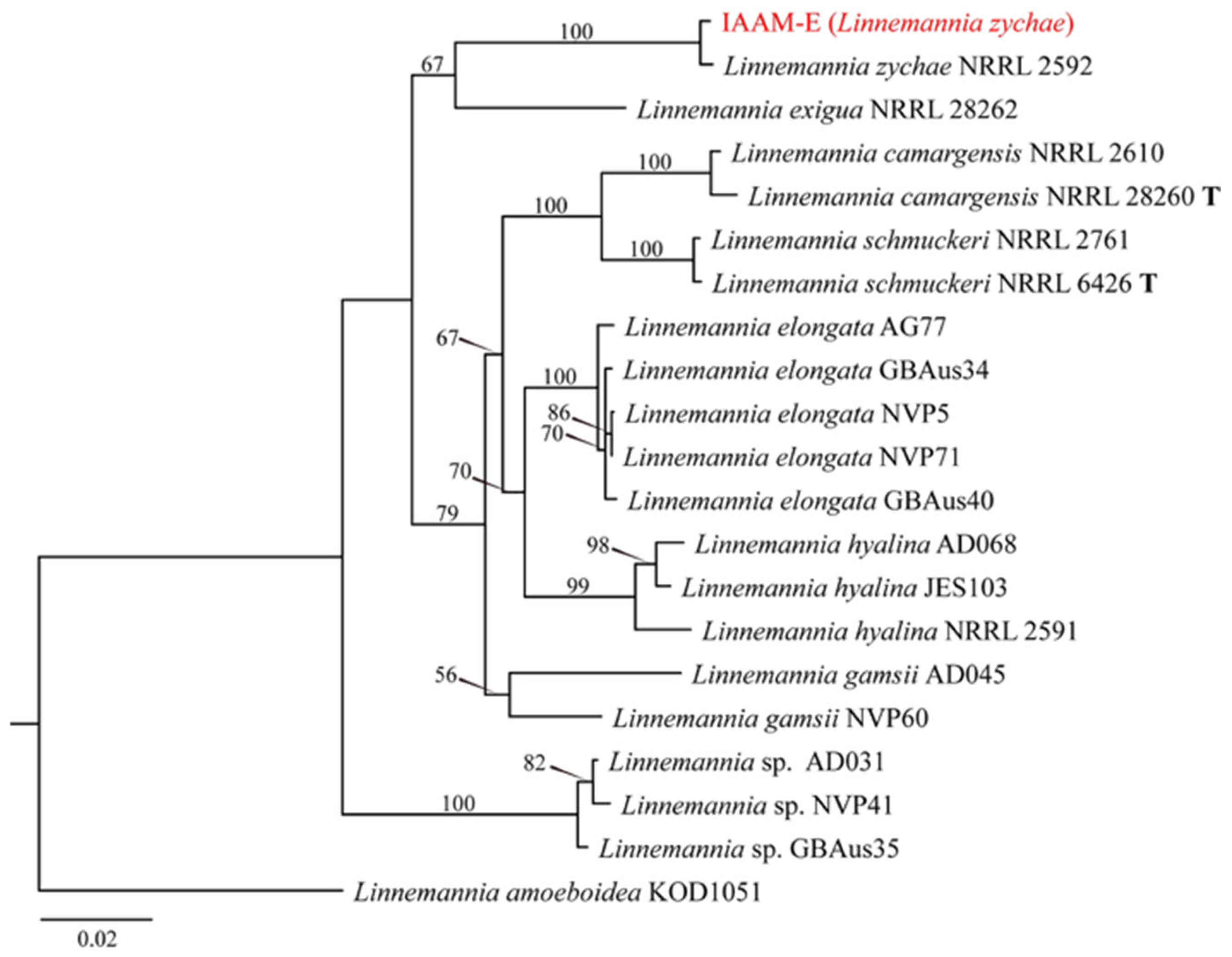
- Identification of Putative Pathogenic Fungus IAAM-E
- (8)
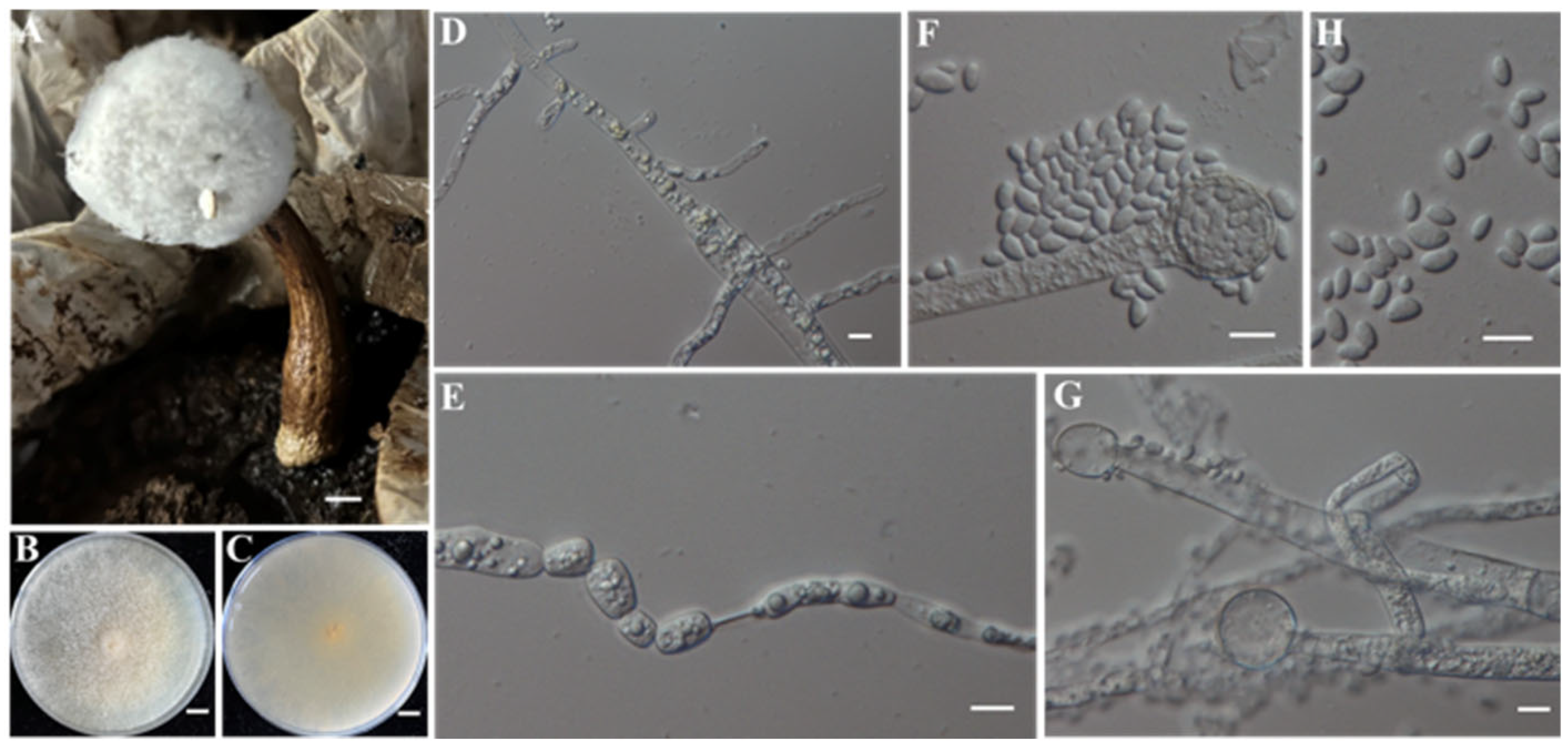
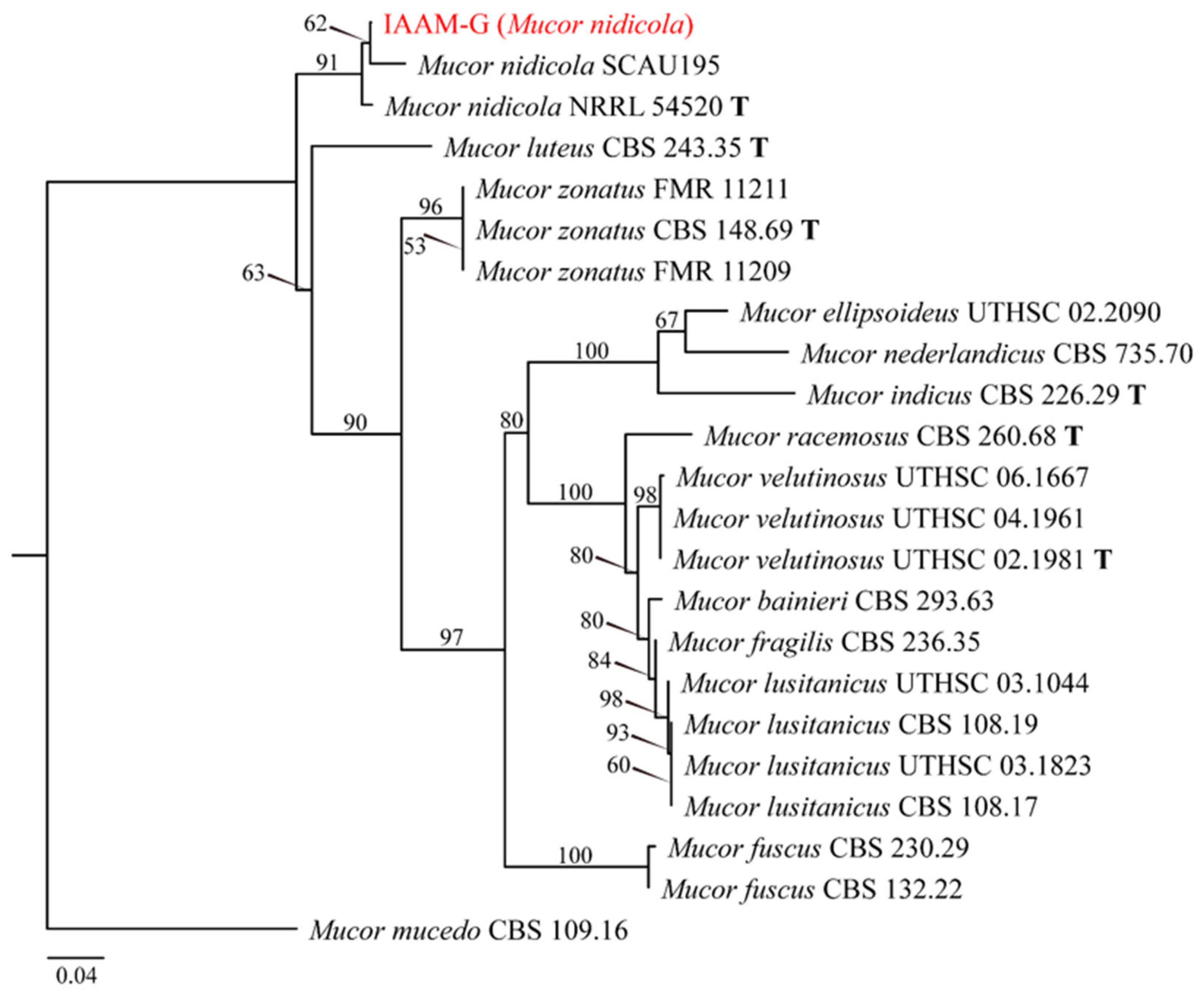
- Identification of Putative Pathogenic Fungus IAAM-G
- (9)
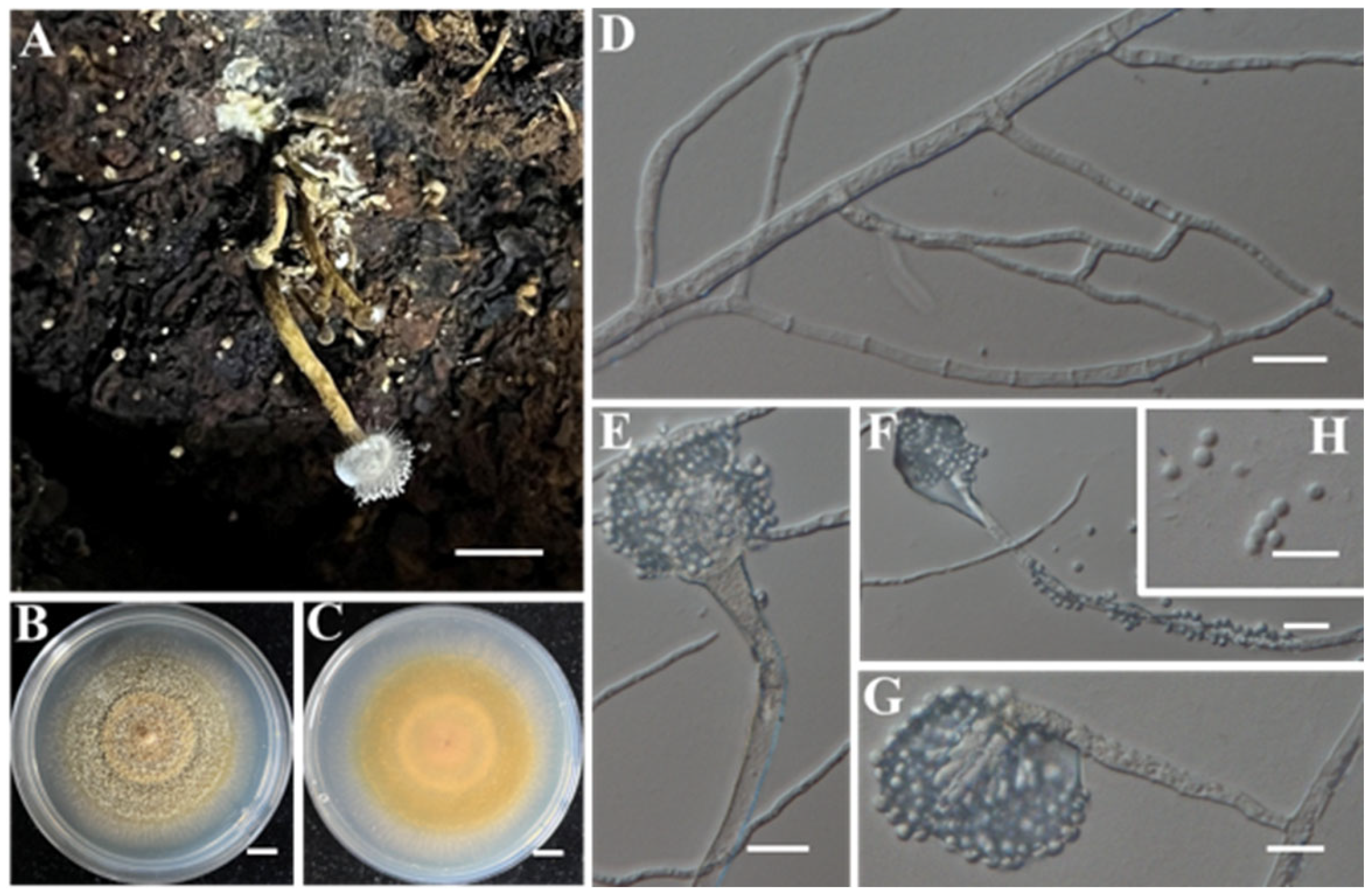
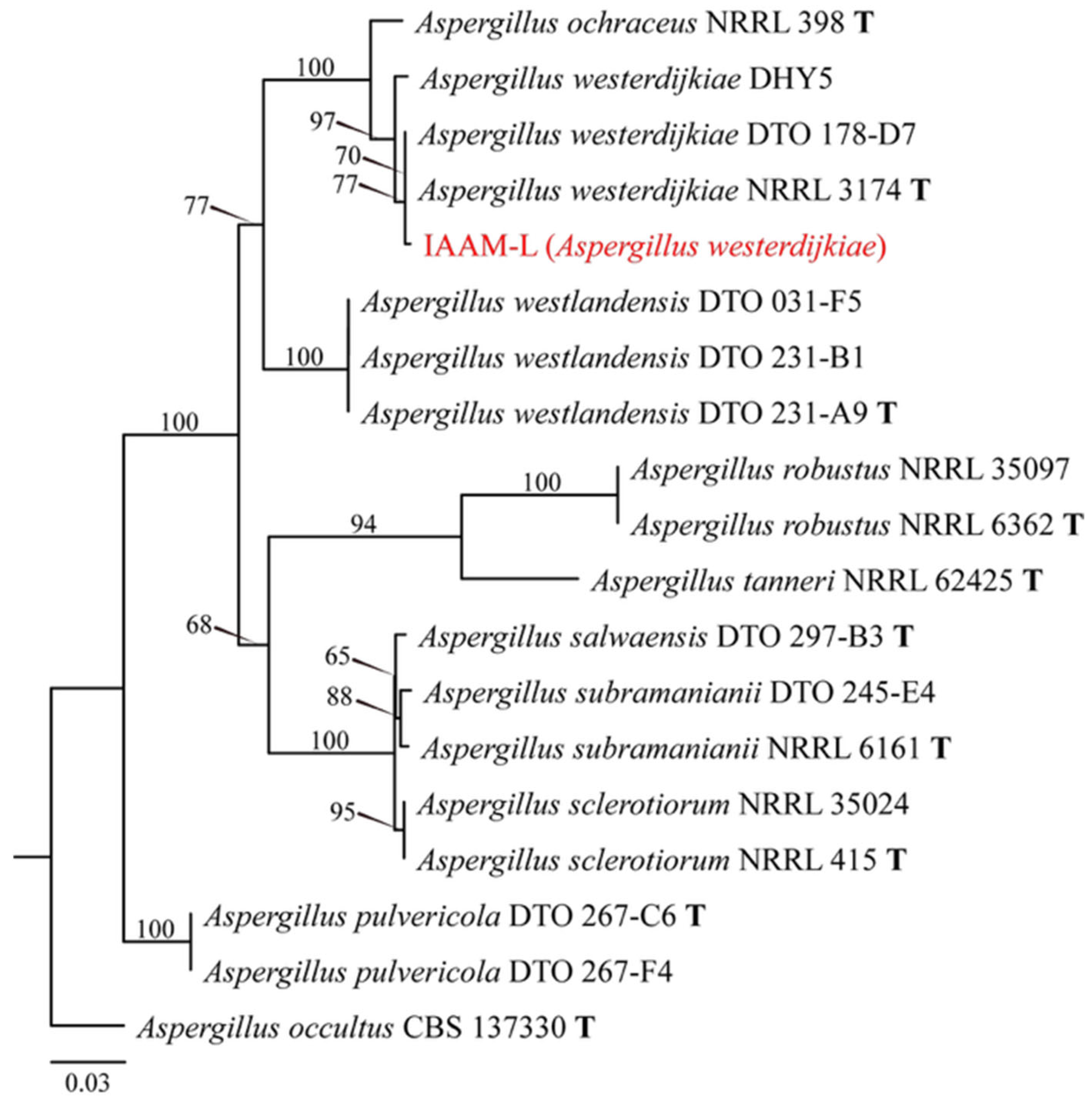
- Identification of Putative Pathogenic Fungus IAAM-L
- (10)
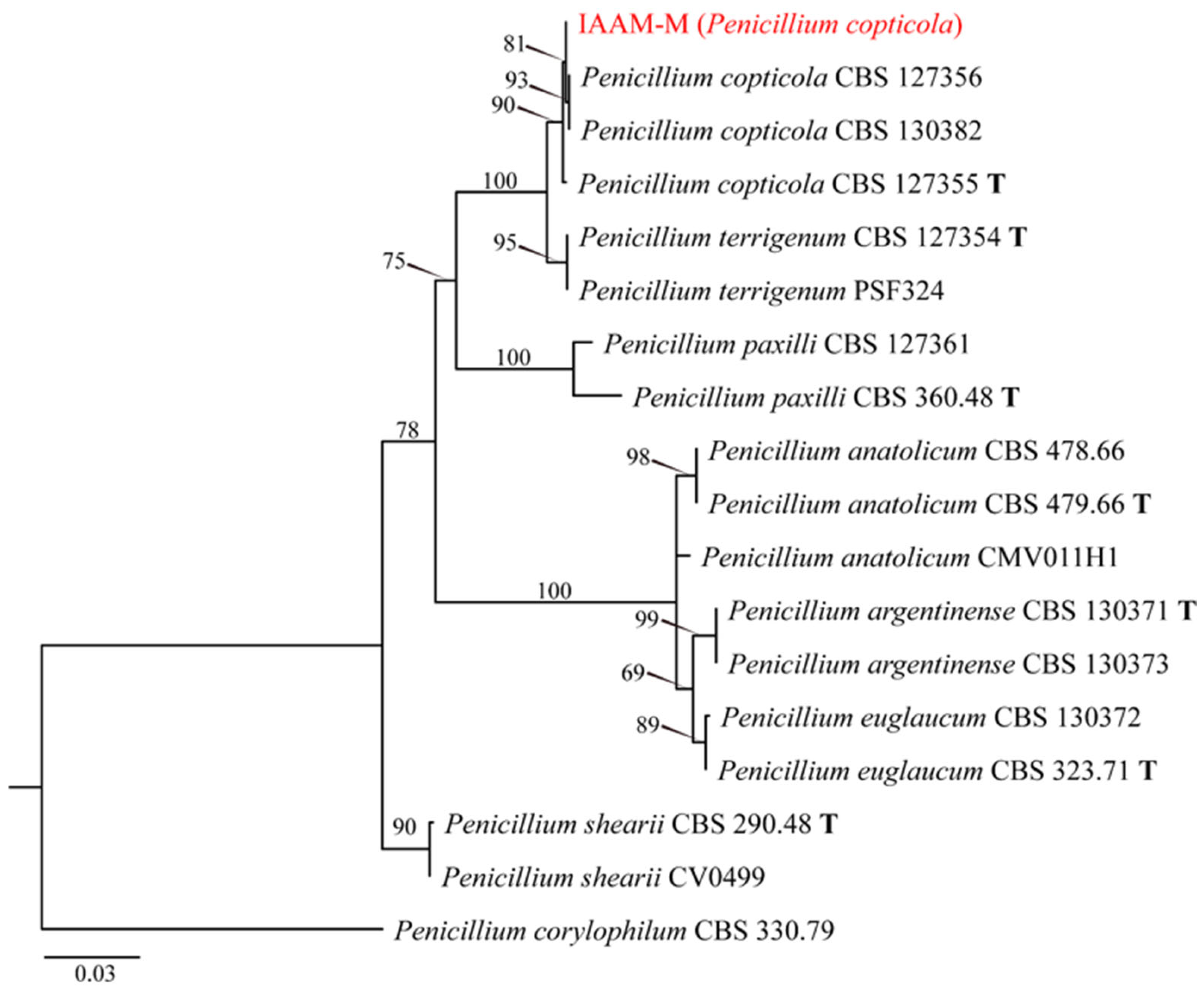
- Identification of Putative Pathogenic Fungus IAAM-M
- (11)
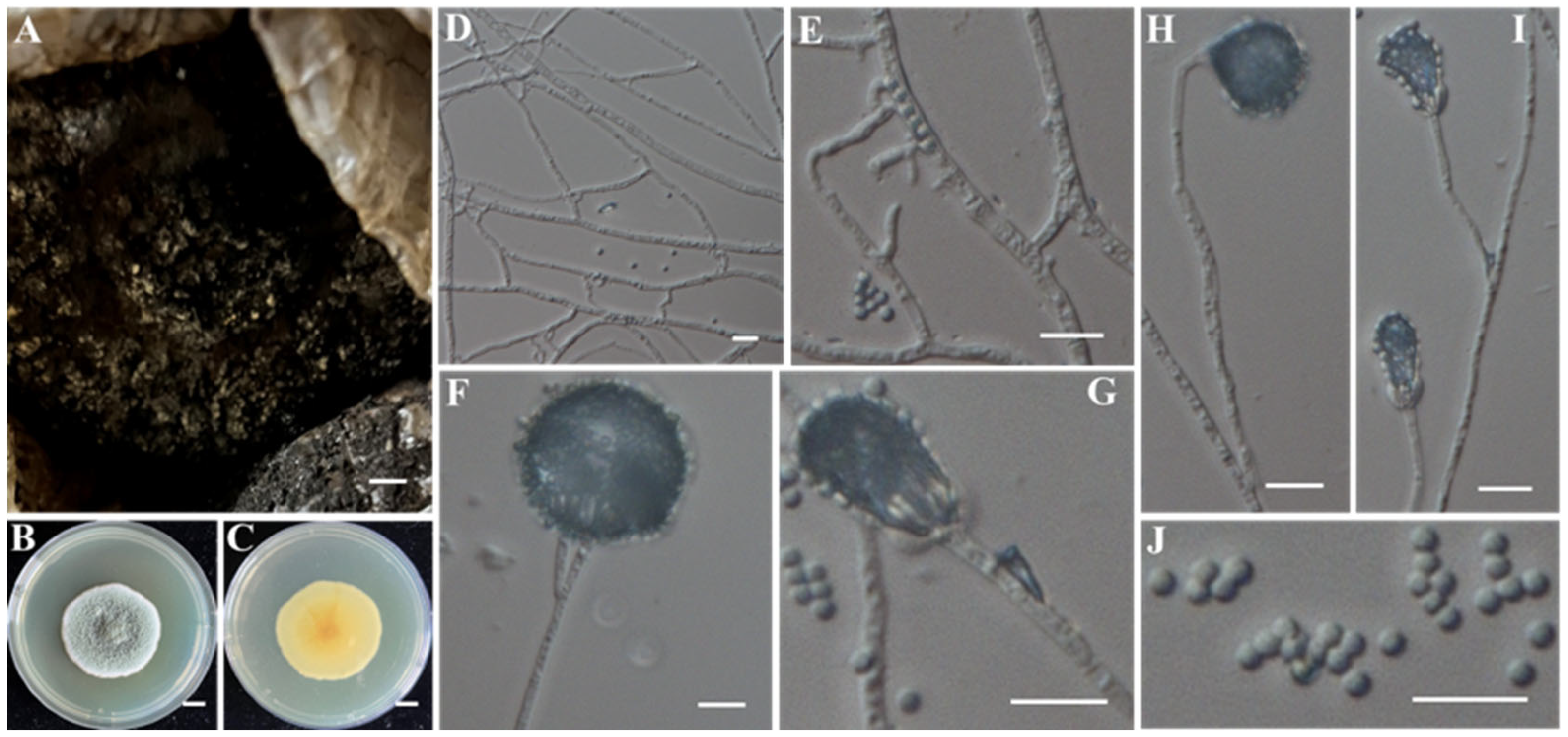
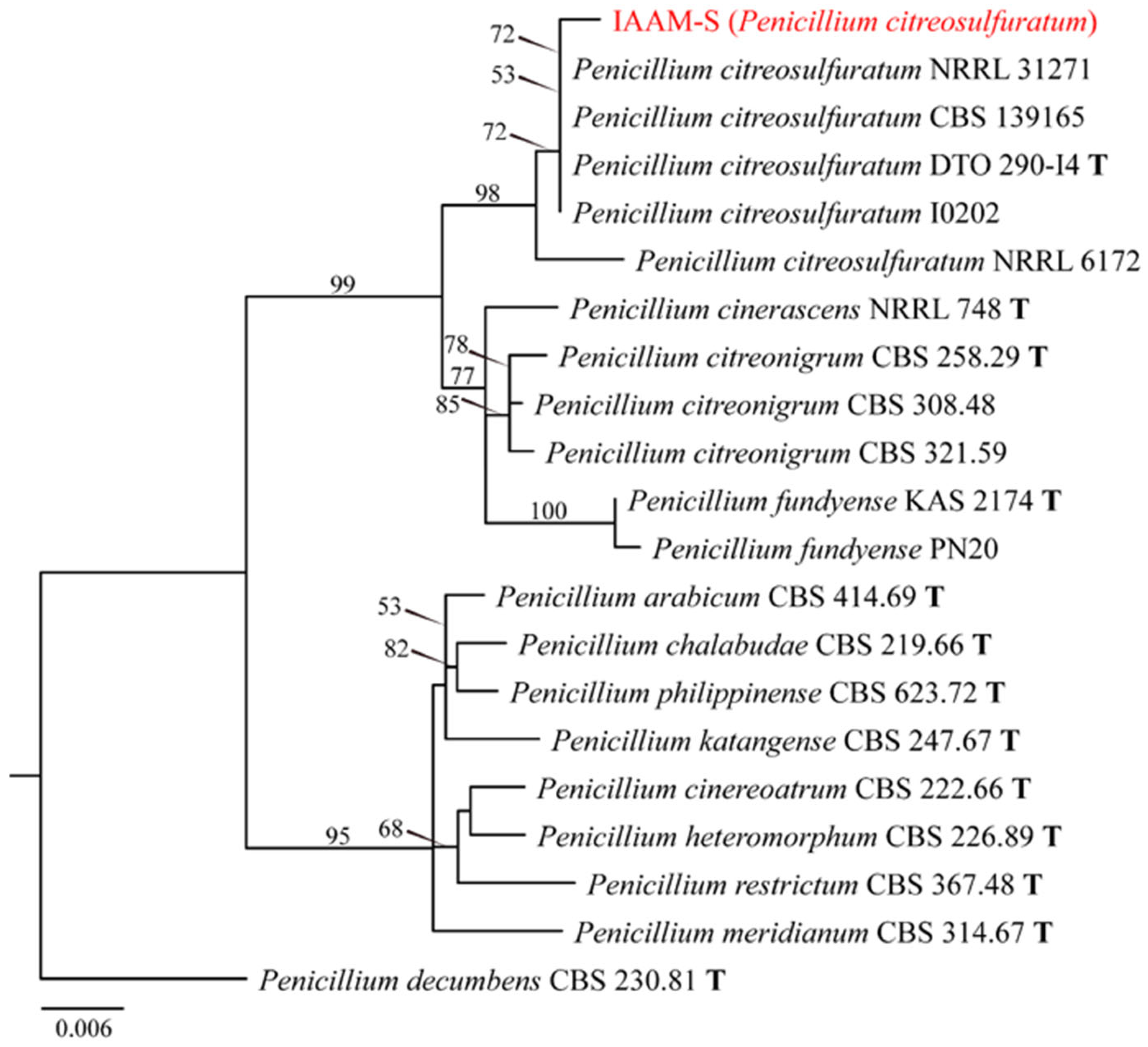
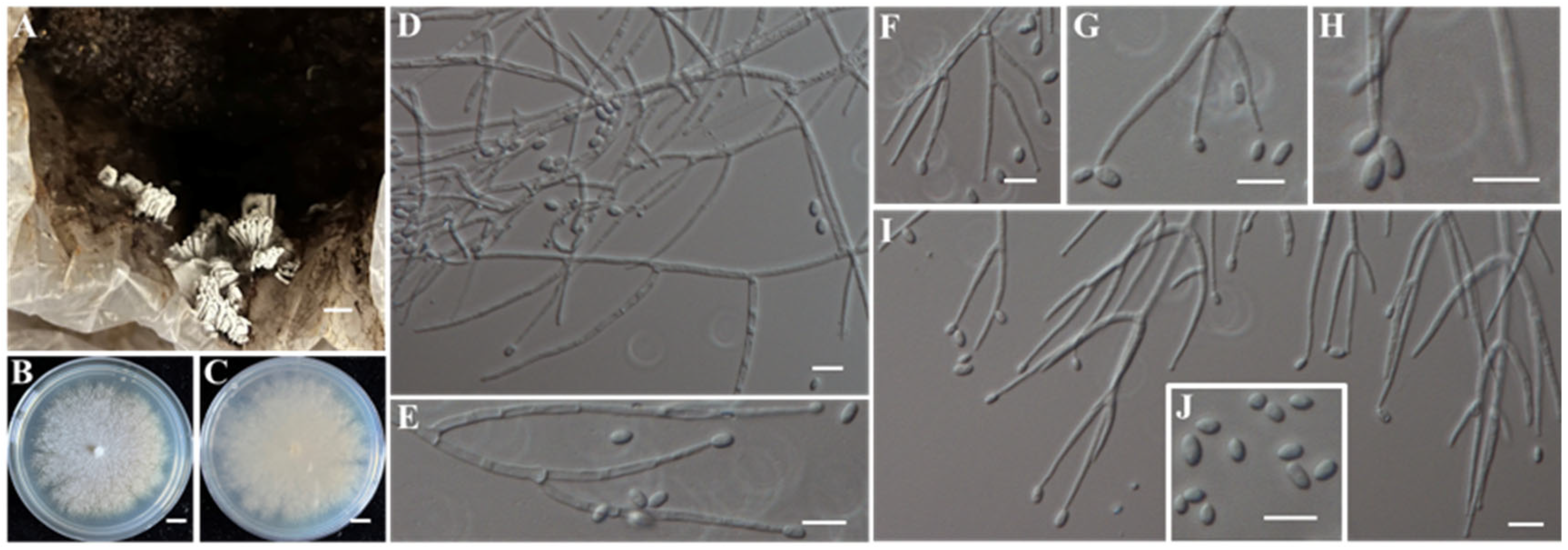
- Identification of Putative Pathogenic Fungus IAAM-S
- (12)
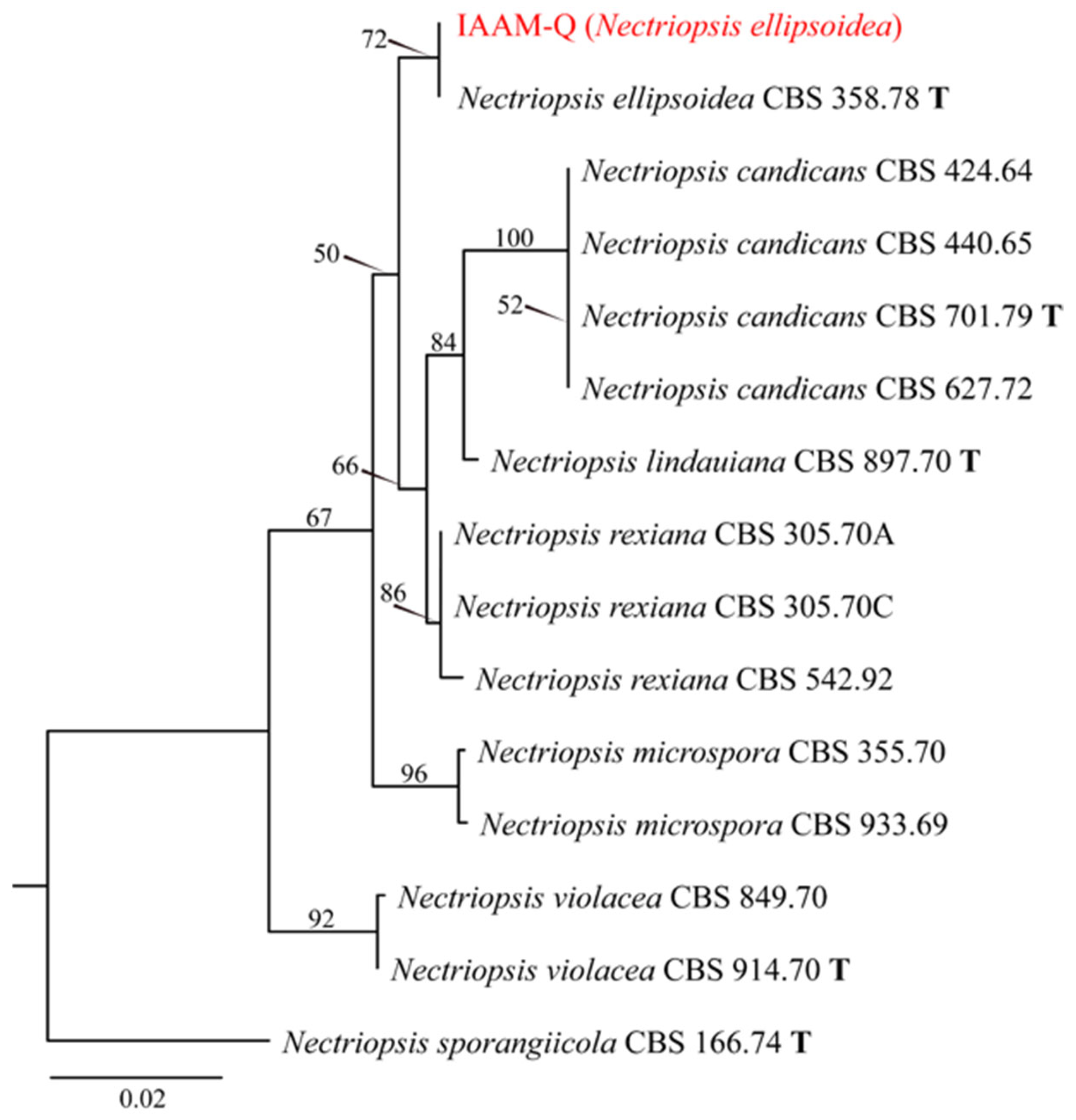
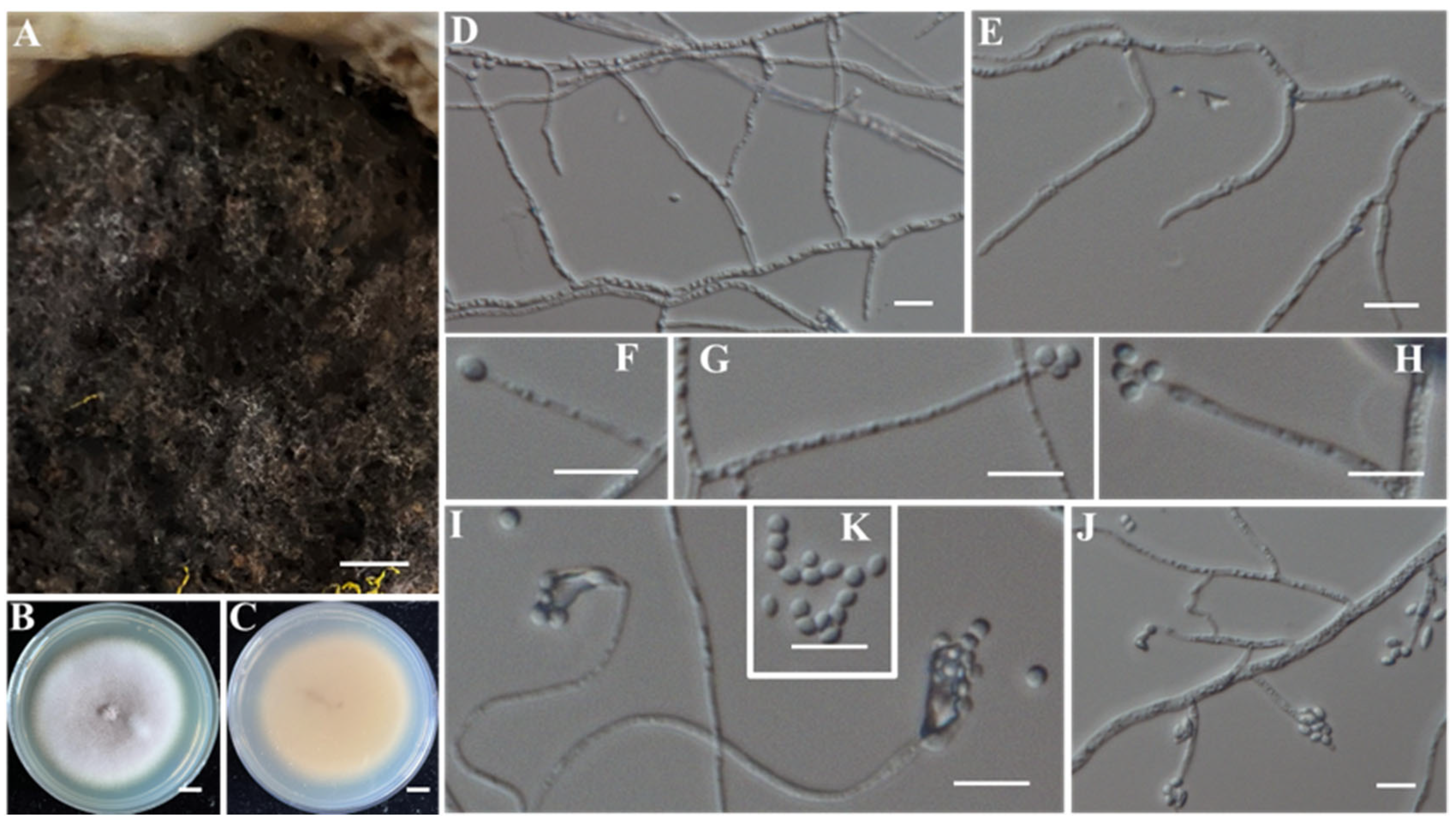
- Identification of Putative Pathogenic Fungus IAAM-Q
- (13)
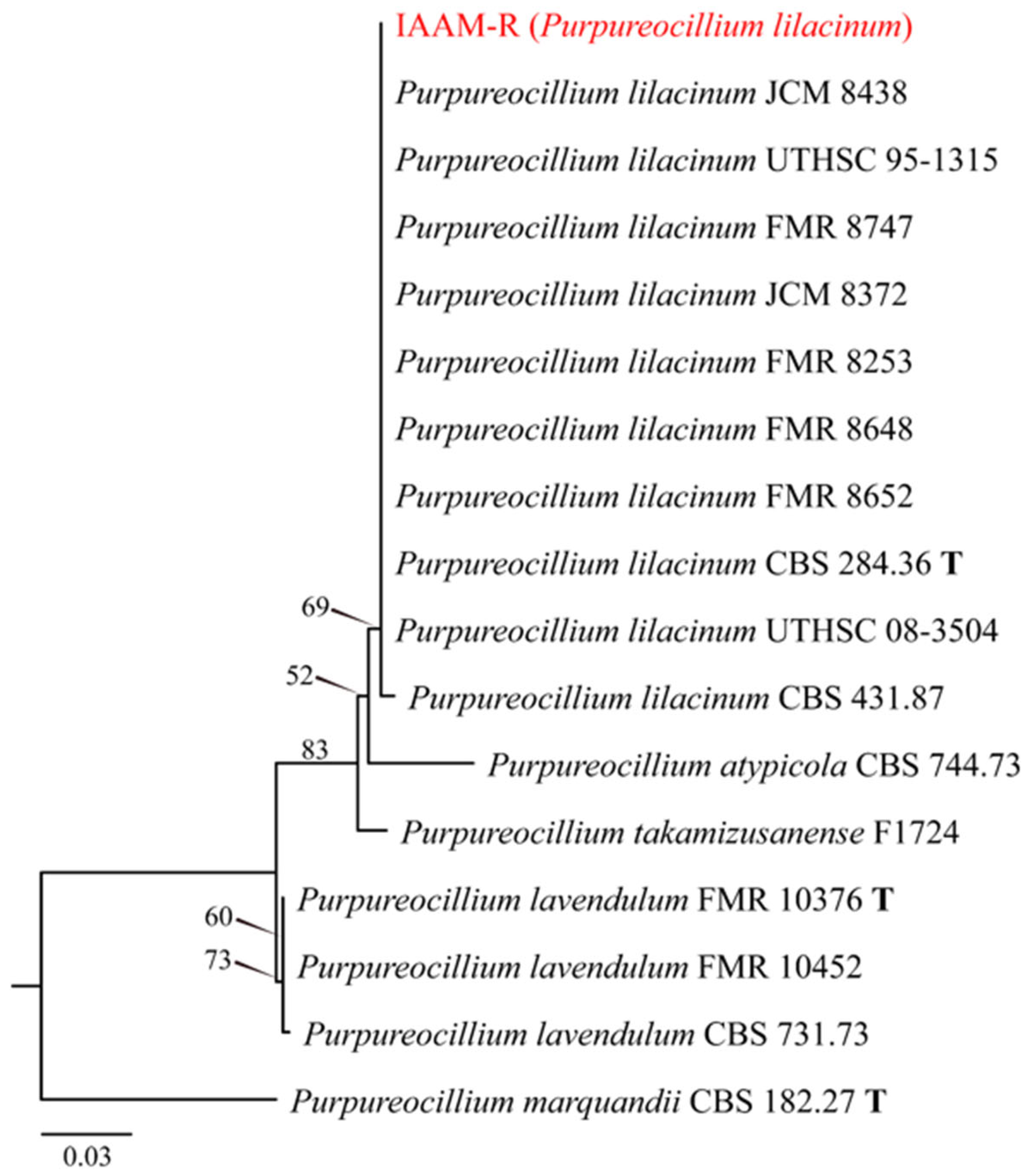
- Identification of Putative Pathogenic Fungus IAAM-R
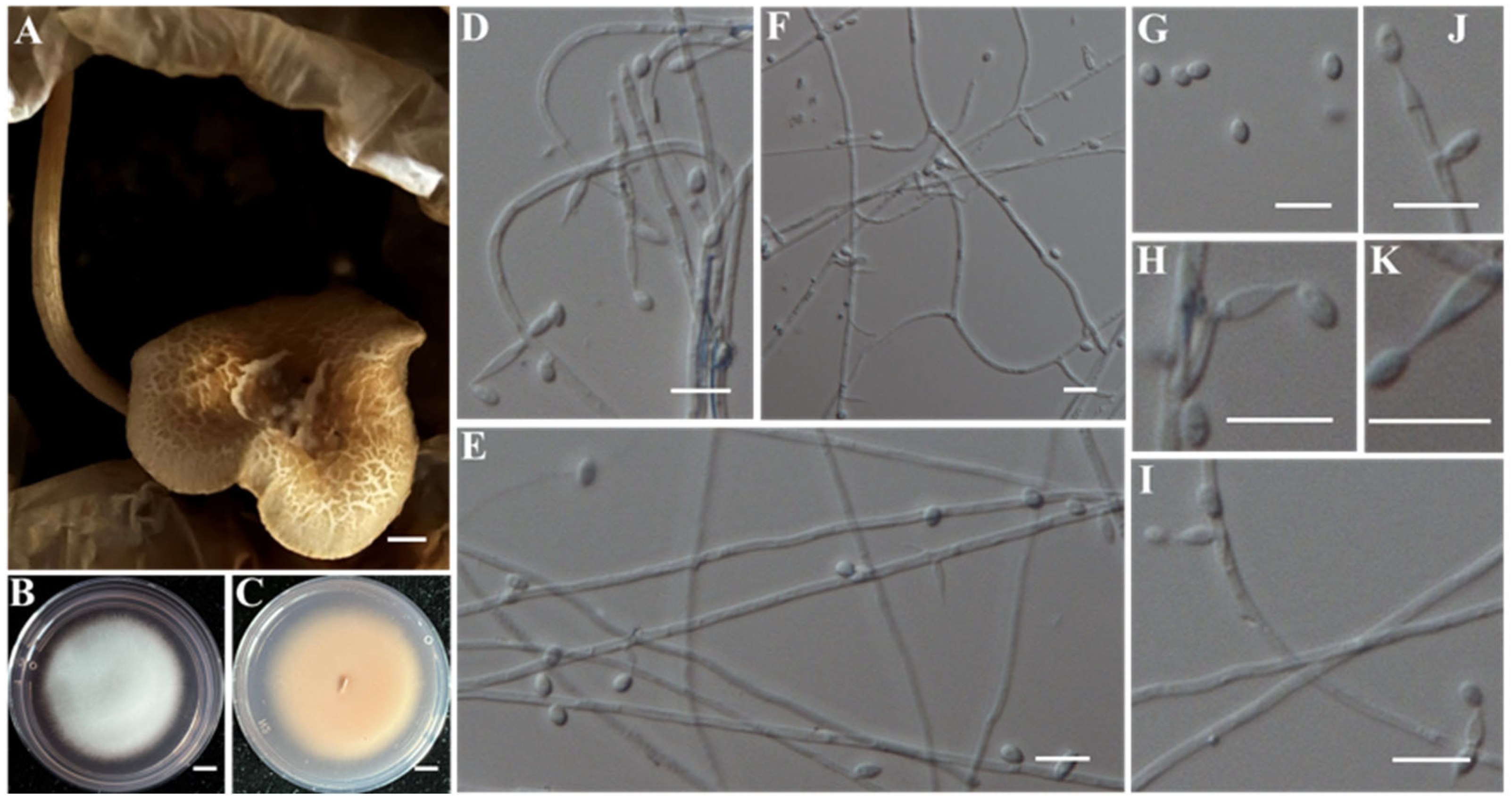
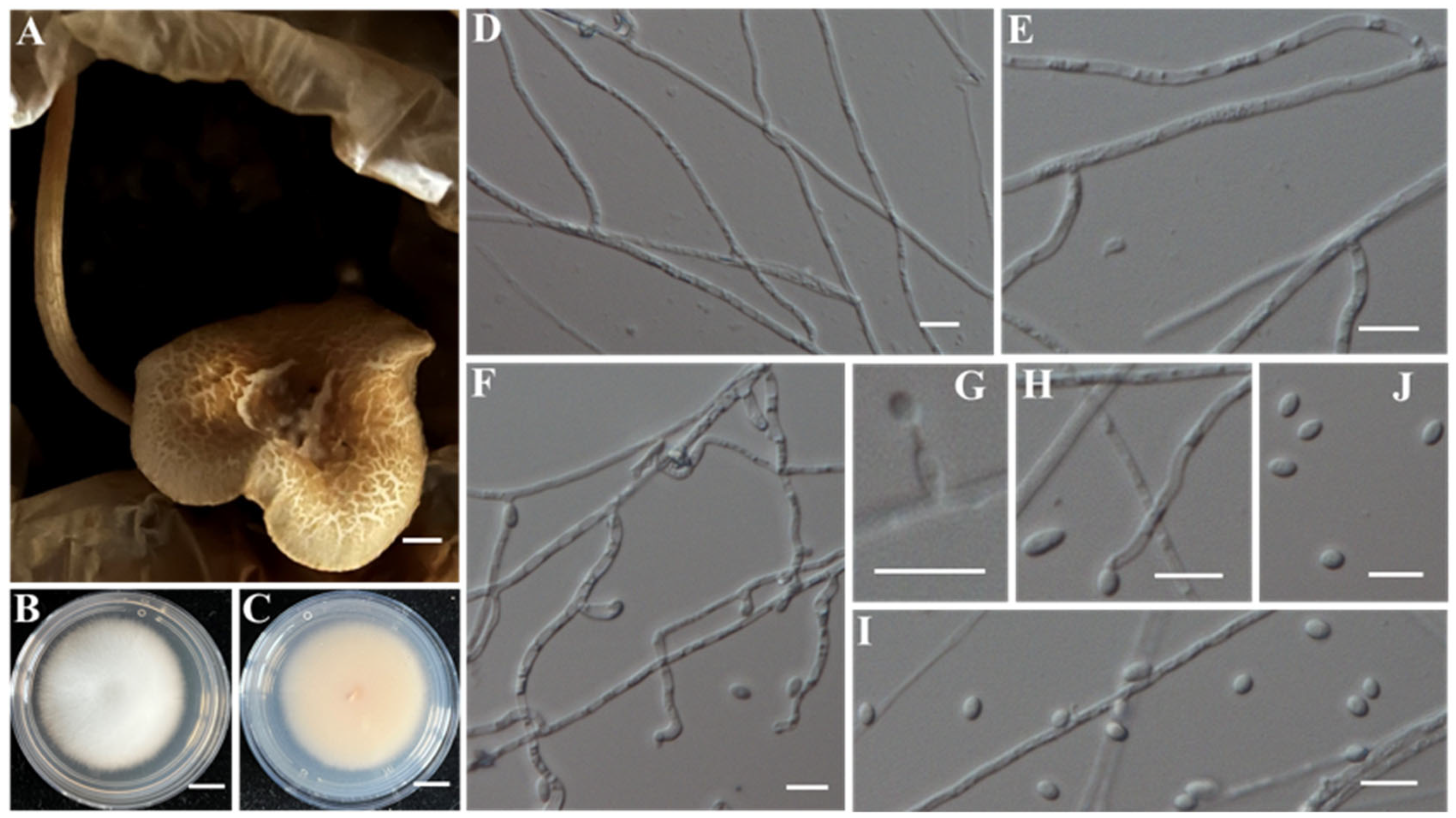
- (14)
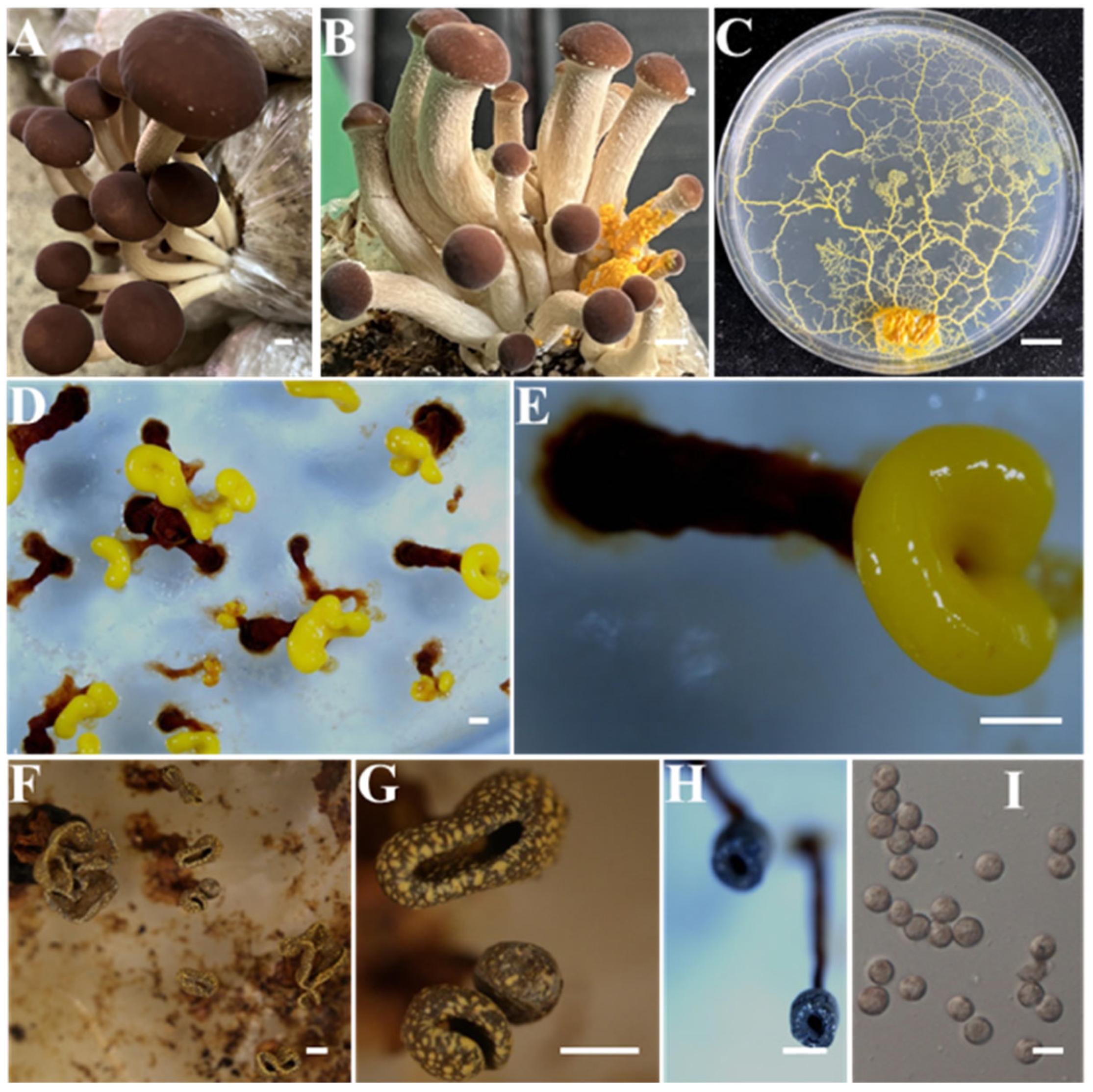
- Identification of Putative Pathogen IAAM-W0001
- (15)
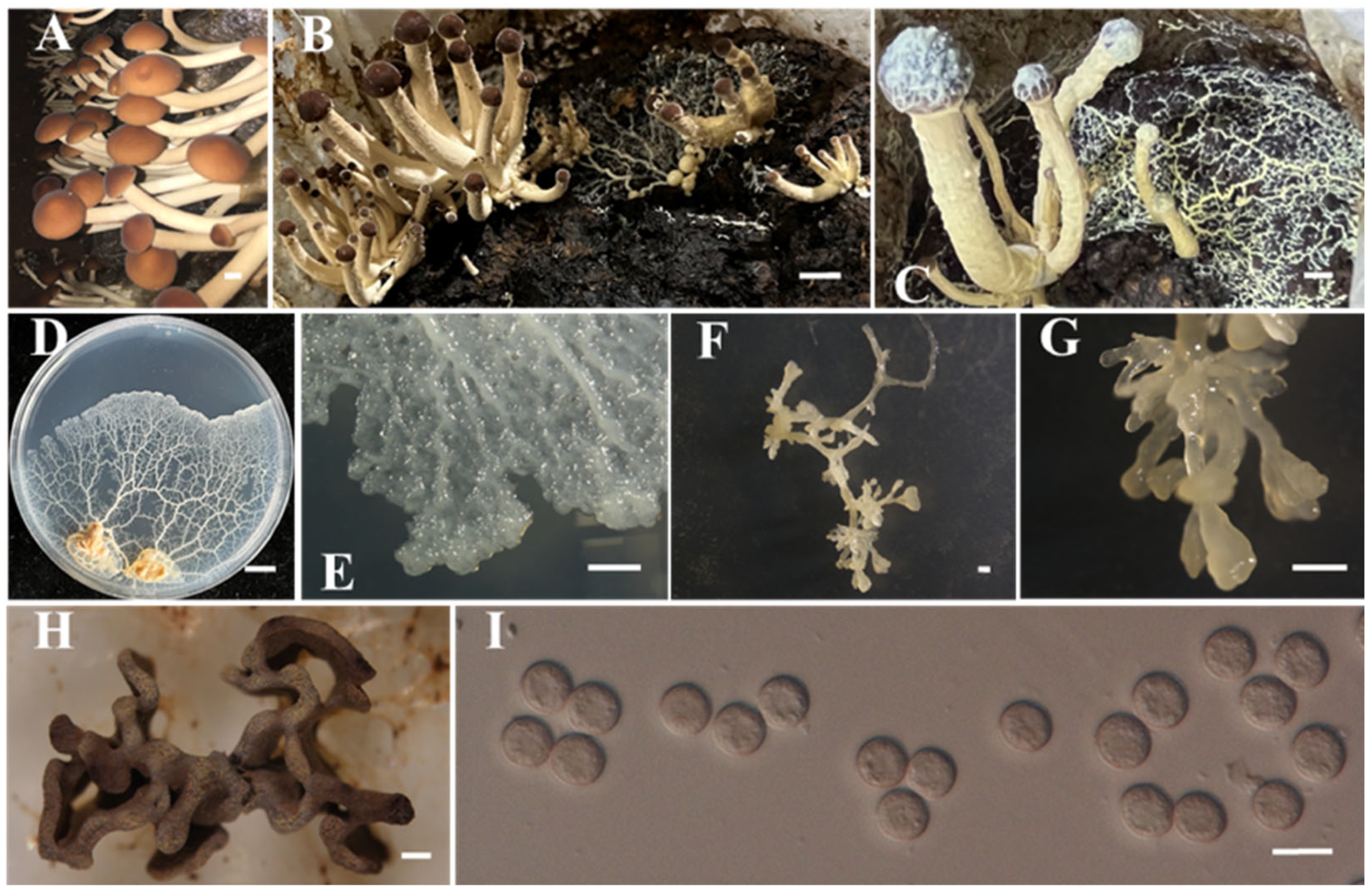
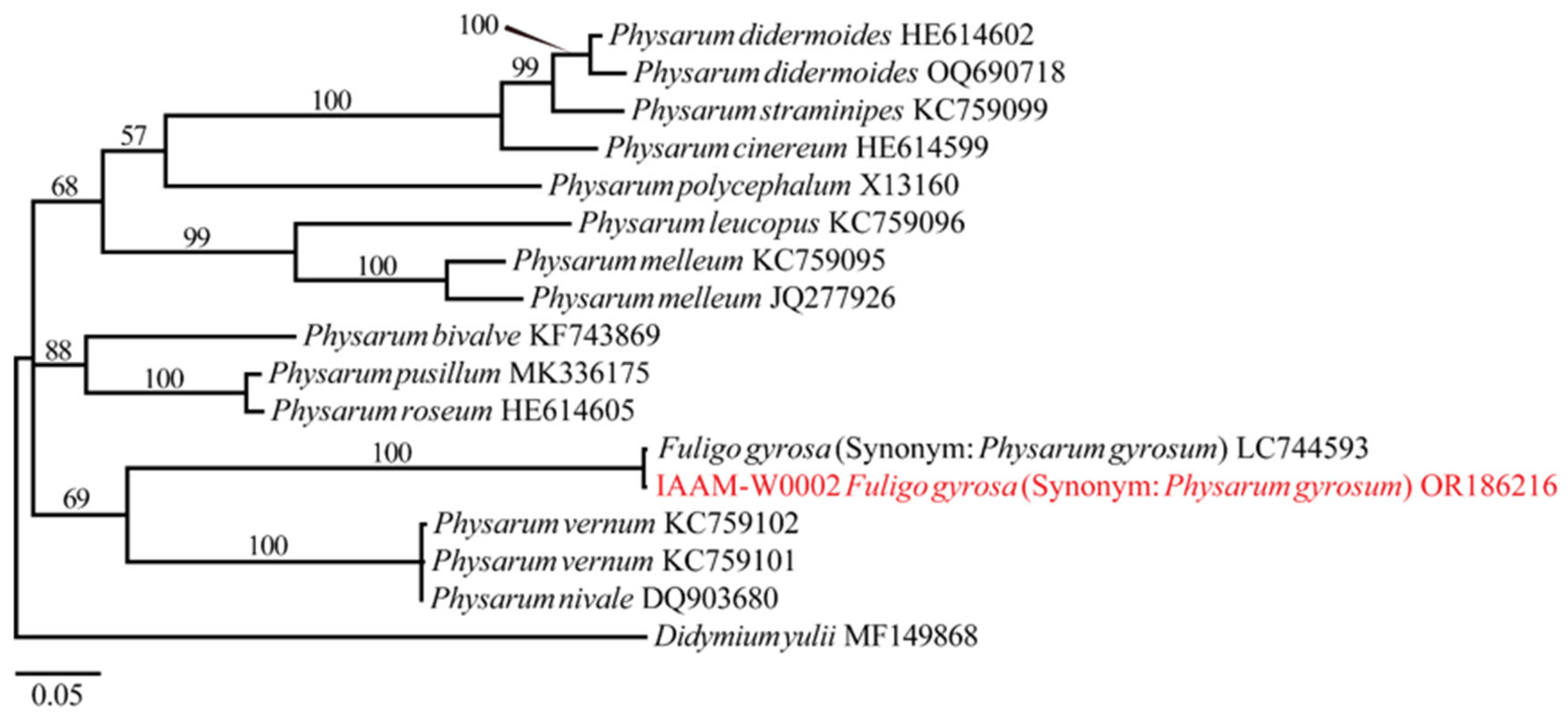
- Identification of Putative Pathogen IAAM-W0002
3.2. Pathogenicity Assessment of the Primary Putative Pathogen of A. chaxingu
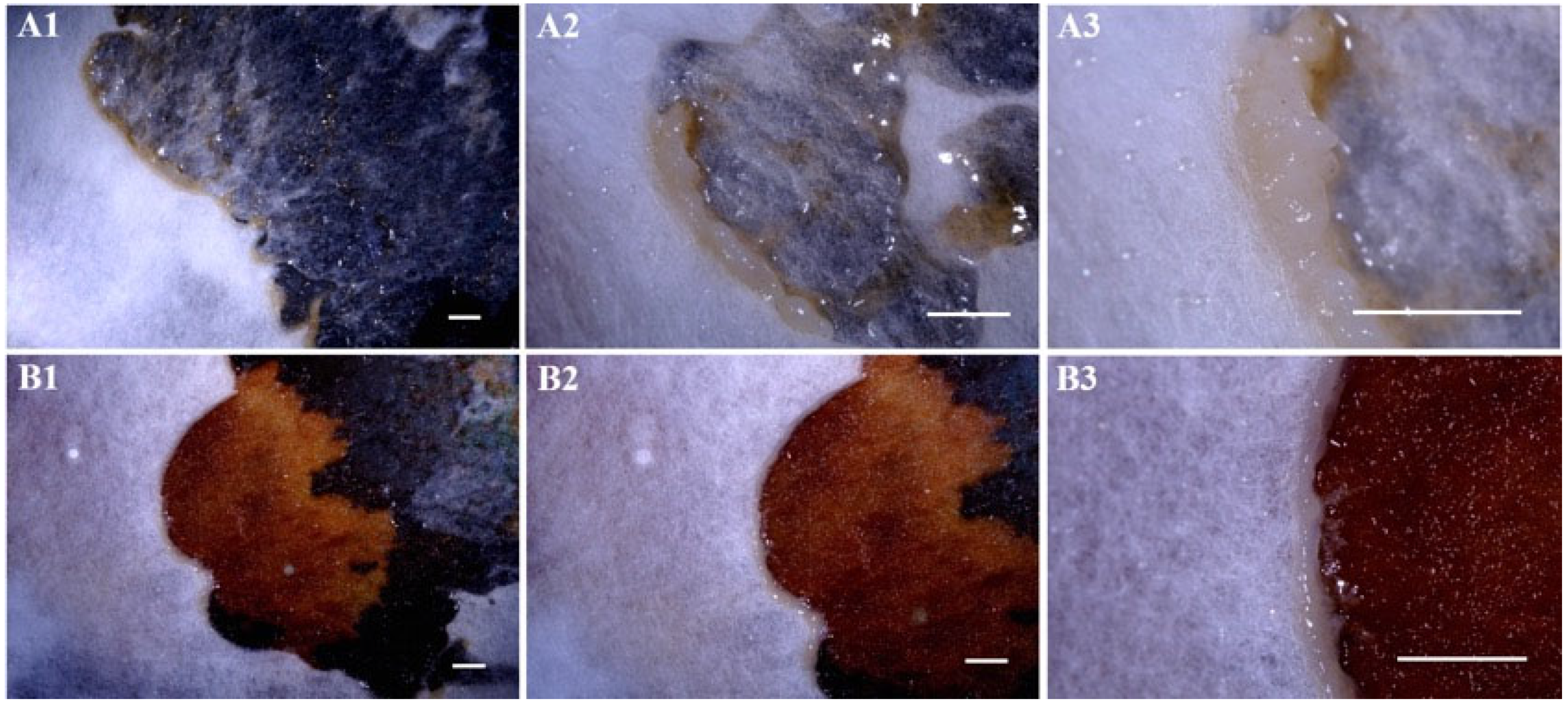
3.2.1. Pathogenicity of F. gyrosa on the Fruiting Bodies of A. chaxingu
3.2.2. Pathogenicity of F. gyrosa on the Mycelium of A. chaxingu
3.3. Infection Process of F. gyrosa on A. chaxingu Mycelia
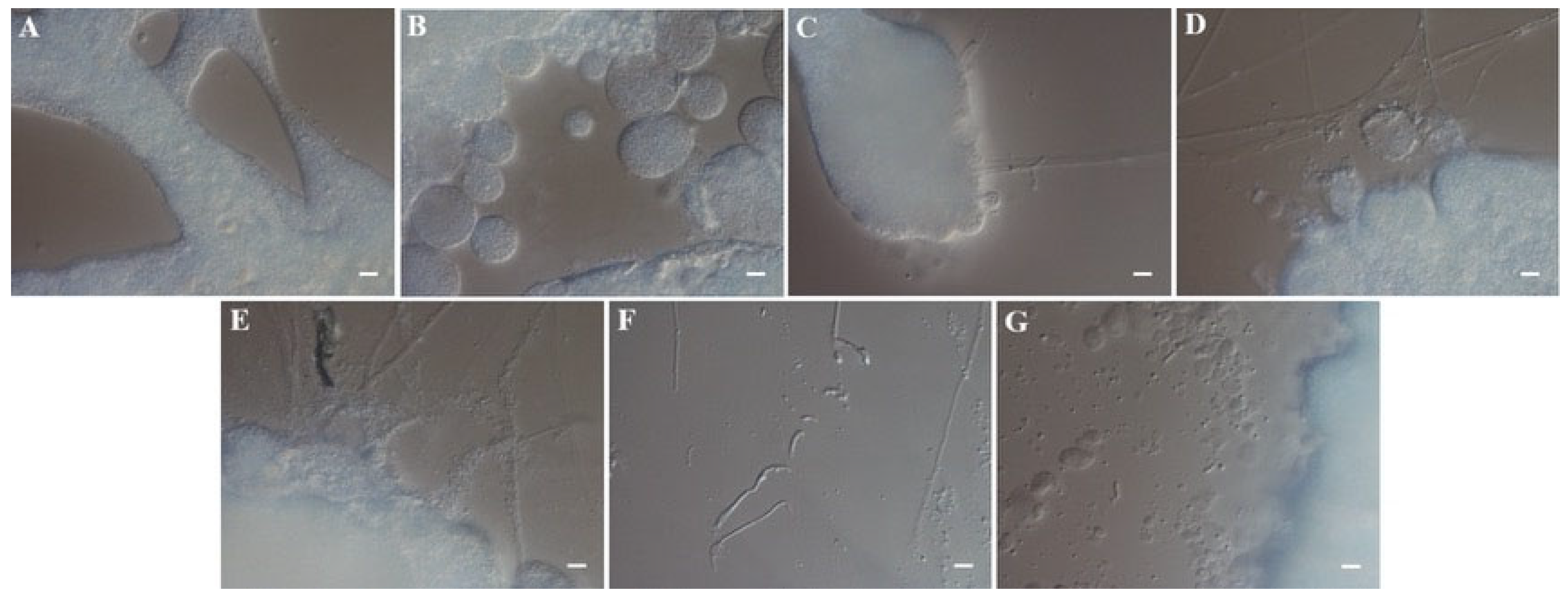
3.3.1. Stereomicroscopic Examination of F. gyrosa Infection on A. chaxingu Mycelia
3.3.2. Light Microscopic Observation of F. gyrosa Infection in A. chaxingu Mycelia
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hu, Q.X.; Liao, C.Z.; Zou, Y.J. Current development status and prospects of rare edible mushrooms in China. Agric. Compr. Dev. China 2022, 4, 50–54. (In Chinese) [Google Scholar]
- Wang, M.; Gu, B.L.; Huang, J.; Jiang, S.; Chen, Y.J.; Yin, Y.L.; Pan, Y.F.; Yu, G.J.; Li, Y.M.; Wong, B.H.C.; et al. Transcriptome and proteome exploration to provide a resource for the study of Agrocybe aegerita. PLoS ONE 2013, 8, e56686. [Google Scholar] [CrossRef]
- Zhang, M.; Xing, S.H.; Qian, C.L.; Liu, J.; Kan, J.; Jin, C.H. Analysis of the nutritional components and antioxidant properties of seven species of edible mushrooms. Food Sci. Technol. 2022, 47, 120–126. (In Chinese) [Google Scholar]
- Wang, Z.W.; Chen, K.Y.; Zhang, K.; He, K.H.; Zhang, D.D.; Guo, X.H.; Huang, T.W.; Hu, J.L.; Zhou, X.T.; Nie, S.P. Agrocybe cylindracea fucoglucogalactan induced lysosome-mediated apoptosis of colorectal cancer cell through H3K27ac-regulated cathepsin D. Carbohyd. Polym. 2023, 319, 121208. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Chen, E.B.; Fu, A.Z.; Liu, Y.; Bi, S. Formation of key aroma compounds in Agrocybe aegerita during hot air drying: Amino acids and reducing sugars identified as flavor precursors. Food Chem. 2025, 465, 141975. [Google Scholar] [CrossRef] [PubMed]
- Zadrazil, F. Cultivation of Agrocybe aegerita (Brig.) Sing. on Lignocellulose Containing Wastes 1. Factors Affecting Saprophytic Colonization and Decomposition of Substrate. Int. Soc. Mushroom Sci. 1989, 12, 357–368. Available online: https://www.pubhort.org/isms/12/2/v12_p2_a38.htm (accessed on 28 August 2025).
- Huang, N.L.; Lin, Z.B.; Chen, G.L. Medicinal and Edible Fungi; Shanghai Scientific and Technical Literature Press: Shanghai, China, 2010; pp. 607–608. (In Chinese) [Google Scholar]
- Bian, Y.B.; Xiao, Y.; Guo, M.P. Research progress on disease control of edible fungi. Acta Edulis Fungi 2021, 28, 121–131. (In Chinese) [Google Scholar]
- Choi, I.Y.; Choi, J.N.; Sharma, P.K.; Lee, W.H. Isolation and identification of mushroom pathogens from Agrocybe aegerita. Mycobiology 2010, 38, 310–315. [Google Scholar] [CrossRef]
- Li, W.Z. Several major diseases and pests of Agrocybe aegerita. Edible Fungi 2002, 2, 40. (In Chinese) [Google Scholar]
- Ding, H.G. Infectious diseases of Agrocybe chaxingu and pollution-free control techniques. Yunnan Agric. Sci. Technol. 2006, 3, 53–54. (In Chinese) [Google Scholar]
- Lu, D.X. Occurrence and prevention and control of common diseases of Agrocybe chaxingu (cylindracea). J. Fungal Res. 2008, 2, 114–118. (In Chinese) [Google Scholar]
- Ma, Y.; Yang, Y.H.; Lyu, Y.F.; Cui, S.Q.; Xu, J.Y.; Li, M.L.; Shen, J.D.; Xie, H.T.; Fang, X.H. Common diseases and insect pests in Agrocybe aegerita production and non-pollution comprehensive control technology. Edible Fungi China 2021, 40, 113–116. (In Chinese) [Google Scholar]
- Liu, Y.G.; Lin, R.K. Symptoms and control techniques of slime mold disease in Agrocybe aegerita. Edible Fungi 2003, 3, 40. (In Chinese) [Google Scholar]
- Chen, S.Z. Reserach on comprehensive prevention and control for edible fungus Ⅱ: Occurrence and control of Polyma disease in Agrocybe chaxingu. J. South. Agric. 2008, 3, 317–319. (In Chinese) [Google Scholar]
- Guan, S.G. Control experiment of slime mold disease in Agrocybe aegerita. Edible Fungi 2004, 4, 41–42. (In Chinese) [Google Scholar]
- He, X.S. Slime molds on the fruiting bodies of Auricularia cornea and Pleurotus ostreatus. Edible Fungi 1991, 1, 39. (In Chinese) [Google Scholar]
- Li, Y.C.; Li, Z.X. Separation and identification and biological characteristics of myxomycetes lyzing the fruitbodies of Auricularia auricula and A. polytricha. J. Guangxi Agric. Coll. 1992, 11, 8–15. (In Chinese) [Google Scholar]
- Li, S.S.; Qian, X.C.; Xu, J.Z. Common contaminant fungi and their control in Auricularia heimuer and Lentinula edodes cultivation in Qinba mountain area. Edible Fungi China 1992, 11, 25–26. (In Chinese) [Google Scholar]
- Lin, X.M. Slime mold contamination in edible fungus cultivation in Henan province. Edible Fungi 1993, 1, 38. (In Chinese) [Google Scholar]
- Hu, J.X.; Xu, M.; Gao, B.; Wang, F.; Jiang, Y.J.; Zhang, J.X. Identification and preliminary control of a slime mold on Pleurotus pulmonarius. Edible Fungi 2023, 45, 62–65. (In Chinese) [Google Scholar]
- Dai, D.; Sun, P.; Wu, X.Y.; Hu, J.; Zhang, B.; Wei, Y.H.; Li, Y. First report of yellow rot caused by Physarum galbeum in Grifola frondosa in China. Plant Dis. 2023, 10, 1094. [Google Scholar] [CrossRef]
- Zhao, J.; Fu, J.H. Pest and disease control in Dictyophora indusiata cultivation. Hunan Agric. 2024, 11, 14. (In Chinese) [Google Scholar]
- Raza, M.; Zhang, Z.F.; Hyde, K.D.; Diao, Y.Z.; Cai, L. Culturable plant pathogenic fungi associated with sugarcane in southern China. Fungal Divers. 2019, 99, 1–104. [Google Scholar] [CrossRef]
- Crous, P.W.; Lombard, L.; Sandoval-Denis, M.; Seifert, K.A.; Schroers, H.J.; Chaverri, P.; Gené, J.; Guarro, J.; Hirooka, Y.; Bensch, K.; et al. Fusarium: More than a node or a foot-shaped basal cell. Stud. Mycol. 2021, 98, 100116. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.P.; Li, Y. Life cycle of Physarella oblonga. Mycosystema 2004, 23, 381–387. (In Chinese) [Google Scholar]
- Hossen, R.; Courtney, M.; Sim, A.; Khan, M.A.A.K.; Verbruggen, H.; Bringloe, T. An optimized CTAB method for genomic DNA extraction from green seaweeds (Ulvophyceae). Appl. Plant Sci. 2024, 13, e11625. [Google Scholar] [CrossRef] [PubMed]
- White, T.J.; Bruns, T.; Lee, S.J.W.T.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In Pcr Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- O’Donnell, K.; Kistler, H.C.; Cigelnik, E.; Ploetz, R.C. Multiple evolutionary origins of the fungus causing panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA 1998, 95, 2044–2049. [Google Scholar] [CrossRef]
- O’Donnell, K.; Sutton, D.A.; Rinaldi, M.G.; Sarver, B.A.; Balajee, S.A.; Schroers, H.J.; Summerbell, R.C.; Robert, V.A.; Crous, P.W.; Zhang, N.; et al. Internet-accessible DNA sequence database for identifying fusaria from human and animal infections. J. Clin. Microbiol. 2010, 48, 3708–3718. [Google Scholar] [CrossRef]
- Reeb, V.; Lutzoni, F.; Roux, C. Contribution of RPB2 to multilocus phylogenetic studies of the Euascomycetes (Pezizomycotina, Fungi) with special emphasis on the lichen-forming Acarosporaceae and evolution of polyspory. Mol. Phylogenet. Evol. 2004, 32, 1036–1060. [Google Scholar] [CrossRef]
- Roux, J.; Steenkamp, E.T.; Marasas, W.F.; Wingfield, M.J.; Wingfield, B.D. Characterization of Fusarium graminearum from Acacia and Eucalyptus using β-tubulin and histone gene sequences. Mycologia 2001, 93, 704–711. [Google Scholar] [CrossRef]
- O’Donnell, K.; Cigelnik, E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylogenet. Evol. 1997, 7, 103–116. [Google Scholar] [CrossRef]
- Cubeta, M.A.; Echandi, E.; Abernethy, T.; Vilgalys, R. Characterization of anastomosis groups of binucleate Rhizoctonia species using restriction analysis of an amplified ribosomal RNA gene. Phytopathology 1991, 81, 1395–1400. [Google Scholar] [CrossRef]
- Rusk, S.A.; Spiegel, F.W.; Lee, S.B. Design of polymerase chain reaction primers for amplifying nuclear ribosomal DNA from slime molds. Mycologia 1995, 87, 140–143. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.T.; Von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Han, S.L.; Wang, M.M.; Ma, Z.Y.; Raza, M.; Zhao, P.; Liang, J.M.; Gao, M.; Li, Y.J.; Wang, J.W.; Hu, D.M.; et al. Fusarium diversity associated with diseased cereals in China, with an updated phylogenomic assessment of the genus. Stud. Mycol. 2023, 104, 87–148. [Google Scholar] [CrossRef]
- Kwon, S.L.; Cho, M.; Lee, Y.M.; Kim, C.; Lee, S.M.; Ahn, B.J.; Lee, H.; Kim, J.J. Two unrecorded Apiospora species isolated from marine substrates in Korea with eight new combinations (A. piptatheri and A. rasikravindrae). Mycobiology 2022, 50, 46–54. [Google Scholar] [CrossRef]
- Zhou, Y.M.; Zou, X.; Zhi, J.R.; Xie, J.Q.; Jiang, T. Fast recognition of Lecanicillium spp., and its virulence against Frankliniella occidentalis. Front. Microbiol. 2020, 11, 561381. [Google Scholar] [CrossRef] [PubMed]
- Vandepol, N.; Liber, J.; Desirò, A.; Na, H.; Kennedy, M.; Barry, K.; Grigoriev, I.V.; Miller, A.N.; O’Donnell, K.; Stajich, J.E.; et al. Resolving the Mortierellaceae phylogeny through synthesis of multi-gene phylogenetics and phylogenomics. Fungal Divers. 2020, 104, 267–289. [Google Scholar] [CrossRef] [PubMed]
- Madden, A.A.; Stchigel, A.M.; Guarro, J.; Sutton, D.; Starks, P.T. Mucor nidicola sp. nov., a fungal species isolated from an invasive paper wasp nest. Int. J. Syst. Evol. Microbiol. 2012, 62, 1710–1714. [Google Scholar] [CrossRef] [PubMed]
- Sandoval-Denis, M.; Lombard, L.; Crous, P.W. Back to the roots: A reappraisal of Neocosmospora. Persoonia 2019, 43, 90–185. [Google Scholar] [CrossRef]
- Yang, J.; Li, S.J.; Li, X.M.; Qin, Y.H.; Gao, S.X.; Wen, Y.; Lu, S.H.; Lu, C.T.; Wang, F. First report of Aspergillus westerdijkiae causing leaf blight on Rehmannia glutinosa in China. Plant Dis. 2024, 10, 1094. [Google Scholar] [CrossRef]
- Houbraken, J.; Frisvad, J.C.; Samson, R.A. Taxonomy of Penicillium section Citrina. Stud. Mycol. 2011, 70, 53–138. [Google Scholar] [CrossRef]
- Hou, L.W.; Giraldo, A.; Groenewald, J.Z.; Rämä, T.; Summerbell, R.C.; Huang, G.Z.; Cai, L.; Crous, P.W. Redisposition of acremonium-like fungi in Hypocreales. Stud. Mycol. 2023, 105, 23–203. [Google Scholar] [CrossRef]
- Perdomo, H.; Cano, J.; Gené, J.; García, D.; Hernández, M.; Guarro, J. Polyphasic analysis of Purpureocillium lilacinum isolates from different origins and proposal of the new species Purpureocillium lavendulum. Mycologia 2013, 105, 151–161. [Google Scholar] [CrossRef]
- Visagie, C.M.; Seifert, K.A.; Houbraken, J.; Samson, R.A.; Jacobs, K. A phylogenetic revision of Penicillium sect. Exilicaulis, including nine new species from fynbos in South Africa. IMA Fungus 2016, 7, 75–117. [Google Scholar]
- Zhao, Z.H.; Lu, G.Z. Fusarium kyushuense, a newly recorded species for China. Mycotaxon 2007, 102, 119–126. [Google Scholar]
- O’Donnell, K.; Ward, T.J.; Geiser, D.M.; Corby Kistler, H.; Aoki, T. Genealogical concordance between the mating type locus and seven other nuclear genes supports formal recognition of nine phylogenetically distinct species within the Fusarium graminearum clade. Fungal Genet. Biol. 2004, 41, 600–623. [Google Scholar] [CrossRef] [PubMed]
- Zare, R.; Gams, W. The genera Lecanicillium and Simplicillium gen. nov. Nova Hedwigia 2001, 73, 1–50. [Google Scholar] [CrossRef]
- Wang, M.M.; Crous, P.W.; Sandoval-Denis, M.; Han, S.L.; Liu, F.; Liang, J.M.; Duan, W.J.; Cai, L. Fusarium and allied genera from China: Species diversity and distribution. Persoonia 2022, 48, 1–53. [Google Scholar] [CrossRef]
- He, J.; Li, D.W.; Cui, W.L.; Zhu, L.H.; Huang, L. Morphological and phylogenetic analyses reveal three new species of Fusarium (Hypocreales, Nectriaceae) associated with leaf blight on Cunninghamialanceolata in China. MycoKeys 2024, 101, 45–80. [Google Scholar] [CrossRef]
- Wagner, L.; Stielow, B.; Hoffmann, K.; Petkovits, T.; Papp, T.; Vágvölgyi, C.; de Hoog, G.S.; Verkley, G.; Voigt, K. A comprehensive molecular phylogeny of the Mortierellales (Mortierellomycotina) based on nuclear ribosomal DNA. Persoonia 2013, 30, 77–93. [Google Scholar] [CrossRef]
- Frisvad, J.C.; Frank, J.M.; Houbraken, J.A.M.P.; Kuijpers, A.F.; Samson, R.A. New ochratoxin a producing species of Aspergillus section Circumdati. Stud. Mycol. 2004, 50, 23–43. [Google Scholar]
- Luangsa-Ard, J.; Houbraken, J.; van Doorn, T.; Hong, S.B.; Borman, A.M.; Hywel-Jones, N.L.; Samson, R.A. Purpureocillium, a new genus for the medically important Paecilomyces lilacinus. FEMS Microbiol. Lett. 2011, 321, 141–149. [Google Scholar] [CrossRef]
- Shi, L.P.; Li, Y. Life cycle of Physarum gyrosum. Mycosystema 2005, 24, 292–296. (In Chinese) [Google Scholar]
- Chen, X.T.; Liu, Q.; Meng, G.L.; Peng, X.H.; Dong, C.H.; Wei, Y.H.; Huo, G.H. First report of white mucus disease caused by Fuligo gyrosa on Agrocybe chaxingu in China. Plant Dis. 2024, 108, 1884. [Google Scholar] [CrossRef]
- Agra, L.A.N.N.; Seixas, C.D.S.; Dianese, J.C. False bean smut caused by slime mold. Plant Dis. 2018, 102, 507–510. [Google Scholar] [CrossRef]
- Vu, D.; Groenewald, M.; Szöke, S.; Cardinali, G.; Eberhardt, U.; Stielow, B.; de Vries, M.; Verkleij, G.J.; Crous, P.W.; Boekhout, T.; et al. DNA barcoding analysis of more than 9 000 yeast isolates contributes to quantitative thresholds for yeast species and genera delimitation. Stud. Mycol. 2016, 85, 91–105. [Google Scholar] [CrossRef]
- Vu, D.; Groenewald, M.; de Vries, M.; Gehrmann, T.; Stielow, B.; Eberhardt, U.; Al-Hatmi, A.; Groenewald, J.Z.; Cardinali, G.; Houbraken, J.; et al. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 2019, 92, 135–154. [Google Scholar] [CrossRef] [PubMed]
- Verkley, G.J.M.; Quaedvlieg, W.; Shin, H.D.; Crous, P.W. A new approach to species delimitation in Septoria. Stud. Mycol. 2013, 75, 213–305. [Google Scholar] [CrossRef]
- Mohanta, T.K.; Bae, H. The diversity of fungal genome. Biol. Proced. Online 2015, 17, 8. [Google Scholar] [CrossRef] [PubMed]
- Irinyi, L.; Serena, C.; Garcia-Hermoso, D.; Arabatzis, M.; Desnos-Ollivier, M.; Vu, D.; Cardinali, G.; Arthur, I.; Normand, A.C.; Giraldo, A.; et al. International society of human and animal mycology (ISHAM)-ITS reference DNA barcoding database--the quality controlled standard tool for routine identification of human and animal pathogenic fungi. Med. Mycol. 2015, 53, 313–337. [Google Scholar] [CrossRef]
- Sung, G.H.; Hywel-Jones, N.L.; Sung, J.M.; Luangsa-Ard, J.J.; Shrestha, B.; Spatafora, J.W. Phylogenetic classification of Cordyceps and the clavicipitaceous fungi. Stud. Mycol. 2007, 57, 5–59. [Google Scholar] [CrossRef]
- Kepler, R.M.; Luangsa-Ard, J.J.; Hywel-Jones, N.L.; Quandt, C.A.; Sung, G.H.; Rehner, S.A.; Aime, M.C.; Henkel, T.W.; Sanjuan, T.; Zare, R.; et al. A phylogenetically-based nomenclature for Cordycipitaceae (Hypocreales). IMA Fungus 2017, 8, 335–353. [Google Scholar] [CrossRef]
- Rodríguez-Ezpeleta, N.; Zinger, L.; Kinziger, A.; Bik, H.M.; Bonin, A.; Coissac, E.; Emerson, B.C.; Lopes, C.M.; Pelletier, T.A.; Taberlet, P.; et al. Biodiversity monitoring using environmental DNA. Mol. Ecol. Resour. 2021, 21, 1405–1409. [Google Scholar] [CrossRef]
- Steele, M.P.; Neaves, L.E.; Klump, B.C.; St Clair, J.J.H.; Fernandes, J.R.S.M.; Hequet, V.; Shaw, P.; Hollingsworth, P.M.; Rutz, C. DNA barcoding identifies cryptic animal tool materials. Proc. Natl. Acad. Sci. USA 2021, 118, e2020699118. [Google Scholar] [CrossRef]






| Genes/DNA Regions | Primers | PCR Amplification Procedures | Amplicon Length (bp) | |
|---|---|---|---|---|
| Name | Primer Name | Primer Sequence (5′→3′) | ||
| 18S ribosomal RNA (18S rRNA) | SMNUR101 | CTGGTTGATCCTGCCAGTAG | 95 °C 5 min; (95 °C 30 s, 55 °C 30 s, 72 °C 10 s) × 35 cycles; 72 °C 5 min; 10 °C | 1000–1500 |
| NS4 | CTTCCGTCAATTCCTTTAAG | |||
| Internal transcribed spacer region of the rDNA (ITS) | ITS4 | TCCTCCGCTTATTGATATGC | 95 °C 5 min; (95 °C 30 s, 52 °C 30 s, 72 °C 10 s) × 35 cycles; 72 °C 5 min; 10 °C | 500–800 |
| ITS5 | GGAAGTAAAAGTCGTAACAAGG | |||
| Translation elongation factor 1-alpha (tef1) | EF1-728F | CATCGAGAAGTTCGAGAAGG | 94 °C 90 s; (94 °C 45 s, 55 °C 45 s, 72 °C 15 s) × 35 cycles; 72 °C 10 min; 10 °C | 600–800 |
| EF-1αF | CTTGCCACCCTTGCCATCG | |||
| EF-1αR | AACGTCGTCGTTATCGGACAC | |||
| EF-1 | ATGGGTAAGGARGACAAGAC | |||
| EF-2 | GGARGTACCAGTSATCATG | |||
| RNA polymerase second largest subunit (rpb2) | 5f2 | GGGGWGAYCAGAAGAAGGC | 94 °C 90 s; (94 °C 45 s, 57 °C 45 s, 72 °C 20 s) × 35 cycles; 72 °C 5 min; 10 °C | 800–1100 |
| 7cr | CCCATRGCTTGYTTRCCCAT | |||
| RNA polymerase largest subunit (rpb1) | F7 | CRACACAGAAGAGTTTGAAGG | 95 °C 5 min; (95 °C 2 min, 58 °C 45 s, 72 °C 20 s) × 5 cycles; (95 °C 2 min, 57 °C 45 s, 72 °C 20 s) × 5 cycles; (95 °C 2 min, 56 °C 45 s, 72 °C 20 s) × 35 cycles; 72 °C 10 min; 10 °C | 1000–1200 |
| G2R | GTCATYTGDGTDGCDGGYTCDCC | |||
| RPB1-EF | TCACGWCCTCCCATGGCGT | |||
| RPB1-ER | AAGGAGGGTCGTCTTCGTGG | |||
| RPB1-F | CAYCCWGGYTTYATCAAGAA | |||
| RPB1-R | CCNGCDATNTCRTTRTCCATRTA | |||
| Histone (H3) | H3-la | ACTAAGCAGACCGCCCGCAGG | 96 °C 2 min; (92 °C 1 min, 60 °C 1 min, 72 °C 10 s) × 30 cycles; 72 °C 5 min; 10 °C | 400–600 |
| H3-lb | GCGGGCGAGCTGGATGTCCTT | |||
| Beta tubulin (tub2) | T1 | AACATGCGTGAGATTGTAAGT | 95 °C 3 min; (94 °C 30 s, 54 °C 45 s, 72 °C 15 s) × 35 cycles; 72 °C 10 min; 10 °C | 300–600 |
| T2 | TAGTGACCCTTGGCCCAGTTG | |||
| Bt-2a | GGTAACCAAATCGGTGCTGCTTTC | |||
| Bt-2b | ACCCTCAGTGTAGTGACCCTTGGC | |||
| TUB2Fd | GTBCACCTYCARACCGGYCARTG | |||
| TUB4Rd | CCRGAYTGRCCRAARACRAAGTTGTC | |||
| 28S large subunit ribosomal RNA gene (LSU) | LR0R | ACCCGCTGAACTTAAGC | 95 °C 3 min; (94 °C 30 s, 48 °C 50 s, 72 °C 90 s) × 35 cycles; 72 °C 10 min; 10 °C | 800–1100 |
| LR5 | ATCCTGAGGGAAACTTC | |||
| Sample ID | Strain ID | Species Name | Incidence | Location | Molecular Identification | References |
|---|---|---|---|---|---|---|
| Ac-01 | IAAM-A | Fusarium kyushuense | 2.6% | Guangchang, Fuzhou; Ganxian, Ganzhou | ITS, tef1, rpb1, rpb2, H3 | [42] |
| Ac-02 | IAAM-B | Apiospora rasikravindrae | 1.7% | ITS, LSU, tef1, tub2 | [43] | |
| Ac-03 | IAAM-C | Apiospora sp. | 1.2% | ITS, LSU, tef1, tub2 | [43] | |
| Ac-04 | IAAM-D | Lecanicillium aphanocladii | 2.2% | ITS, rpb1 | [44] | |
| Ac-05 | IAAM-E | Linnemannia zychae | 1.5% | ITS, tef1, rpb1 | [45] | |
| Ac-06 | IAAM-F | Fusarium asiaticum | 0.8% | ITS, tef1, rpb1, rpb2, H3 | [42] | |
| IAAM-P | Fusarium paranisikadoi | ITS, tef1, rpb1, rpb2 | [42] | |||
| Ac-07 | IAAM-G | Mucor nidicola | 0.9% | ITS | [46] | |
| Ac-08 | IAAM-H | Lecanicillium aphanocladii | 1.9% | ITS, rpb1 | [44] | |
| IAAM-T | Lecanicillium aphanocladii | ITS, rpb1 | [44] | |||
| Ac-09 | IAAM-I | Lecanicillium psalliotae | 2.7% | ITS, rpb1 | [44] | |
| Ac-10 | IAAM-J | Fusarium parceramosum | 1.3% | ITS, tef1, rpb2 | [47] | |
| Ac-11 | IAAM-K | Fusarium fujikuroi | 2.9% | ITS, tef1, tub2, rpb2 | [42] | |
| IAAM-N | Fusarium fujikuroi | ITS, tef1, tub2, rpb2 | [42] | |||
| Ac-12 | IAAM-L | Aspergillus westerdijkiae | 1.8% | ITS, tub2 | [48] | |
| Ac-13 | IAAM-M | Penicillium copticola | 2.2% | ITS, tub2 | [49] | |
| Ac-14 | IAAM-O | Apiospora arundinis | 0.6% | ITS, LSU, tef1, tub2 | [43] | |
| Ac-15 | IAAM-Q | Nectriopsis ellipsoidea | 1.4% | ITS, LSU | [50] | |
| Ac-16 | IAAM-R | Purpureocillium lilacinum | 2.8% | ITS, tef1 | [51] | |
| Ac-17 | IAAM-S | Penicillium citreosulfuratum | 2.1% | ITS, tub2 | [52] | |
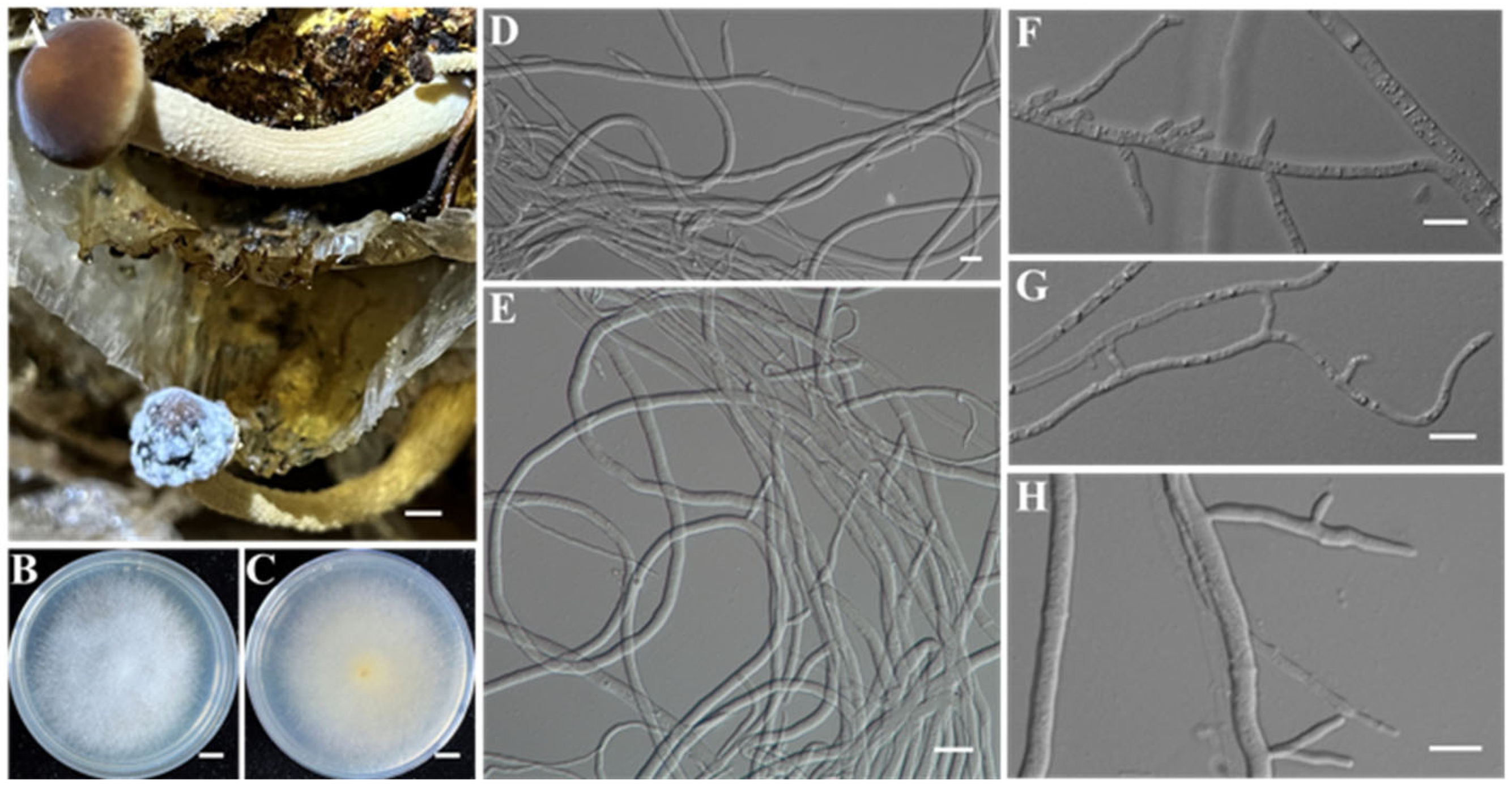
| Ac-18 | IAAM-W0001 | Physarella oblonga | 4.6% | 18S rRNA | [28,35] | |
| Ac-19 | IAAM-W0002 | Fuligo gyrosa | 17.3% | 18S rRNA | [28,35] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, X.; Meng, G.; Liu, M.; Dai, J.; Huo, G.; Dong, C.; Wei, Y. Pathogen Survey in Agrocybe chaxingu and Characterization of the Dominant Pathogen Fuligo gyrosa. Horticulturae 2025, 11, 1190. https://doi.org/10.3390/horticulturae11101190
Chen X, Meng G, Liu M, Dai J, Huo G, Dong C, Wei Y. Pathogen Survey in Agrocybe chaxingu and Characterization of the Dominant Pathogen Fuligo gyrosa. Horticulturae. 2025; 11(10):1190. https://doi.org/10.3390/horticulturae11101190
Chicago/Turabian StyleChen, Xutao, Guoliang Meng, Mengqian Liu, Jiancheng Dai, Guanghua Huo, Caihong Dong, and Yunhui Wei. 2025. "Pathogen Survey in Agrocybe chaxingu and Characterization of the Dominant Pathogen Fuligo gyrosa" Horticulturae 11, no. 10: 1190. https://doi.org/10.3390/horticulturae11101190
APA StyleChen, X., Meng, G., Liu, M., Dai, J., Huo, G., Dong, C., & Wei, Y. (2025). Pathogen Survey in Agrocybe chaxingu and Characterization of the Dominant Pathogen Fuligo gyrosa. Horticulturae, 11(10), 1190. https://doi.org/10.3390/horticulturae11101190







