Abstract
The management of Meloidogyne incognita often depends on chemical nematicides, which pose environmental and health risks. This study investigated the potential of bacterial strains isolated from uncultivated native soil as biocontrol agents and plant growth-promoting rhizobacteria (PGPR) in tomato plants artificially infected with this nematode. Fifteen strains were screened in vitro for nematicidal and ovicidal activity, and four promising strains (307, GB16, GB24, and GB29) were selected for greenhouse trials. All strains reduced the nematode reproduction factor and the number of nematodes/g of root. Strains 307 and GB24 showed the highest reductions, 61.39 and 57.24%, respectively. Despite some positive physiological trends, Bacillus spp. did not promote a significant increase in plant growth. Metabolomic analysis revealed that the strains produced a wide range of primary metabolites with potential nematicidal activity. All strains also secreted proteases and chitinases, enzymes linked to nematode cuticle degradation. Preliminary identification based on the 16S rRNA gene and phylogenetic analysis grouped the four strains into the Bacillus subtilis group (strains GB16, GB29 and 307) or Bacillus cereus group (strain GB24); however, genome sequencing will be required in future studies. Overall, strains 307 and GB24 demonstrated strong biocontrol potential, supporting their use as sustainable and complementary alternatives to chemical nematicides.
1. Introduction
Meloidogyne incognita is recognized as a major plant-parasitic nematode due to its wide host range and high reproductive capacity [1]. Global economic losses associated with this species are estimated at approximately USD 157 billion annually [1]. In tomato plants, the damage is particularly severe, with yield reductions ranging from 10 to 85%, depending on cultivation conditions and infestation intensity [2]. Parasitism mainly affects the root system, where the nematode induces gall formation and impairs water and nutrient uptake, ultimately resulting in reduced plant growth and yield [1].
Several management strategies have been applied to control M. incognita in tomato, including the use of resistant cultivars carrying Mi genes [3], crop rotation [4], and the application of chemical nematicides, both fumigant and non-fumigant [5]. However, many of these practices show limited efficacy, high costs, or environmental restrictions, which hinder their long-term adoption [1]. In this context, there is growing interest in sustainable and environmentally safer alternatives, particularly biological control.
Among microbial agents, species of the genus Bacillus have shown great promise due to their ability to produce a wide range of bioactive compounds and enzymes with direct effects on nematode eggs and juveniles [6]. These microorganisms can also indirectly influence parasitism dynamics by modulating nematode behavior, competing for nutrients, and forming biofilms on roots that disrupt plant–pathogen interactions [6,7]. Many Bacillus spp. strains also act as PGPR, improving nutrient uptake through the production of phytohormones (auxins, cytokinins, and gibberellins), phosphorus solubilization, siderophore synthesis, and the induction of systemic resistance, contributing to overall plant health and resilience [8].
Traditionally, studies have focused on investigating secondary metabolites, such as volatile organic compounds [9] and cyclic lipopeptides [10], that act against nematode through direct effects, causing mortality and inhibition of juvenile and egg development, and through indirect effects, by inducing defense responses in the host plant. Extracellular enzymes, such as proteases and chitinases [7], also play important roles: while proteases degrade intestinal tissues and the cuticle of M. incognita juveniles [11], chitinases compromise the integrity of the eggshell, disrupting embryonic development [12].
In addition to the classical bioactive compounds, Bacillus spp. also secrete primary metabolites, such as amino acids, organic acids, and fatty acids, which may exhibit nematicidal activity [13]. Although these metabolites are primarily involved in essential cellular functions, they can also interfere with nematode physiology. Metabolomic analyses are still rarely reported, particularly regarding the identification of primary metabolites with potential nematicidal activity [14]. Since these metabolites are directly involved in essential processes of microbial survival and interaction, understanding their functions may reveal new biochemical mechanisms for nematode suppression and expand the repertoire of strategies for sustainable biological management [14].
Another aspect to be considered in this study is the origin of the Bacillus isolates. The strains evaluated were obtained from uncultivated native soils, since comparative analyses between cultivated ecosystems and natural environments have shown that microorganisms associated with wild areas have greater ecological and functional diversity [15]. This increased diversity may be reflected in richer metabolic profiles and a differentiated adaptive capacity, which makes these isolates particularly promising as biocontrol agents [16].
Based on this premise, this study selected strains of Bacillus spp. from uncultivated native soil, with the aim of exploring their metabolic diversity and evaluating their potential for biological control of M. incognita, both in vitro and in plant, as well as to assess whether Bacillus-mediated biocontrol could mitigate the negative effects of the nematode on plant growth and photosynthetic parameters. In addition, metabolomic analyses were carried out with an emphasis on the identification of primary metabolites and lytic enzymes involved in the Bacillus–nematode–plant interaction, aiming to contribute to advancing the understanding of the role of primary metabolism in Bacillus-mediated biocontrol.
2. Materials and Methods
2.1. Bacterial Strains, Media and Culture Conditions
The bacteria were isolated from soil samples collected in a native forest area in the municipality of Patos de Minas, Minas Gerais, Brazil (18°36′41.2″ S 46°29′22.7″ W), according to the methodology of Valicente and Barreto [17], and stored in Luria–Bertani (LB) broth medium with 20% glycerol at −80 °C until further analysis. In this study, fifteen bacterial strains (307, GB01, GB05, GB08, GB10, GB11, GB13, GB16, GB20, GB24, GB25, GB26, GB27, GB28, and GB29) were selected and sent for analysis to the Research Laboratory of NOOA, Company of Agricultural Science and Technology (Patos de Minas, Brazil). In order to isolate spore forming bacteria, the soil samples were heat treated at 80 °C for 12 min [18]. Purity was verified by culturing the strains on nutrient agar (NA) at 30 °C for 72 h. A single colony from each purified isolate was then transferred to 70 mL of LB broth and incubated at 30 °C under shaking (200 rpm) for 48 h. The cultures were monitored every 24 h under an optical microscope Olympus CX41 (Olympus Optical do Brasil, Barueri, São Paulo, Brazil) to observe spore formation. Finally, serial dilutions followed by plating on NA were performed to quantify the bacterial inocula for in vitro and in planta tests.
2.2. Preparation of Meloidogyne incognita Inoculum
The nematode M. incognita was obtained from the roots of bell pepper plants (Capsicum annuum) grown in sterile soil and under controlled greenhouse conditions at an average temperature of 25 ± 2 °C and relative humidity close to 70%. Eggs were extracted from bell pepper roots by agitating in 0.5% NaOCl for 30 s and then collected and rinsed with water using sieves with 150 µm and 25 µm mesh sizes [19]. To collect the second-stage juveniles (J2), bell pepper roots infected with the nematode were crushed in a blender with 500 mL of water, and the material was transferred to sieves with 150 µm and 25 µm mesh sizes. The contents of the sieve were washed with water and transferred to a Baermann funnel. After 24 h of incubation in the dark at 25 °C, the J2 were transferred from the funnel to a beaker.
2.3. In Vitro Nematicidal Activity of Bacterial Strains Against Meloidogyne incognita
The nematicidal potential of the bacterial strains was investigated by analyzing their ability to inhibit egg hatching and increase the mortality rate of J2. The selected strains were tested (307, GB01, GB05, GB08, GB10, GB11, GB13, GB16, GB20, GB24, GB25, GB26, GB27, GB28 and GB29) along with a negative control (0.85% NaCl solution) and a positive control (QUARTZO®, a biological commercial product based on B. subtilis and B. licheniformis).
The bacterial suspension was prepared as previously described. The concentration of bacterial cells was determined by measuring optical density at 540 nm using a spectrophotometer SP220 (Biospectro, Curitiba, Paraná, Brazil) and adjusted with sterile water to a final concentration of 1 × 108 CFU mL−1. All the experiments in vitro were performed in five replicates and repeated twice. The most promising strains were selected for the in planta tests.
2.3.1. Meloidogyne incognita Eggs Hatching Test
The experiment was conducted in sterile Petri dishes containing 3.5 mL of the bacterial suspension and 3.5 mL of egg suspension (eggs suspended in 3.5 mL sterile 0.85% NaCl solution). For the negative control, the egg suspension was immersed in 3.5 mL of sterile 0.85% NaCl solution, while for the positive control, the egg suspension was mixed with 3.5 mL of a commercial biological product Quartzo® (FMC Química do Brasil Ltda, Campinas, São Paulo, Brazil). The plates were sealed with Parafilm, wrapped in aluminum paper and kept at 25 °C in the absence of light. The percentage of egg hatching was assessed using an optical microscope Olympus CX41 (Olympus Optical do Brasil, Barueri, São Paulo, Brazil) at 1, 6 and 12 days of incubation. There were five replicates and the test was carried out twice. A total of 170 eggs were used for the preparation of the egg suspension in the first test, and 200 eggs in the second.
2.3.2. Mortality Test on Second-Stage Juveniles
An aliquot of 3.5 mL of the bacterial suspension and 3.5 mL of the J2 suspension (J2 + 3.5 mL sterile 0.85% NaCl solution) were transferred to sterile Petri dishes. In the negative and positive controls, the J2 were immersed in 3.5 mL of sterile 0.85% NaCl solution and 3.5 mL of a commercial product solution Quartzo® (FMC Química do Brasil Ltda, Campinas, São Paulo, Brazil), respectively. The plates were sealed with parafilm, wrapped in aluminum paper and kept at 25 °C in the absence of light. The percentage of dead J2 was assessed by adding 800 μL of NaOH (1N), after 1 and 2 days of incubation, and those that remained completely distended and motionless for 3 min after the addition of the base were considered dead. The experiment was conducted in a completely randomized design, with five replicates per treatment, and was carried out twice: in the first assay, the suspension contained 50 J2, while in the second, 80 J2.
The number of eggs and J2 evaluated in assays 1 and 2 varied because nematode collection was performed at different times, reflecting the quantities available in bell pepper roots during each sampling period. However, this variation did not compromise the consistency of the analyses, since the results were expressed as percentages of egg hatching inhibition and J2 mortality, rather than absolute values. This normalization approach allowed for an adequate comparison between treatments and ensured the statistical robustness of the inferences made.
2.4. Bioassays In Planta
2.4.1. Soil Preparation and Inoculation with Meloidogyne incognita Eggs
The soil analysis and necessary adjustments were carried out and the soil was then autoclaved twice at 120 °C for 1 h, with a 24 h interval between sterilizations. Autoclaved soil was distributed into 56 five-liter pots, and 5000 M. incognita eggs (extracted as described in Section 2.2) were incorporated and thoroughly homogenized within the soil of each pot to ensure uniform nematode distribution.
2.4.2. Preparation of Bacterial Suspensions and Soil Treatment
Strains 307, GB16, GB24 and GB29 showed the best results in the in vitro tests and were selected for the in planta analyses. The bacterial suspension was prepared according to the procedure described in Section 2.3. After inoculation with M. incognita eggs, 70 mL of the bacterial suspension was applied to the soil using a pressurized hand sprayer, with the application rate adjusted to the equivalent of 1 L ha−1.
In addition to the treatments with bacterial suspensions, experimental controls were included: water (negative control), a commercial Bacillus-based biological product QUARTZO® (FMC Química do Brasil Ltda, Campinas, São Paulo, Brazil) - positive biological control, and a chemical nematicide NIMITZ® (Adama Agricultural Solutions, Londrina, Paraná, Brazil) - positive chemical control. The commercial products used in the positive controls were prepared according to the manufacturer’s recommendations. A volume of 70 mL of water was applied to the negative control, and 70 mL of the respective commercial formulations were applied to the positive controls. To enhance the penetration of bacterial cells into the soil and their contact with nematode eggs, 200 mL of sterile water were added to the soil surface after the preparation of all treatments.
2.4.3. Plant Material and Growing Conditions
Tomato seedlings (Solanum lycopersicum L.) of the Santa Cruz variety were grown for 40 days in 15 g of commercial substrate Carolina Soil (40% sphagnum peat, 20% toasted rice husk, 15% perlite, 20% coconut fiber, 5% hydrofiber and trace amounts of macro and micronutrients) in 128-cell polystyrene trays measuring 2.8 cm in height, 2.8 cm in width, and 5.5 cm in depth per cell.
Two days after preparing the soils for the bacterial and nematode treatments, tomato seedlings were transplanted into 5 L pots. At the time of transplanting, each pot received 100 mL of a nutrient solution composed of 30 g L−1 monoammonium phosphate (MAP), 0.139 g L−1 boric acid, 0.185 g L−1 copper(II) sulfate pentahydrate, 0.225 g L−1 ferric chloride hexahydrate, 0.395 g L−1 manganese(II) chloride tetrahydrate, 0.010 g L−1 sodium molybdate dihydrate, and 0.527 g L−1 zinc sulfate heptahydrate. The plants were kept in a greenhouse at an average temperature of 25 °C ± 2 °C, relative humidity close to 70% for 45 days, and were irrigated daily as needed. After this period, the effect of bacterial treatments on photosynthetic variables, tomato growth promotion and nematode control was analyzed. All the experiments in planta were conducted in a randomized block design with eight replicates, each pot with one plant representing an experimental unit, and repeated twice.
2.4.4. Photosynthetic Measurements
The fluorescence attributes and gas exchange of tomato plants were evaluated with a portable infrared analyzer (LICOR 6400XT, Li-COR, Lincoln, NE, USA) on fully expanded leaves, 45 days after planting. Net CO2 assimilation (A), stomatal conductance (gs) and the internal and external CO2 concentration ratio (Ci/Ca) were determined in an open system under saturated artificial light (1000 μmol m−2 s−1 of photons) and simultaneously, the efficiency of energy capture by the photosystem II reaction centers (Fv’/Fm’) was determined.
2.4.5. Tomato Growth Promotion Assay
The stems of the plants were cut at soil level and the material was weighed to determine the fresh mass of the aerial part (FMAP). Subsequently, all of this material was dried in an oven at 70 °C for 48 h to obtain the dry mass of the aerial part (DMAP). The fresh mass of the roots (FMR) was also determined. However, since nematode extraction from the roots required a destructive analysis, it was not possible to measure root dry weight.
2.4.6. Quantification of M. incognita in Tomato Plants
After extracting nematodes from the roots (extracted as described in Section 2.2) and quantifying the number of eggs and juveniles (J2) per plant, the following formulas were used to calculate the number of nematodes per gram of root (g/root).
The reproduction factor (RF) of the nematode in the different treatments was calculated, according to the formula described by Oostenbrink [20]:
where FP is the final population (number of eggs and juveniles extracted from the roots at the end the experimental period) and IP is the initial population (number of eggs and juveniles inoculated at the beginning of the experiment).
2.5. Bacterial Metabolic and Enzymatic Profile
2.5.1. Analysis of Metabolites by GC-MS
The bacterial strains were cultured in 750 mL of nutrient broth at 30 °C, with agitation (200 rpm) for 72 h. After incubation, the samples were centrifuged at 13,000× g for 15 min, and the supernatant was collected for metabolite extraction. Aliquots of 200 mL of supernatant were extracted with 100 mL of methanol. This process was repeated three times, and the organic phases of the successive extractions were combined, dried with anhydrous sodium sulfate and rotary evaporated at 60 °C, at 100 rpm. The resulting concentrate was resuspended in 10 mL of methanol and subjected to derivatization.
For derivatization, the samples were subjected to methoxyamination by adding 50 μL of pyridine containing 15 mg mL−1 methoxyamine hydrochloride at 30 °C for 30 min. Trimethylsilylation was then carried out by adding 50 μL of MSTFA with 1% TMCS reagent, followed by incubation at 37 °C for 50 min. The liquid-derivatized samples were transferred into glass vials for GCMS analysis.
Metabolite analysis was performed using a gas chromatography–mass spectrometry system (GCMS-QP2010, Shimadzu, Kyoto, Japan) equipped with a Rtx-5MS capillary column (30 m × 250 μm ID, 0.25 μm film thickness; Restek). The oven temperature was initially set at 80 °C for 2 min, then gradually raised to 315 °C at a rate of 5 °C min−1, and maintained at 315 °C for 12 min. Helium was used as the carrier gas at a constant flow rate of 1.0 mL min−1. The interface and ion source temperatures were both set to 240 °C for optimal performance. A 1 μL injection volume with a 10:1 split ratio was used to ensure effective sample introduction and consistent results throughout the GC-MS analysis. Mass spectral data were acquired in electron ionization (EI) mode at 70 eV, covering a mass scan range of 50–650 m/z at a rate of 5 scans/s. A solvent cut time of 4 min, based on the pyridine solvent retention time, was implemented. To ensure measurement accuracy and retention index calculations, a standard alkane mix (C7-C30) was used.
Metabolite annotation was conducted by comparing the conformity of the retention time (RT), retention index (RI), and mass spectrum values with reference metabolites available in the AllPublic-KovatsRI-VS2 library. National Institute of Standards and Technology (NIST 2014) [21] mass spectral library was used to confirm the annotated compounds using a similarity of ≥80%.
For quantification, chromatographic peak areas were determined by integrating the total ion chromatogram (TIC) reconstruction from the full scan, using for each compound the ion basis (m/z intensity 100%).
2.5.2. Protein Quantification and Determination of Protease and Chitinase Activities
A single colony of each strain was transferred to three flasks containing 50 mL of BHI medium and kept at 30 °C, 120 rpm. Aliquots of 700 µL were transferred to microtubes after 24 and 48 h of incubation. To obtain the enzymatic extract, the microtubes were centrifuged at 12,000× g for 10 min, and the supernatant was used for the determination of protein concentration and protease activity.
Protein concentration was determined according to Bradford [22], using bovine serum albumin (BSA) as a standard at concentrations of 0 to 0.2 mg mL−1. The protease activity was determined using the method described by Charney and Tomarelli [23]. The reaction was initiated by incubating 60 μL of enzymatic extract with 50 μL of 0.2% azocasein solution at 37 °C for 30 min. The reaction control consisted of a tube containing 50 μL of 0.1 M Tris–HCl buffer (pH 8.0) and 60 μL of enzymatic extract, without the substrate. A blank was prepared in a separate tube containing 50 μL of 0.2% azocasein solution and 60 μL of 0.1 M Tris–HCl buffer (pH 8.0), without the enzyme extract. The reaction was stopped by adding 240 μL of 10% trichloroacetic acid, followed by vortexing and keeping the tubes on ice for 15 min. The sample was then centrifuged at 8000× g for 5 min and 240 μL of the supernatant was transferred to another microtube, followed by the addition of 280 μL of 1 M NaOH solution. The absorbance of the samples was measured at 440 nm using a UV/VIS-spectrofotometer SP220 (Biospectro, Curitiba, Paraná, Brazil).
The production of chitinase by the bacterial strains was evaluated using the dinitrosalicylic acid (DNS) method [24], which is based on determining the concentration of N-acetyl-glucosamine (NAG), the final product of the hydrolysis of the colloidal chitin solution. The colloidal chitin was prepared by adding 5 g of powdered chitin (Sigma-Aldrich, St. Louis, Missouri, USA) to 60 mL of 37% HCl, which was maintained at room temperature under vigorous agitation for 1 h. The material was filtered through glass wool to retain undissolved fractions of chitin, followed by the addition of 200 mL of 50% ethanol with vigorous stirring. Finally, 1 L of distilled water was added for chitin washing. After chitin precipitation, the distilled water was changed continuously until reaching a pH close to 7. The colloidal chitin was stored at 4 °C. For the preparation of the enzymatic extract, one CFU of each strain was initially cultured in 10 mL of BHI medium and maintained at 30 °C, 120 rpm, for 24 h. Then, 100 µL of the culture was transferred to the surface of Yamaguchi agar (containing 0.4% yeast extract, 0.2% tryptone, 0.5% MgSO4, 0.12% KH2PO4, 0.28% K2HPO4, 1.5% colloidal chitin and 1.5% agar) followed by incubation at 30 °C for 24 h. After grown, one pure colony was transferred to 10 mL of Yamaguchi liquid medium followed by incubation at 30 °C, 120 rpm for 24 h. This pre-inoculum was then transferred to 90 mL of Yamaguchi liquid medium and incubated again a 30 °C, 120 rpm, for 48 h. The control consisted of sterile Yamaguchi liquid medium without bacterial inoculation. For determination of enzymatic activity, 5 mL of bacterial culture and control were collected after 24 and 48 h, followed by centrifugation at 10,000× g for 15 min and the supernatant was retained (enzymatic extract). The standard curve was prepared using the DNS reagent, with N-acetylglucosamine (Sigma Aldrich) concentrations of 50–500 μg mL−1. The activity values were expressed in units (U), where 1 U represents 1 μmol of released reducing sugar per hour of reaction. A mixture containing 1 mL of enzymatic extract and 1 mL of a 0.5% suspension of colloidal chitin in phosphate buffer (50 mM, pH 7.4) was vortexed and incubated at 37 °C in a water bath for 6 h. After this period, 200 μL of the reaction was transferred to microtubes containing 1 mL of DNS reagent and heated at 100 °C for 10 min. A blank consisted of 1 mL of sterile Yamaguchi medium and 1 mL of colloidal chitin solution. The absorbance of the samples was measured at 550 nm using a UV/VIS-spectrofotometer SP220 (Biospectro, Curitiba, Paraná, Brazil).
2.6. Molecular Identification of Bacterial Strains
2.6.1. PCR Amplification, Sequencing and Analysis of 16S rRNA Gene
For the molecular identification of strains 307, GB29, GB16 and GB24, the most promising strains in both in vitro and in planta assays, total genomic DNA was extracted using the Extract-N-Amp Tissue PCR Kit (Sigma-Aldrich). The 16S rRNA gene region was amplified with primers fD1 (5′–AGAGTTTGATCCTGGCTCAG–3′) and rP1 (5′–ACGGTTACCTTGTTACGACTT–3′) [25] and 2× GoTaq Green Master Mix (Promega). The amplification of the 16S rRNA gene was performed in a Bio-Rad T100™ Thermal Cycler (Bio-Rad Laboratories, Hercules, CA, USA) with the following program: initial denaturation at 94 °C for 4 min; 40 cycles of 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 90 s, and a final extension at 72 °C for 4 min. The amplified products were purified by polyethylene glycol precipitation according to the protocol described by Schmitz and Riesner [26]. The DNA amplicons were sequenced by Phytopathological Diagnostic Laboratory, Biological Institute of São Paulo (Brazil) using the chain-termination method and a 3500xL Genetic Analyzer capillary sequencer Applied Biosystems (Thermo Fisher Scientific, Waltham, MA, USA). All 16S rRNA gene sequences were compared with nucleotide sequences available in the GenBank database (http://blast.ncbi.nlm.nih.gov/Blast.cgi) on 8 May 2025 to assess homologous hits using the Basic Local Alignment Search Tool (BLAST) [27], with default parameters. The partial 16S rRNA gene sequences of the strains in this study were deposited in GenBank under the accession numbers from PV804181 to PV804184.
2.6.2. 16S rRNA Gene Phylogenetic Analysis
A phylogenetic tree based on the partial 16S rRNA gene sequences of 21 strains was constructed using MEGA version 10.2 [28]. The analysis included the sequences of 16 reference Bacillus type strains and the four Bacillus strains in this study. The NCBI GenBank accession numbers of 16S rRNA gene sequences of these reference Bacillus type strains are listed in Supplementary Table S1. Escherichia coli NBRC 102203T was used as the outgroup. The 16S rRNA gene sequences of reference Bacillus type strains were downloaded from the List of Prokaryotic names with Standing in Nomenclature (LPSN) (https://lpsn.dsmz.de/). The sequences were aligned using the MUSCLE algorithm in MEGA version 10.2. The evolutionary history was inferred by using the Maximum Likelihood method and Tamura 3-parameter model [29], identified as the optimal substitution model. A discrete Gamma distribution was used to model evolutionary rate differences among sites (5 categories (+G, parameter = 0.4101)). All positions with less than 95% site coverage were eliminated (partial deletion option), retaining a total of 1294 bp in the final dataset. The reliability of the tree obtained was statistically evaluated by bootstrap analysis (1000 replicates).
2.7. Statistical Analysis
The data obtained were subjected to the Shapiro–Wilks and O’Neill–Mathews tests to verify the normality and homogeneity of variances, respectively, and subsequently the analysis of variance was performed. The means were compared using Tukey’s test at a significance level of p ≤ 0.05. Statistical analysis of the data was performed using R version 4.0.0.
3. Results
3.1. Inhibition of M. incognita Eggs Hatching In Vitro
The hatching of M. incognita eggs exposed to bacterial suspensions was assessed at three time points, as shown in Figure 1. After 1 day of egg exposure to Bacillus spp., significant inhibition was observed following treatments C+ (commercial biological product), GB01, GB13, GB16, GB24, GB27, and GB29 (Figure 1A). At 6 days (Figure 1B) and 12 days (Figure 1C), significant egg hatching inhibition persisted only in treatments C+, 307, GB16, GB24, and GB29. It was also observed that the inhibitory effect decreased over time, with the inhibition rate not exceeding 30% at 12 days (Figure 1C).
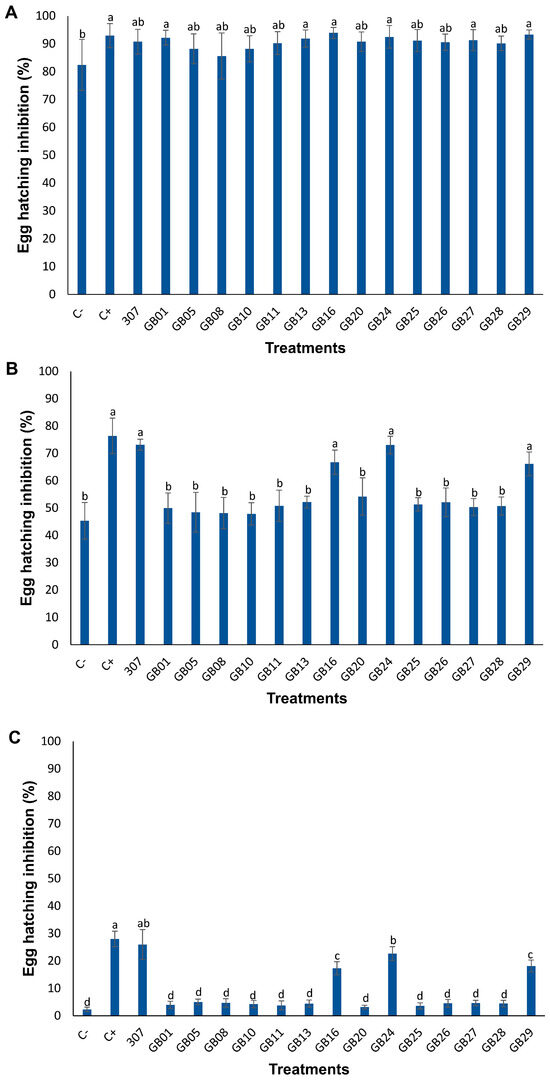
Figure 1.
Inhibition of M. incognita egg hatching exposed to bacterial suspension for 1 (A), 6 (B) and 12 days (C) at 25 °C in the dark. The bars represent the mean values of 10 replicates. Different letters within the same graphic indicate significant differences by Tukey’s test (p ≤ 0.05). C: negative control (0.85% NaCl solution). C+: positive control (commercial biological product QUARTZO® based on B. subtilis and B. licheniformis).
3.2. Mortality of Second-Stage Juveniles
All bacterial treatments caused significantly higher J2 mortality when compared to C-, in both periods evaluated, 24 h (Figure 2A) and 48 h (Figure 2B). However, four strains (307, GB16, GB24, and GB29) stood out for exhibiting a nematicidal effect similar to C+ and significantly increased as compared to the other treatments (Figure 2). At 24 h, J2 mortality was significantly higher in treatments C+ (47.22%), 307 (51.28%), GB16 (44.55%), GB24 (40.90%), and GB29 (41.92%) compared to the negative control (0.45%) (Figure 2A). In the other treatments, mortality ranged from 9.53% (GB28) to 23.09% (GB08). Similar results were observed at 48 h, but with an increased percentage of J2 mortality: C+ (58.12%), 307 (52.77%), GB16 (48.02%), GB24 (41.12%), and GB29 (43.40%) (Figure 2B). Mortality in the other treatments ranged from 11.62% (GB25) to 23.50% (GB08).
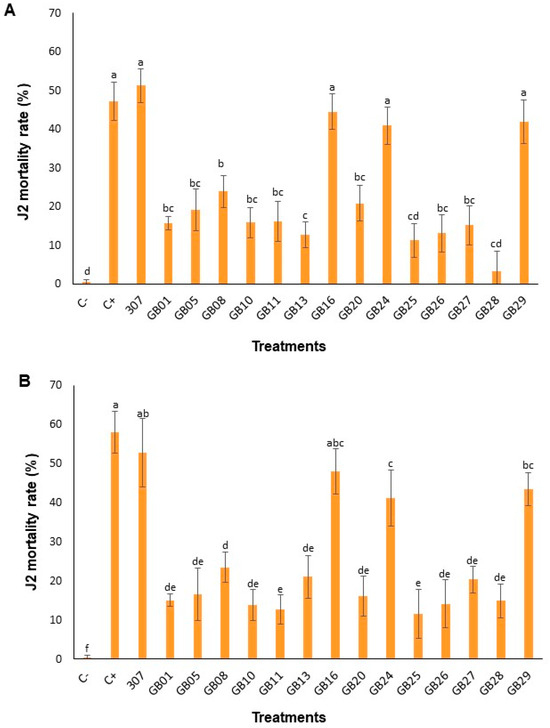
Figure 2.
J2 mortality of M. incognita exposed to bacterial suspensions of the tested strains for 24 (A) and 48 h (B) at 25 °C in the absence of light. The bars represent the mean values of 10 replicates. Different letters within the same graphic indicate significant differences by Tukey’s test (p ≤ 0.05). C-: negative control (0.85% NaCl solution). C+: positive control (commercial biological product QUARTZO® based on B. subtilis and B. licheniformis).
3.3. Effect of Bacterial Treatments on the Photosynthetic Parameters of Tomato Plants Under M. incognita Infection
In plants treated with strains GB16, the photosynthetic parameters A and Fv’/Fm’ were higher than in the other treatments. The ratio between internal and external CO2 concentration (Ci/Ca) also varied among treatments, being significantly higher in the GB24 treatment. Stomatal conductance (Gs) did not differ significantly among treatments (Table 1).

Table 1.
Gas exchange parameters of tomato plants 45 days after exposure to the root-knot nematode M. incognita and the bacterial strains in this study.
3.4. Effect of Bacterial Treatments on the Growth Promotion of Tomato Plants Under M. incognita Infection
No significant differences were observed between the treatments evaluated in relation to the growth of the aerial part and root system of tomato plants (Table 2).

Table 2.
Effect of bacterial treatments on growth promotion in tomato plants grown under greenhouse conditions and artificially infected with the root-knot nematode M. incognita.
3.5. Effect of Bacterial Treatments on the Control of M. incognita in Tomato Plants
All bacterial strains exhibited a significant effect (p < 0.05) in controlling M. incognita on tomato plants (Table 3). Strains 307 and GB24 demonstrated the highest efficacy, as they determined the highest percentage of total nematode population control among the treatments evaluated. However, no treatment surpassed the average reduction percentage achieved by the chemical nematicide (84.12%). The percentage reduction in the number of eggs ranged from 37.56 to 65.76%, observed in treatments with GB29 and 307, respectively. The number of J2 following bacterial treatments was also significantly lower than in the negative control, with reduction percentages ranging from 35.14 (GB29) to 60.21% (GB24).

Table 3.
Effect of bacterial treatments on control of M. incognita population in tomato plants under greenhouse conditions.
The results of the reproduction factor of M. incognita (Figure 3) suggest that all tested strains reduced the nematode ability to reproduce in tomato plants. RF values ranged from 0.87 (C+ Chemical) to 9.66 (C−), and among the strains, GB24 showed the lowest RF value (3.02).
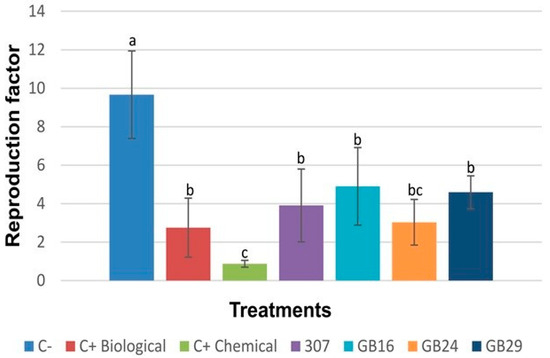
Figure 3.
Reproduction factor of M. incognita in the tomato plants subjected to bacterial treatments evaluated at 45 days after inoculation (DAI). Different letters indicate significant differences by Tukey’s test (p ≤ 0.05). C-: negative control (water). C+ (Biological): positive control (commercial biological product QUARTZO® based on B. subtilis and B. licheniformis). C+ (Chemical): commercial chemical nematicide NIMITZ®.
3.6. Bacterial Metabolic Profile and Enzyme Production
A total of 36 metabolites were detected in the methanolic extracts of the tested strains (Table 4). Most compounds were classified as amino acids (30.5%), followed by fatty acids (19.44%) and organic acids (19.44%). In lower proportions (5.55%), non-protein amino acids, alcohols, cyclic polyols, and monoacylglycerols were identified. Metabolites belonging to the classes of azoles, nitrogenous base, and sugars were detected in minor amounts, accounting for 2.77% of the total. The methanolic extract of strain 307 contained 21 metabolites, distributed across all classes, except azoles. Thirteen metabolites were identified in the extracts of strains GB24 and GB29, while only five metabolites were detected in the extract of strain GB16 (Table 4).

Table 4.
Metabolites identified in the methanolic extracts of the bacterial strains in this study analyzed by GC-MS.
Protease and chitinase activities were assessed in the bacterial suspensions after 24 and 48 h of incubation at 30 °C. The control consisted of only nutrient agar (NA) medium. All strains produced and secreted proteases into the culture medium at both time points (Table 5). However, after 48 h, the strain GB16 showed a significantly higher specific protease activity compared to the other strains tested. After 24 h of growth, strains 307 and GB24 exhibited significantly higher chitinase activity as compared to control (Table 5). At 48 h, all strains produced chitinases, with strain GB16 again showing the highest activity.

Table 5.
Activity of hydrolytic enzymes, protease and chitinase, produced by bacterial strains after 24 and 48 h of growth in NA medium at 30 °C.
3.7. Molecular Identification and Phylogeny of Bacterial Strains
The four most promising strains (307, GB29, GB16, GB24) in the in vitro and in planta assays were preliminarily identified at the molecular level by amplification and sequencing of 16S rRNA gene using primers fD1 and rP1. BLASTn analysis showed that they belong to the genus Bacillus. In addition, the partial 16S rRNA gene sequences of strains GB29, GB16 and 307 shared, respectively, a 99.86%, 99.71% and 100% identity value with strains belonging to B. subtilis group species (E-value = 0.0), while strain GB24 16S rRNA gene sequence exhibited a 98.90% identity with strains from Bacillus cereus group (E-value = 0.0). A phylogenetic tree was then constructed from the 16S rRNA gene sequences of 20 Bacillus strains, including the four strains in this study and reference Bacillus type strains (Figure 4). The tree delineated two distinct clades; the first clade (10 strains; 95% bootstrap value) included Bacillus strains GB29, GB16 and 307, that grouped close to type strains of species belonging to B. subtilis group, namely B. siamensis KCTC 13613T and B. velezensis NRRL B41580T (Figure 4). Bacillus strain GB24 grouped in the second clade (10 strains; 96% bootstrap value), comprising species affiliated with B. cereus group, with B. thuringiensis IAM 12077T being the closest strain (Figure 4).

Figure 4.
Phylogenetic relationships of 16 type strains of reference Bacillus species and the four Bacillus strains in this study (in bold) inferred from the alignment of the 16S rRNA gene sequence. The tree was constructed using MEGA software version 10.2 [28]. The evolutionary history was inferred by using the Maximum Likelihood method and Tamura 3-parameter model [29]. The tree with the highest log likelihood (-3388.71) is shown. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. T, type strain. Escherichia coli NBRC 102203T was used as outgroup.
4. Discussion
In this study, we evaluated the effects of Bacillus spp. strains isolated from native soil on the control of M. incognita, as well as assessing whether Bacillus-mediated biocontrol could mitigate the negative effects of the nematode on plant growth and photosynthetic parameters. Four strains—307, GB16, GB24, and GB29—exhibited ovicidal and nematicidal activity both in vitro and in planta, with efficacy comparable to that observed for the commercial products evaluated. Improvements in photosynthetic performance were observed in plants treated with strains GB16 and GB24; however, none of the strains promoted plant growth. Furthermore, metabolomic and biochemical analyses of the culture medium revealed the presence of primary metabolites and enzymes potentially associated with the nematicidal activity observed. Preliminary molecular analyses grouped strains 307, GB16, and GB29 into the B. subtilis group, close to the species B. siamensis and B. velezensis. Strain GB24 was grouped into the B. cereus group, with B. thuringiensis being the closest strain. We consider these strains to be promising for future use in commercial formulations, and thus complete genome sequencing will be necessary.
In vitro assays demonstrated that strains 307, GB16, GB24, and GB29 exhibited ovicidal (above 90%) and nematicidal (above 40%) activity against M. incognita. Similar results have been reported in studies evaluating 662 PGPR strains for the biocontrol of M. incognita, where approximately 33% of the tested bacteria caused high J2 mortality and egg hatching inhibition under in vitro conditions, with the genus Bacillus standing out for its nematicidal activity [30]. Our findings are also consistent with previous reports highlighting the nematicidal effects of species such as B. altitudinis, B. cereus, and B. subtilis against M. incognita [31,32,33]. Concerning this, B. cereus strains BCM2 and SZ5 and B. altitudinis strain CCM7 caused 100% mortality of J2 [31], while the fermentation broth of B. cereus strain Bc-cm103 induced complete juvenile mortality and reduced egg hatching by 40.06% [32]. Similarly, Cao [33] reported a significant decrease in egg hatching rates and 100% J2 mortality following treatment with the crude fermentation broth of B. subtilis strain Bs-1 under in vitro conditions.
Studies have also shown that the nematicidal effectiveness of Bacillus strains is directly related to the nematode incubation period [32,34]. The exposure time to the crude extract of B. amyloliquefaciens significantly increased the mortality of J2 and decreased the egg hatching of M. incognita over time [34]. In another study, Basyony and Zaid [35] evaluated the effect of different B. subtilis B10 isolate fractions (culture broth, cell suspension, and supernatant) on egg hatching and J2 mortality of M. incognita. The initial applications showed strong effects; however, the efficacy varied depending on the form of the inoculant and the timing of application, suggesting that the bioactive compounds lose potency with prolonged incubation. Similar results were observed in our study, where J2 mortality increased with longer incubation periods, while egg hatch inhibition reached approximately 90%, 60%, and less than 30% after 1, 6, and 12 days of incubation, respectively. This effect can be attributed to the continuous and cumulative production of bioactive bacterial compounds, such as lipopeptides (e.g., surfactin, iturin) [36], toxins (e.g., Cry proteins) [37], siderophores, and hydrolytic enzymes, including proteases and chitinases [38]. These metabolites exert direct action on J2 juveniles by damaging the cuticle, causing osmotic imbalance and physiological dysfunctions that ultimately lead to cell death. However, as the incubation period extends, some of the bacterial compounds may undergo degradation or lose effectiveness, which can favor the partial resumption of egg hatching, although in many cases the hatched juveniles exhibit low viability [6]. These results suggest that the efficacy of biological control by Bacillus is dependent on the duration of exposure and the stability of nematicidal compounds in the environment.
The presence of Meloidogyne affects not only the external morphology of plants, but also disrupts essential physiological and metabolic processes such as photosynthesis [39]. On the other hand, certain Bacillus species function as PGPR and some are capable of enhancing photosynthetic activity [40]. Our results show that plants inoculated with strain GB16 exhibited enhanced carbon assimilation and photosystem II efficiency, while those treated with the strain GB24 exhibited an increase in the Ci/Ca ratio, suggesting improved balance between CO2 uptake and utilization. Previous studies have also reported enhanced photosynthetic capacity in Capsicum annuum plants inoculated with Bacillus spp. [41]. Moreover, Capsicum chinense plants infected with PepGMV and inoculated with Bacillus spp. maintained a functional photosynthetic mechanism, with improvements in both photochemical parameters and gas exchange [40]. These findings support the hypothesis that Bacillus spp. inoculation can prime plants to better cope with both biotic and abiotic stresses while preserving photosynthetic efficiency.
Despite the fact that several species within the Bacillus genus are widely recognized as PGPR, it cannot be generalized that all of them perform this function. In this study, the treatment with Bacillus spp. was effective in controlling nematodes, but did not affect the growth of infested tomato plants. A similar result was reported by Magalhães [42], who evaluated the combined application of two strains closely related to B. velezensis (BMH and INV) in tomato plants. Although the combination effectively suppressed nematode development, it did not promote plant growth. These findings reinforce that growth-promoting activity is highly dependent on the characteristics of the specific Bacillus strain. In addition to their nematicidal capacity, mediated by their genetic profile and ecological competitiveness in the soil microbiome, PGPR strains should also be able to enhance nutrient uptake through the production of phytohormones, phosphorus solubilization, and siderophore synthesis [43].
One of the main findings of this study was that inoculation of these Bacillus spp. strains in tomato plants resulted in suppression of M. incognita populations, comparable to the commercial biological control, as evidenced by a reduction in the nematode reproduction factor. In order to elucidate the possible compounds involved in the nematicidal and ovicidal activity observed, the metabolic profile and the activity of lytic enzymes secreted in the bacterial culture medium were analyzed. Amino acids such as leucine and valine were detected in all the cultures. These two amino acids in particular are recognized as precursors in the biosynthesis of CLPs, such as surfactin and iturin, whose deleterious effects on the nematode include damage to the cuticle and induction of osmotic imbalance in the J2 [10,36]. In addition, L-phenylalanine and DL-phenylalanine were identified exclusively in the culture medium of strain 307, this amino acid being an important precursor of phenolic compounds and salicylic acid, key molecules in the activation of induced defense mechanisms in plants [44]. The direct action of amino acids on Meloidogyne was reported by Amdadul [45], who showed that six amino acids, including valine and phenylalanine, induced significant effects on the growth of tomato plants, accompanied by a significant reduction in the number of galls, adult females, egg masses and juveniles of M. javanica. Among the compounds analyzed, DL-phenylalanine was noticeable for its effectiveness in inhibiting egg hatching and reducing the viability of juveniles under in vitro conditions.
Organic compounds, such as alcohols and acids, were also identified in the Bacillus spp. cultures in this study. Glycerol, present in the culture of strain 307, showed nematicidal potential by enhancing the biocontrol efficacy against M. javanica, promoting tomato plant growth, and favoring rhizospheric and endophytic colonization by beneficial bacteria [46]. Organic acids, including acetic, lactic, succinic, and citric acids, are frequently produced by Bacillus and can alter the environmental pH, compromising the viability of eggs and J2, or disrupt nematode cuticle integrity, increasing their susceptibility to other bioactive compounds [47]. Organic acids produced by Bacillus altitudinis AMCC 1040, including acetic acid, 3-methylbutyric acid, and 2-methylbutyric acid, have demonstrated strong nematicidal activity against M. incognita, causing vacuolization and severe damage to nematode body integrity [48].
The nitrogenous base uracil, detected in the culture of strain 307, has also been associated with nematicidal activity. According to Oliveira [49], metabolites produced by B. cereus and B. subtilis with nematicidal effects against the plant-parasitic nematode M. exigua were identified as uracil, 9H-purine, and dihydrouracil. The results showed that dihydrouracil, in particular, exhibited greater control potential than the chemical nematicide carbofuran, as evidenced by its lower lethal concentration (LC50) value in comparative bioassays. These findings suggest that pyrimidine derivatives, such as uracil, may play a significant role in the suppression of root-knot nematodes. Similarly, short- and medium-chain fatty acids, such as octanoic acid, have been associated with membrane destabilization and metabolic disturbances in M. incognita juveniles [50]. Research attributes the biological activity of some fatty acids to specific structural features, such as carbon chain length (particularly C8–C11 fatty acids) and the presence of double bonds.
Proteolytic and chitinolytic activities were detected in all evaluated Bacillus spp. cultures. These findings suggest that the secretion of lytic enzymes may have contributed to the observed nematicidal effect. According to Hu [11], B. cereus BCM2 is capable of adhering to the surface of the eggshell and J2 of M. incognita, secreting extracellular proteases such as alkaline serine protease and neutral protease. Ultrastructural analyses using scanning and transmission electron microscopy have shown that extracellular proteases produced by B. cereus BCM2 cause severe damage to the body of M. incognita juveniles and to the eggshell structure. These enzymes do not affect the internal organization of the nematode but degrade its cuticle, leading to lysis and leakage of cellular contents [11]. Another important group of enzymes potentially involved in the ovicidal activity of the evaluated Bacillus spp. is represented by chitinase. Soliman [51] demonstrated that eggs treated with chitinase produced by rhizobacteria such as B. cereus and B. subtilis exhibited an increased number and size of vacuoles in the chitin layer, along with a higher mortality rate of J2 M. incognita in vitro assays. Another relevant study investigated the relationship between the biocontrol efficacy of 12 Bacillus strains and their respective proteolytic and chitinolytic activities. Enzymatic analyses revealed a strong positive correlation between protease activity and efficacy against root-knot nematodes, whereas chitinase activity showed little to no correlation. These findings suggest that in vitro protease activity may serve as a promising parameter for the screening and selection of biological control agents with nematicidal potential [52].
5. Conclusions
The results obtained in this study reinforce that the use of Bacillus spp. represents a viable alternative for the integrated management of plant-parasitic nematodes. All strains were effective in controlling M. incognita, with strains 307 and GB24 showing the highest percentages of nematode population reduction. The observed nematicidal activity may be associated with the production of bioactive metabolites, as well as proteases and chitinases secreted by these bacteria into the culture medium. Despite some positive physiological trends, Bacillus spp. did not promote a significant increase in plant growth. The phylogenetic and metabolic distinction between strains 307 and GB24 suggests that these microorganisms may act through complementary mechanisms of action. This hypothesis suggests the relevance of evaluating the potential of combined formulations, which may result in a more pronounced synergistic effect in the control of M. incognita.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae11101189/s1, Table S1: NCBI accession numbers of 16S rRNA gene of type strains of the species used for the construction of the phylogenetic tree.
Author Contributions
M.V.C.: conceptualization, validation, formal analysis, data curation, visualization, writing—original draft. L.M.S.: methodology, formal analysis. Writing—original draft E.A.L.: conceptualization, methodology, investigation, resources, visualization, writing—original draft. W.V.d.C.: investigation, resources, visualization, writing—original draft. V.C.: methodology, Funding acquisition, visualization, writing—original draft. G.D.: methodology, formal analysis, data curation, visualization, writing—original draft. L.E.V.: conceptualization, methodology, validation, investigation, resources, data curation, writing—original draft preparation, supervision, writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
The original contributions presented in this study are included in the article and supplementary material. Further inquiries can be directed to the corresponding author.
Acknowledgments
This work was carried out as part of the master’s thesis of the first author. We would like to express our sincere gratitude to NOOA Brasil-Ciência e Tecnologia Agrícola for kindly providing their facilities and infrastructure, which were essential for the development of this study. During the preparation of this manuscript, the authors used SciSpace (version 1.5.1) and Elicit (Ought Inc.)for the purposes of assisting in the search and organization of relevant scientific literature, and ChatGPT (Open IA) for improving the clarity and fluency of the manuscript written in English. The authors have reviewed and edited the output and take full responsibility for the content of this publication.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Sikandar, A.; Zhang, M.Y.; Wang, Y.Y.; Zhu, X.F.; Liu, X.Y.; Fan, H.Y.; Xuan, Y.H.; Chen, L.J.; Duan, Y.X. Meloidogyne incognita (Root-Knot Nematode) a Risk to Agriculture. Appl. Ecol. Environ. Res. 2020, 18, 1679–1690. [Google Scholar] [CrossRef]
- Nagachandrabose, S.; Shanthi, M.; Shanmugam, S.P.; Elaiyabharathi, T.; Sharmila, R.; Devrajan, K.; Manickam, R.; Srinivasan, R. Integrating Grafting and Bio-Inputs for Sustainable Management of Root Knot Nematode, Meloidogyne incognita, in Tomato Cultivation. Front. Plant Sci. 2025, 16, 1623444. [Google Scholar] [CrossRef] [PubMed]
- El-Sappah, A.H.; Islam, M.M.; El-awady, H.H.; Yan, S.; Qi, S.; Liu, J.; Cheng, G.; Liang, Y. Tomato Natural Resistance Genes in Controlling the Root-Knot Nematode. Genes 2019, 10, 925. [Google Scholar] [CrossRef] [PubMed]
- Fullana, A.M.; Expósito, A.; Escudero, N.; Cunquero, M.; Loza-Alvarez, P.; Giné, A.; Sorribas, F.J. Crop Rotation with Meloidogyne-Resistant Germplasm Is Useful to Manage and Revert the (a)Virulent Populations of Mi1.2 Gene and Reduce Yield Losses. Front. Plant Sci. 2023, 14, 1133095. [Google Scholar] [CrossRef] [PubMed]
- Grabau, Z.J.; Liu, C.; Sandoval-Ruiz, R. Meloidogyne incognita Management by Nematicides in Tomato Production. J. Nematol. 2021, 53, e2021-55. [Google Scholar] [CrossRef]
- Vasantha-Srinivasan, P.; Park, K.B.; Kim, K.Y.; Jung, W.J.; Han, Y.S. The Role of Bacillus Species in the Management of Plant-Parasitic Nematodes. Front. Microbiol. 2025, 15, 1510036. [Google Scholar] [CrossRef]
- Moreira, A.C.S.; Lopes, E.A.; Visôtto, L.E.; Soares, M.S.; Londe, M.L.A.; Ribeiro, L.B.; Terra, W.C.; Moreira, S.I.; Pedroso, M.P.; Pereira, L.F.; et al. Bacillus amyloliquefaciens Strain BaNCT02: An Antagonist with Multiple Mechanisms of Action against Meloidogyne incognita. Plant Pathol. 2025, 74, 320–329. [Google Scholar] [CrossRef]
- Sun, W.; Shahrajabian, M.H. Biostimulant and Beyond: Bacillus spp., the Important Plant Growth-Promoting Rhizobacteria (PGPR)—Based Biostimulant for Sustainable Agriculture. Earth Syst. Environ. 2025, 9, 1465–1498. [Google Scholar] [CrossRef]
- Deng, X.; Wang, X.; Li, G. Nematicidal Effects of Volatile Organic Compounds from Microorganisms and Plants on Plant-Parasitic Nematodes. Microorganisms 2022, 10, 1201. [Google Scholar] [CrossRef]
- Chavarria-Quicaño, E.; De La Torre-González, F.; González-Riojas, M.; Rodríguez-González, J.; Asaff-Torres, A. Nematicidal Lipopeptides from Bacillus paralicheniformis and Bacillus subtilis: A Comparative Study. Appl. Microbiol. Biotechnol. 2023, 107, 1537–1549. [Google Scholar] [CrossRef]
- Hu, H.; Gao, Y.; Li, X.; Chen, S.; Yan, S.; Tian, X. Identification and Nematicidal Characterization of Proteases Secreted by Endophytic Bacteria Bacillus cereus BCM2. Phytopathology 2020, 110, 336–344. [Google Scholar] [CrossRef] [PubMed]
- Pawar, P.; Doshi, J.; Patil, S.G.; Dandekar, P.; Poornima, K. The Characterization of Chitinolytic Soil Bacterial Isolates for their Antagonistic Activity against Root Knot Nematode Meloidogyne incognita: An Effort towards Developing ‘Green’ Nematicidal Agents. BioControl 2023, 68, 511–524. [Google Scholar] [CrossRef]
- Engelbrecht, G.; Horak, I.; Rensburg, P.J.J.; Claassens, S. Bacillus-Based Bionematicides: Development, Modes of Action and Commercialisation. Biocontrol Sci. Technol. 2018, 28, 629–653. [Google Scholar] [CrossRef]
- Horak, I.; Engelbrecht, G.; Rensburg, P.J.J.; Claassens, S. Microbial Metabolomics: Essential Definitions and the Importance of Cultivation Conditions for Utilizing Bacillus Species as Bionematicides. J. Appl. Microbiol. 2019, 127, 326–343. [Google Scholar] [CrossRef] [PubMed]
- Kačergius, A.; Sivojienė, D.; Gudiukaitė, R.; Bakšienė, E.; Masevičienė, A.; Žičkienė, L. Comparison of the Structure of Soil Microbial Communities of Different Ecosystems Using the Microbiome Sequencing Approach. Soil Syst. 2023, 7, 70. [Google Scholar] [CrossRef]
- Cao, X.; Yuan, Q.; Hu, C.; Zhang, H.; Sun, X.; Yan, B.; Ma, X.; Zhang, L.; Huang, L.; Li, S.; et al. Wild Wisdom Meets Cultivation: Comparative Rhizomicrobiome Analysis Unveils the Key Role of Paraburkholderia in Growth Promotion and Disease Suppression in Coptis chinensis. Microbiome 2025, 13, 150. [Google Scholar] [CrossRef]
- Valicente, F.H.; Barreto, M.R. Bacillus thuringiensis Survey in Brazil: Geographical Distribution and Insecticidal Activity against Spodoptera frugiperda (J.E. Smith) (Lepidoptera: Noctuidae). Neotrop. Entomol. 2003, 32, 639–644. [Google Scholar] [CrossRef]
- Logan, N.A.; Vos, P.D. Bacillus. In Bergey’s Manual of Systematics of Archaea and Bacteria; Whitman, W.B., Ed.; Wiley: Hoboken, NJ, USA, 2015; pp. 1–163. [Google Scholar] [CrossRef]
- Hussey, R.S.; Barker, K.R. A Comparison of Methods of Collecting Inocula of Meloidogyne spp., Including a New Technique. Plant Dis. Rep. 1973, 57, 1025–1028. [Google Scholar]
- Oostenbrink, M. Major Characteristics of the Relation between Nematodes and Plants. Mededelingen Landbouwhogeschool 1966, 66, 1–46. [Google Scholar]
- National Institute of Standards and Technology (NIST). NIST/EPA/NIH Mass Spectral Library with Search Program—SRD 1A; Version 2.2; U.S. Department of Commerce: Gaithersburg, MD, USA, 2014. Available online: https://chemdata.nist.gov/ (accessed on 11 July 2025).
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Charney, J.; Tomarelli, R.M. A Colorimetric Method for the Determination of the Proteolytic Activity of Duodenal Juice. J. Biol. Chem. 1947, 170, 501–505. [Google Scholar] [CrossRef]
- Miller, G.L. Use of DinitrosaIicyIic Acid Reagent for Determination of Reducing Sugar. Anal. Chem. 1959, 31, 426–428. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S Ribosomal DNA Amplification for Phylogenetic Study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef]
- Schmitz, A.; Riesner, D. Purification of Nucleic Acids by Selective Precipitation with Polyethylene Glycol 6000. Anal. Biochem. 2006, 354, 311–313. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Tamura, K. Estimation of the Number of Nucleotide Substitutions when there are Strong Transition-Transversion and G+C-Content Biases. Mol. Biol. Evol. 1992, 9, 678–687. [Google Scholar] [CrossRef]
- Xiang, N.; Lawrence, K.S.; Kloepper, J.W.; Donald, P.A.; McInroy, J.A.; Lawrence, G.W. Biological Control of Meloidogyne incognita by Spore-Forming Plant Growth-Promoting Rhizobacteria on Cotton. Plant Dis. 2017, 101, 774–784. [Google Scholar] [CrossRef]
- Hu, H.J.; Chen, Y.L.; Wang, Y.F.; Tang, Y.Y.; Chen, S.L.; Yan, S.Z. Endophytic Bacillus cereus Effectively Controls Meloidogyne incognita on Tomato Plants Through Rapid Rhizosphere Occupation and Repellent Action. Plant Dis. 2017, 101, 448–455. [Google Scholar] [CrossRef]
- Yin, N.; Zhao, J.L.; Liu, R.; Li, Y.; Ling, J.; Yang, Y.H.; Xie, B.Y.; Mao, Z.C. Biocontrol Efficacy of Bacillus cereus Strain Bc-Cm103 Against Meloidogyne incognita. Plant Dis. 2021, 105, 2061–2070. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Jiao, Y.; Yin, N.; Li, Y.; Ling, J.; Mao, Z.; Yang, Y.; Xie, B. Analysis of the Activity and Biological Control Efficacy of the Bacillus subtilis Strain Bs-1 against Meloidogyne incognita. Crop Prot. 2019, 122, 125–135. [Google Scholar] [CrossRef]
- Jamal, Q.; Cho, J.-Y.; Moon, J.-H.; Munir, S.; Anees, M.; Kim, K.Y. Identification for the First Time of Cyclo (d-Pro-l-Leu) Produced by Bacillus amyloliquefaciens Y1 as a Nematocide for Control of Meloidogyne incognita. Molecules 2017, 22, 1839. [Google Scholar] [CrossRef]
- Basyony, A.G.; Abo-Zaid, G.A. Biocontrol of the Root-Knot Nematode, Meloidogyne incognita, Using an Eco-Friendly Formulation from Bacillus subtilis, Lab. and Greenhouse Studies. Egypt. J. Biol. Pest Control 2018, 28, 87. [Google Scholar] [CrossRef]
- Nadeem, H.; Niazi, P.; Asif, M.; Kaskavalci, G.; Ahmad, F. Bacterial Strains Integrated with Surfactin Molecules of Bacillus subtilis MTCC441 Enrich Nematocidal Activity against Meloidogyne incognita. Plant Biol. 2021, 23, 1027–1036. [Google Scholar] [CrossRef] [PubMed]
- Peng, D.; Chai, L.; Wang, F.; Zhang, F.; Ruan, L.; Sun, M. Synergistic Activity between Bacillus thuringiensis Cry6Aa and Cry55Aa Toxins against Meloidogyne incognita. Microb. Biotechnol. 2011, 4, 794–798. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Zheng, J.; Zhang, Z.; Peng, D.; Sun, M. Nematicidal Spore-Forming Bacilli Share Similar Virulence Factors and Mechanisms. Sci. Rep. 2016, 6, 31341. [Google Scholar] [CrossRef]
- Lu, P.; Davis, R.F.; Kemerait, R.C.; Iersel, M.W.V.; Scherm, H. Physiological Effects of Meloidogyne incognita Infection on Cotton Genotypes with Differing Levels of Resistance in the Greenhouse. J. Nematol. 2014, 46, 352–359. [Google Scholar] [PubMed]
- Gámez, B.Y.S.; Garruña, R.; Suárez, J.M.T.; Valenzuela, O.A.M.; Ramírez, A.R.; Gough, R.E.V.; Catzim, C.E.A.; Palomar, L.T. Healthy Photosynthetic Mechanism Suggests ISR Elicited by Bacillus spp. in Capsicum chinense Plants Infected with PepGMV. Pathog. 2021, 10, 455. [Google Scholar] [CrossRef]
- Gámez, B.Y.S.; Garruña, R.; Suárez, J.M.T.; Can, J.K.; Ramírez, A.R.; Díaz, L.C. Bacillus spp. Inoculation Improves Photosystem II Efficiency and Enhances Photosynthesis in Pepper Plants. Chil. J. Agric. Res. 2016, 76, 409–416. [Google Scholar] [CrossRef]
- Magalhães, V.C.; Guimarães, R.A.; Silva, J.C.P.; Faria, A.F.; Pedroso, M.P.; Campos, V.P.; Marbach, P.A.S.; Medeiros, F.H.V.; Souza, J.T. The combination of two Bacillus strains supresses Meloidogyne incognita, but does not enhance plant growth. Pest Manag. Sci. 2021, 78, 722–732. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, N.; Bi, X.; Bi, T.; Baloch, F.B.; Miao, J.; Zeng, N.; Li, B.; An, Y. Growth Promotion on Maize and Whole-Genome Sequence Analysis of Bacillus velezensis D103. Microbiol. Spectr. 2024, 12, e01147-24. [Google Scholar] [CrossRef]
- Zou, Z.; Fan, Q.; Zhou, X.; Fu, X.; Jia, Y.; Li, H.; Liao, Y. Biochemical Pathways of Salicylic Acid Derived from l-Phenylalanine in Plants with Different Basal SA Levels. J. Agric. Food Chem. 2024, 72, 2898–2910. [Google Scholar] [CrossRef] [PubMed]
- Amdadul, A.K.M.; Bhuiyan, M.R.; Khan, M.A.I.; Mahmud, A.; Ahmad, M.U. Effect of Amino Acids on Root-Knot Nematode (Meloidogyne javanica) Infecting Tomato Plant. Arch. Phytopathol. Plant Prot. 2013, 47, 1921–1928. [Google Scholar] [CrossRef]
- Siddiqui, I.A.; Shaukat, S.S. Zinc and Glycerol Enhance the Production of Nematicidal Compounds in vitro and Improve the Biocontrol of a in Tomato by Fluorescent Pseudomonads. Lett. Appl. Microbiol. 2002, 35, 212–217. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.J.; Park, A.R.; Kim, S.; Yeon, J.; Yu, N.H.; Ha, S.; Chang, J.Y.; Park, H.W.; Kim, J.-C. Biological Control of Root-Knot Nematodes by Organic Acid-Producing Lactobacillus brevis WiKim0069 Isolated from Kimchi. Plant Pathol. J. 2019, 35, 662–673. [Google Scholar] [CrossRef]
- Ye, L.; Wang, J.Y.; Liu, X.F.; Guan, Q.; Dou, N.X.; Li, J.; Zhang, Q.; Gao, Y.M.; Wang, M.; Li, J.S.; et al. Nematicidal Activity of Volatile Organic Compounds Produced by Bacillus altitudinis AMCC 1040 against Meloidogyne incognita. Arch. Microbiol. 2022, 204, 521. [Google Scholar] [CrossRef]
- Oliveira, D.F.; Santos Júnior, H.M.D.; Nunes, A.S.; Campos, V.P.; Pinho, R.S.C.D.; Gajo, G.C. Purification and Identification of Metabolites Produced by Bacillus cereus and B. subtilis Active against Meloidogyne exigua, and Their in Silico Interaction with a Putative Phosphoribosyltransferase from M. incognita. An. Acad. Bras. Ciênc. 2014, 86, 525–538. [Google Scholar] [CrossRef]
- Wang, J.Y.; Li, Q.Y.; Ren, L.; Guo, C.; Qu, J.P.; Gao, Z.; Wang, H.F.; Zhang, Q.; Zhou, B. Transcriptomic and Physiological Analysis of the Effect of Octanoic Acid on Meloidogyne incognita. Pestic. Biochem. Physiol. 2023, 193, 105432. [Google Scholar] [CrossRef]
- Soliman, G.M.; Ameen, H.H.; Abdel-Aziz, S.M.; El-Sayed, G.M. In Vitro Evaluation of Some Isolated Bacteria against the Plant Parasite Nematode Meloidogyne incognita. Bull. Natl. Res. Cent. 2019, 43, 171. [Google Scholar] [CrossRef]
- Wei, L.H.; Xue, Q.Y.; Wei, B.Q.; Wang, Y.M.; Li, S.M.; Chen, L.F.; Guo, J.H. Screening of Antagonistic Bacterial Strains against Meloidogyne incognita Using Protease AcTivity. Biocont. Sci. Technol. 2010, 20, 739–750. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).