Transcriptomic Analysis Reveals Calcium and Ethylene Signaling Pathway Genes in Response to Cold Stress in Cinnamomum camphora
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Cold Stress Treatments
2.2. RNA Extraction
2.3. Transcriptome Analysis
2.4. Identification of ERF Proteins in C. camphora
2.5. Phylogenetic Analysis
2.6. Reverse Transcription and qRT-PCR Analysis
2.7. Statistical Analyses
3. Results
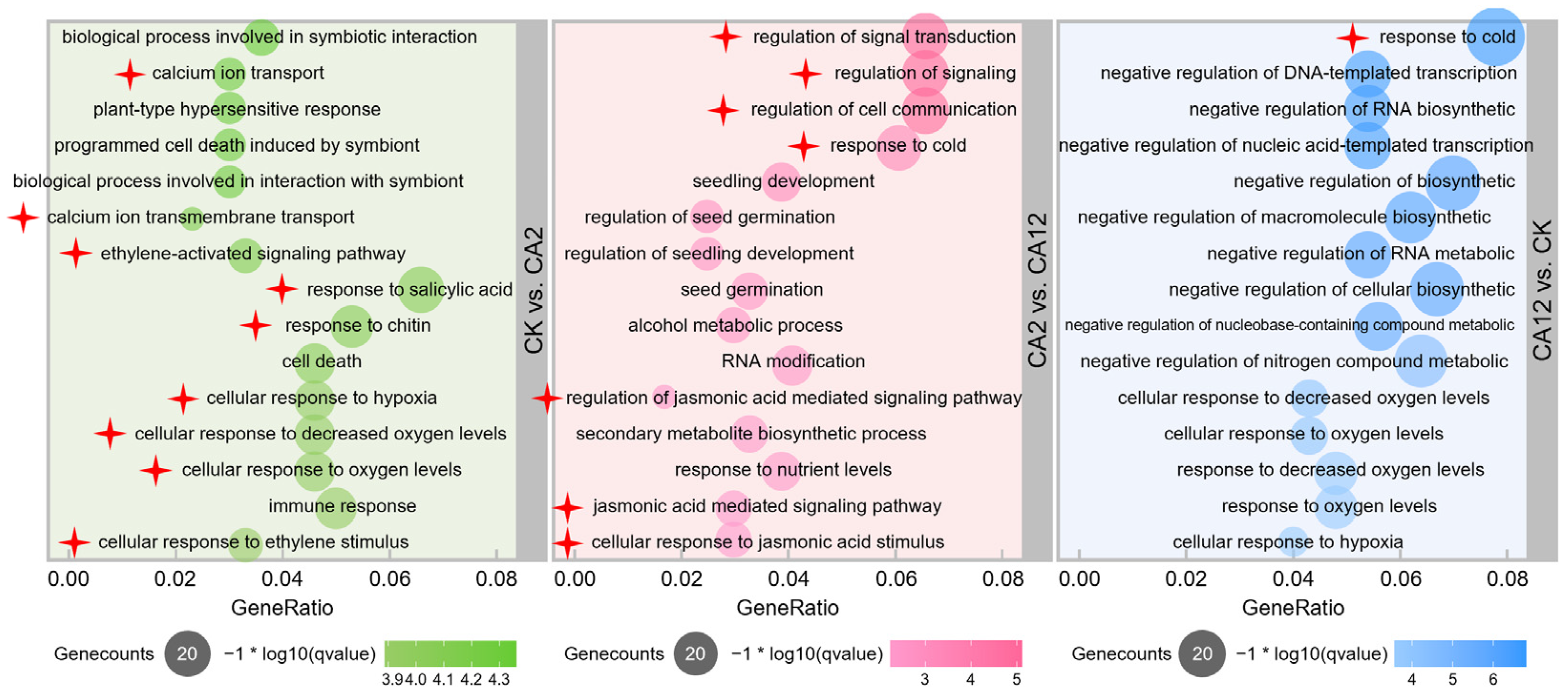
3.1. Functional Enrichment Analysis of DEGs between Different Cold Stress Treatments
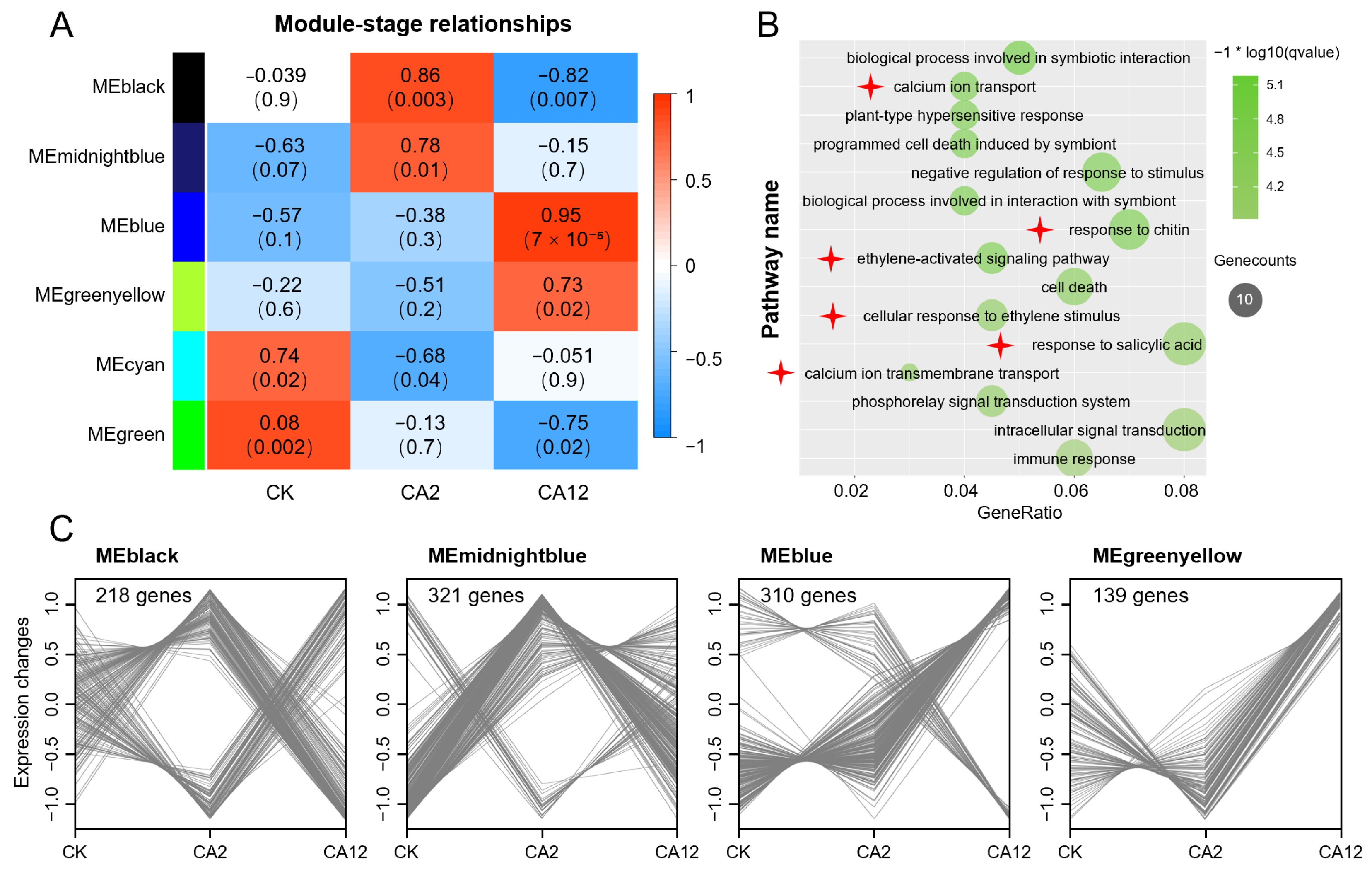
3.2. WGCNA—Revealed Key Pathways and Gene Responses to Cold
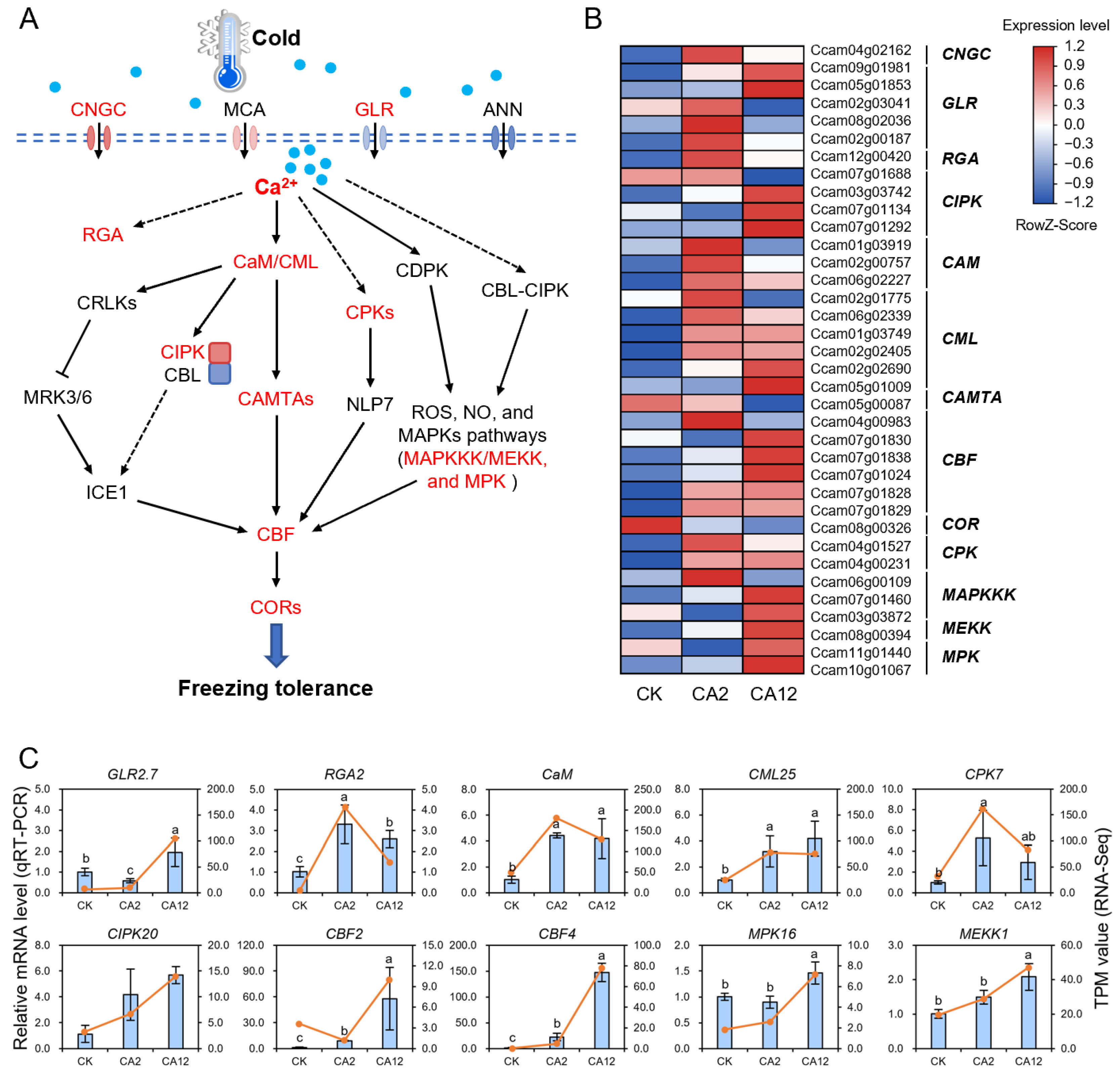
3.3. Impact of Cold Stress Treatment on Calcium Signaling-Related Gene Expression
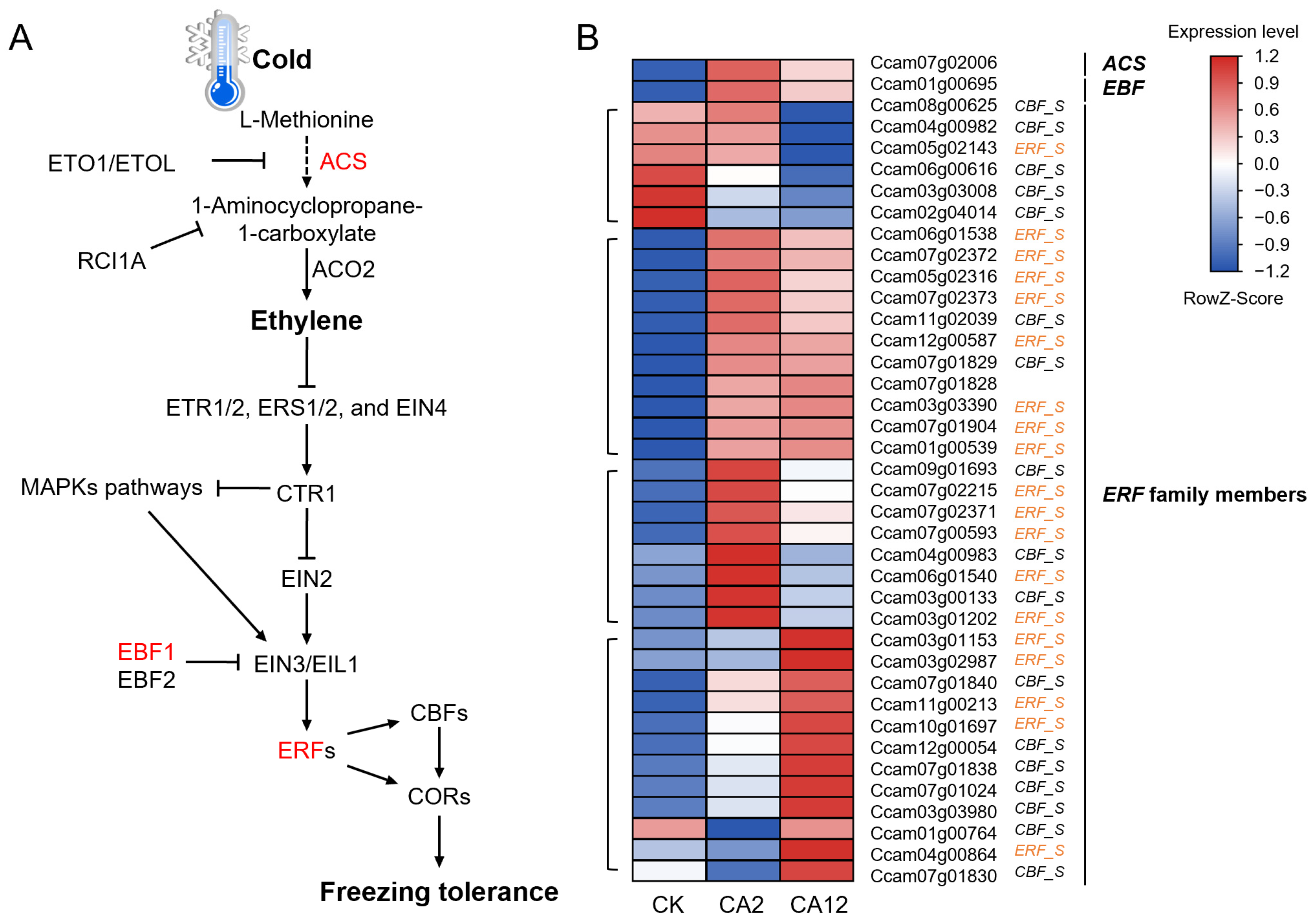
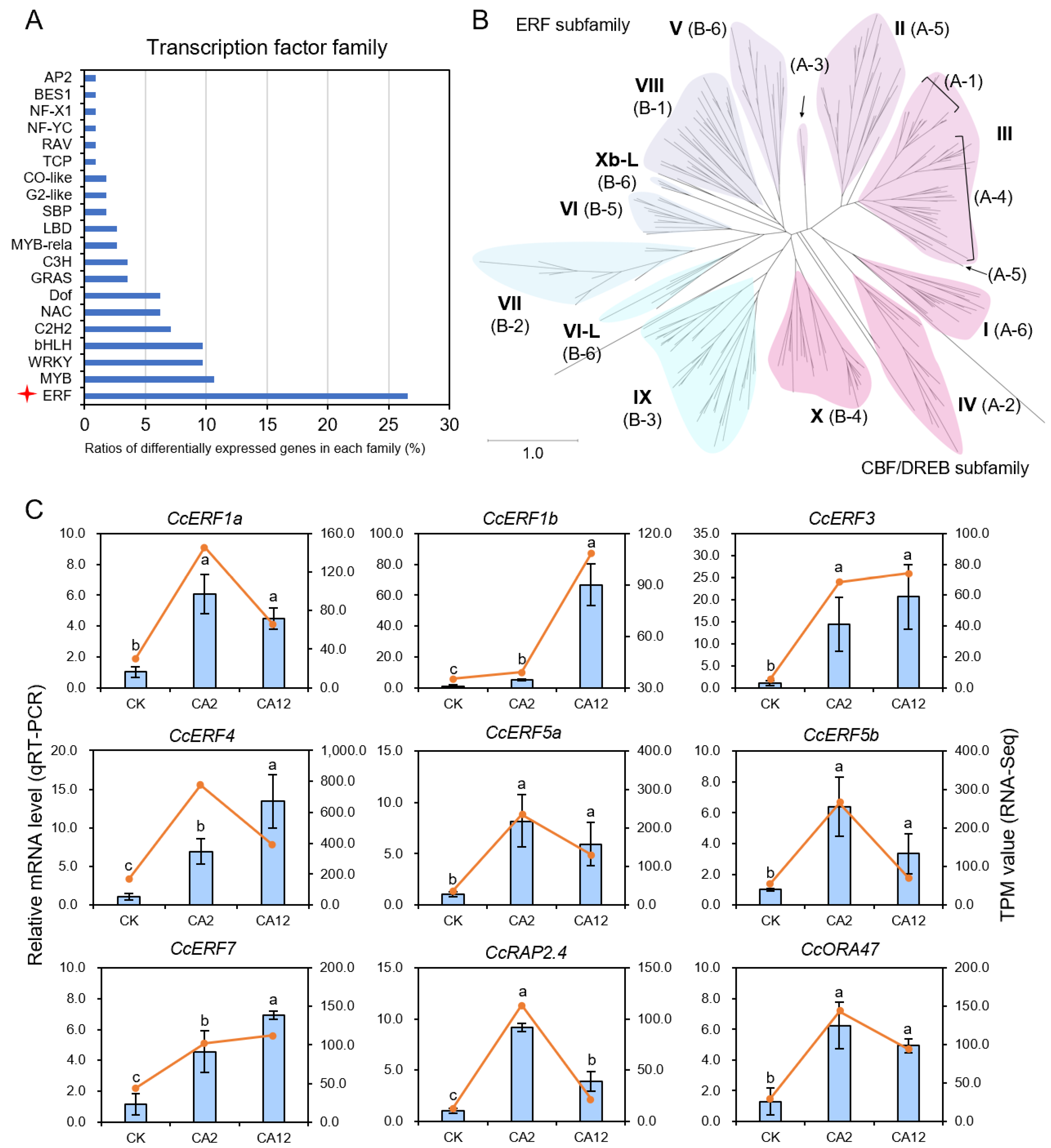
3.4. Alterations in Gene Expression Related to Ethylene Biosynthesis and Signaling Pathways
3.5. Phylogenetic Characterization of ERF Gene Family Members in C. camphora
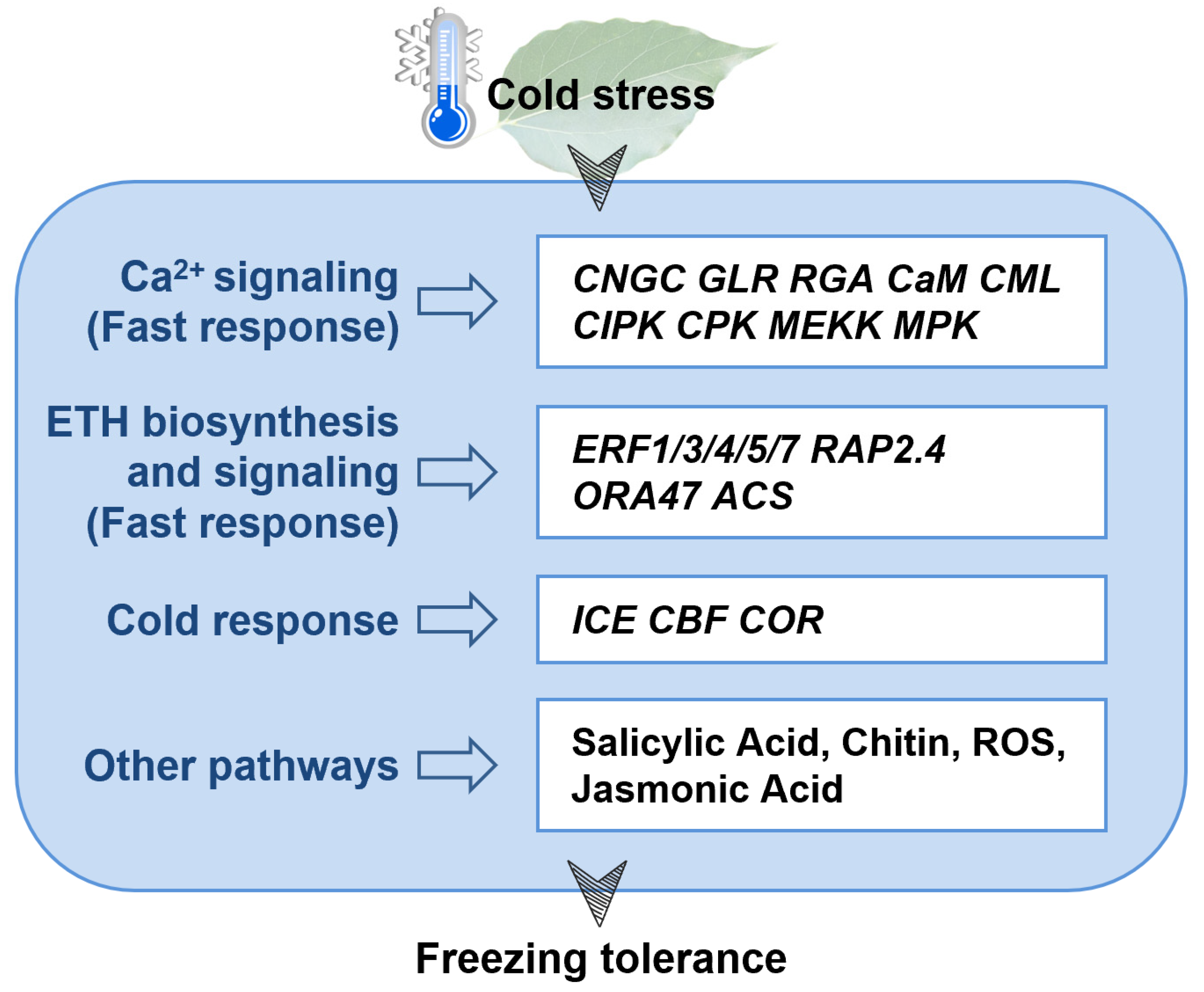
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ding, Y.; Yang, S. Surviving and thriving: How plants perceive and respond to temperature stress. Dev. Cell 2022, 57, 947–958. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Nahar, K.; Hossain, M.S.; Mahmud, J.A.; Rahman, A.; Inafuku, M.; Oku, H.; Fujita, M. Coordinated Actions of Glyoxalase and Antioxidant Defense Systems in Conferring Abiotic Stress Tolerance in Plants. Int. J. Mol. Sci. 2017, 18, 200. [Google Scholar] [CrossRef] [PubMed]
- Sedaghathoor, S.; Baladeh, M.K.; Piri, S. A study on chilling hardiness of three persimmon genotypes by electrolyte leakage parameter. Technol. Hortic. 2023, 3, 10. [Google Scholar] [CrossRef]
- Liu, C.T.; Wang, W.; Mao, B.G.; Chu, C.C. Cold stress tolerance in rice: Physiological changes, molecular mechanism, and future prospects. Yi Chuan = Hered. 2018, 40, 171–185. [Google Scholar] [CrossRef]
- Guo, X.; Liu, D.; Chong, K. Cold signaling in plants: Insights into mechanisms and regulation. J. Integr. Plant Biol. 2018, 60, 745–756. [Google Scholar] [CrossRef]
- Liu, Q.; Ding, Y.; Shi, Y.; Ma, L.; Wang, Y.; Song, C.; Wilkins, K.A.; Davies, J.M.; Knight, H.; Knight, M.R.; et al. The calcium transporter ANNEXIN1 mediates cold-induced calcium signaling and freezing tolerance in plants. EMBO J. 2021, 40, e104559. [Google Scholar] [CrossRef]
- Shao, L.; Xu, T.; Wang, X.; Zhang, R.; Wang, X.; Ren, Z.; Zhang, J.; Xia, Y.; Li, D. Integrative Comparative Assessment of Cold Acclimation in Evergreen and Deciduous Iris Species. Antioxidants 2022, 11, 977. [Google Scholar] [CrossRef]
- Wingler, A.; Tijero, V.; Müller, M.; Yuan, B.; Munné-Bosch, S. Interactions between sucrose and jasmonate signalling in the response to cold stress. BMC Plant Biol. 2020, 20, 176. [Google Scholar] [CrossRef]
- You, J.; Chan, Z. ROS Regulation During Abiotic Stress Responses in Crop Plants. Front. Plant Sci. 2015, 6, 1092. [Google Scholar] [CrossRef]
- Nykiel, M.; Gietler, M.; Fidler, J.; Prabucka, B.; Labudda, M. Abiotic Stress Signaling and Responses in Plants. Plants 2023, 12, 3405. [Google Scholar] [CrossRef]
- Sangwan, V.; Orvar, B.L.; Beyerly, J.; Hirt, H.; Dhindsa, R.S. Opposite changes in membrane fluidity mimic cold and heat stress activation of distinct plant MAP kinase pathways. Plant J. Cell Mol. Biol. 2002, 31, 629–638. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Poovaiah, B.W. Calcium/calmodulin-mediated signal network in plants. Trends Plant Sci. 2003, 8, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Sanders, D.; Pelloux, J.; Brownlee, C.; Harper, J.F. Calcium at the crossroads of signaling. Plant Cell 2002, 14, S401–S417. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.C.; Chung, W.S.; Yun, D.J.; Cho, M.J. Calcium and calmodulin-mediated regulation of gene expression in plants. Mol. Plant 2009, 2, 13–21. [Google Scholar] [CrossRef]
- Liu, J.; Lenzoni, G.; Knight, M.R. Design Principle for Decoding Calcium Signals to Generate Specific Gene Expression Via Transcription. Plant Physiol. 2020, 182, 1743–1761. [Google Scholar] [CrossRef] [PubMed]
- Yuan, P.; Yang, T.; Poovaiah, B.W. Calcium Signaling-Mediated Plant Response to Cold Stress. Int. J. Mol. Sci. 2018, 19, 3896. [Google Scholar] [CrossRef]
- Lv, X.; Li, H.; Chen, X.; Xiang, X.; Guo, Z.; Yu, J.; Zhou, Y. The role of calcium-dependent protein kinase in hydrogen peroxide, nitric oxide and ABA-dependent cold acclimation. J. Exp. Bot. 2018, 69, 4127–4139. [Google Scholar] [CrossRef]
- Huang, J.; Zhao, X.; Bürger, M.; Chory, J.; Wang, X. The role of ethylene in plant temperature stress response. Trends Plant Sci. 2023, 28, 808–824. [Google Scholar] [CrossRef]
- Sun, X.; Zhao, T.; Gan, S.; Ren, X.; Fang, L.; Karungo, S.K.; Wang, Y.; Chen, L.; Li, S.; Xin, H. Ethylene positively regulates cold tolerance in grapevine by modulating the expression of ETHYLENE RESPONSE FACTOR 057. Sci. Rep. 2016, 6, 24066. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, H.; Mao, Z.; Liu, W.; Jiang, S.; Xu, H.; Su, M.; Zhang, J.; Wang, N.; Zhang, Z.; et al. Ethylene increases the cold tolerance of apple via the MdERF1B-MdCIbHLH1 regulatory module. Plant J. Cell Mol. Biol. 2021, 106, 379–393. [Google Scholar] [CrossRef]
- Ritonga, F.N.; Ngatia, J.N.; Wang, Y.; Khoso, M.A.; Farooq, U.; Chen, S. AP2/ERF, an important cold stress-related transcription factor family in plants: A review. Physiol. Mol. Biol. Plants Int. J. Funct. Plant Biol. 2021, 27, 1953–1968. [Google Scholar] [CrossRef] [PubMed]
- Illgen, S.; Zintl, S.; Zuther, E.; Hincha, D.K.; Schmülling, T. Characterisation of the ERF102 to ERF105 genes of Arabidopsis thaliana and their role in the response to cold stress. Plant Mol. Biol. 2020, 103, 303–320. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Huang, R. Enhanced tolerance to freezing in tobacco and tomato overexpressing transcription factor TERF2/LeERF2 is modulated by ethylene biosynthesis. Plant Mol. Biol. 2010, 73, 241–249. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Liu, B.; Davis, C.C.; Yang, Y. Plastome phylogenomics, systematics, and divergence time estimation of the Beilschmiedia group (Lauraceae). Mol. Phylogenet. Evol. 2020, 151, 106901. [Google Scholar] [CrossRef]
- Li, D.Q.; Lin, H.Y.; Wang, X.Y.; Bi, B.; Gao, Y.; Shao, L.M.; Zhang, R.L.; Liang, Y.W.; Xia, Y.P.; Zhao, Y.P.; et al. Genome and whole-genome resequencing of elucidate its dominance in subtropical urban landscapes. BMC Biol. 2023, 21, 192. [Google Scholar] [CrossRef]
- Chen, S.F.; Zhou, Y.Q.; Chen, Y.R.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Kovaka, S.; Zimin, A.V.; Pertea, G.M.; Razaghi, R.; Salzberg, S.L.; Pertea, M. Transcriptome assembly from long-read RNA-seq alignments with StringTie2. Genome Biol. 2019, 20, 278. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, D.; Zhang, D.; Shi, X.; Wu, Y.; Qi, Z.; Ding, H.; Zhu, K.; Xia, Y.; Zhang, J. Improving crucial details and selecting the optimal model for evaluating the chilling requirement of Paeonia lactiflora Pall. at low latitudes during four winters. Sci. Hortic. 2020, 265, 109175. [Google Scholar] [CrossRef]
- Johnson, N.; Rodriguez Diaz, D.; Ganapathy, S.; Bass, J.S.; Kutchan, T.M.; Khan, A.L.; Flavier, A.B. Evaluation of reference genes for qRT-PCR studies in the colchicine producing Gloriosa superba L. Plant Biotechnol. Rep. 2023, 17, 561–571. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Li, W.; Fu, Y.; Lv, W.; Zhao, S.; Feng, H.; Shao, L.; Li, C.; Yang, J. Characterization of the early gene expression profile in Populus ussuriensis under cold stress using PacBio SMRT sequencing integrated with RNA-seq reads. Tree Physiol. 2022, 42, 646–663. [Google Scholar] [CrossRef]
- Wang, J.; Yang, Y.; Liu, X.; Huang, J.; Wang, Q.; Gu, J.; Lu, Y. Transcriptome profiling of the cold response and signaling pathways in Lilium lancifolium. BMC Genom. 2014, 15, 203. [Google Scholar] [CrossRef]
- Ren, H.; Zhang, Y.; Zhong, M.; Hussian, J.; Tang, Y.; Liu, S.; Qi, G. Calcium signaling-mediated transcriptional reprogramming during abiotic stress response in plants. Theor. Appl. Genet. 2023, 136, 210. [Google Scholar] [CrossRef]
- Zhang, Q.; Liu, Y.; Yu, Q.; Ma, Y.; Gu, W.; Yang, D. Physiological changes associated with enhanced cold resistance during maize (Zea mays) germination and seedling growth in response to exogenous calcium. Crop Pasture Sci. 2020, 71, 529–538. [Google Scholar] [CrossRef]
- Chen, S.; Wang, L.; Kang, R.; Liu, C.; Xing, L.; Wu, S.; Wang, Z.; Wu, C.; Zhou, Q.; Zhao, R.J.H. Exogenous Calcium Alleviates the Photosynthetic Inhibition and Oxidative Damage of the Tea Plant under Cold Stress. Horticulturae 2024, 10, 666. [Google Scholar] [CrossRef]
- Wang, J.; Ren, Y.; Liu, X.; Luo, S.; Zhang, X.; Liu, X.; Lin, Q.; Zhu, S.; Wan, H.; Yang, Y.; et al. Transcriptional activation and phosphorylation of OsCNGC9 confer enhanced chilling tolerance in rice. Mol. Plant 2021, 14, 315–329. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Jiang, X.; Lv, X.; Ahammed, G.J.; Guo, Z.; Qi, Z.; Yu, J.; Zhou, Y. Tomato GLR3.3 and GLR3.5 mediate cold acclimation-induced chilling tolerance by regulating apoplastic H(2) O(2) production and redox homeostasis. Plant Cell Environ. 2019, 42, 3326–3339. [Google Scholar] [CrossRef] [PubMed]
- Ye, K.; Shen, W.; Zhao, Y. External application of brassinolide enhances cold resistance of tea plants (Camellia sinensis L.) by integrating calcium signals. Planta 2023, 258, 114. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, C.; Zhu, Y.; Zhang, L.; Chen, T.; Zhou, F.; Chen, H.; Lin, Y. The calcium-dependent kinase OsCPK24 functions in cold stress responses in rice. J. Integr. Plant Biol. 2018, 60, 173–188. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Huang, R.; Zhu, H.; Sun, Y.; Guo, Z. A novel Medicago truncatula calmodulin-like protein (MtCML42) regulates cold tolerance and flowering time. Plant J. Cell Mol. Biol. 2021, 108, 1069–1082. [Google Scholar] [CrossRef]
- Chu, M.; Li, J.; Zhang, J.; Shen, S.; Li, C.; Gao, Y.; Zhang, S. AtCaM4 interacts with a Sec14-like protein, PATL1, to regulate freezing tolerance in Arabidopsis in a CBF-independent manner. J. Exp. Bot. 2018, 69, 5241–5253. [Google Scholar] [CrossRef]
- Ma, X.; Gai, W.X.; Li, Y.; Yu, Y.N.; Ali, M.; Gong, Z.H. The CBL-interacting protein kinase CaCIPK13 positively regulates defence mechanisms against cold stress in pepper. J. Exp. Bot. 2022, 73, 1655–1667. [Google Scholar] [CrossRef]
- Li, J.; Zou, X.; Chen, G.; Meng, Y.; Ma, Q.; Chen, Q.; Wang, Z.; Li, F. Potential Roles of 1-Aminocyclopropane-1-carboxylic Acid Synthase Genes in the Response of Gossypium Species to Abiotic Stress by Genome-Wide Identification and Expression Analysis. Plants 2022, 11, 1524. [Google Scholar] [CrossRef]
- Zhao, R.; Xie, H.; Lv, S.; Zheng, Y.; Yu, M.; Shen, L.; Sheng, J. LeMAPK4 participated in cold-induced ethylene production in tomato fruit. J. Sci. Food Agric. 2013, 93, 1003–1009. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Liu, W.; Xia, X.; Wang, T.; Zhang, W.H. Cold acclimation-induced freezing tolerance of Medicago truncatula seedlings is negatively regulated by ethylene. Physiol. Plant. 2014, 152, 115–129. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Tian, S.; Hou, L.; Huang, X.; Zhang, X.; Guo, H.; Yang, S. Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 2012, 24, 2578–2595. [Google Scholar] [CrossRef]
- Ma, Y.; Zhang, L.; Zhang, J.; Chen, J.; Wu, T.; Zhu, S.; Yan, S.; Zhao, X.; Zhong, G. Expressing a Citrus ortholog of Arabidopsis ERF1 enhanced cold-tolerance in tobacco. Sci. Hortic. 2014, 174, 65–76. [Google Scholar] [CrossRef]
- Li, C.; Mao, B.; Wang, K.; Xu, L.; Fan, L.; Wang, Y.; Li, Y.; Ma, Y.; Wang, L.; Liu, L. RsERF40 contributes to cold stress tolerance and cell expansion of taproot in radish (Raphanus sativus L.). Hortic. Res. 2023, 10, uhad013. [Google Scholar] [CrossRef]
- Lv, K.; Li, J.; Zhao, K.; Chen, S.; Nie, J.; Zhang, W.; Liu, G.; Wei, H. Overexpression of an AP2/ERF family gene, BpERF13, in birch enhances cold tolerance through upregulating CBF genes and mitigating reactive oxygen species. Plant Sci. Int. J. Exp. Plant Biol. 2020, 292, 110375. [Google Scholar] [CrossRef] [PubMed]
- Bolt, S.; Zuther, E.; Zintl, S.; Hincha, D.K.; Schmülling, T. ERF105 is a transcription factor gene of Arabidopsis thaliana required for freezing tolerance and cold acclimation. Plant Cell Environ. 2017, 40, 108–120. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, L.; Wong, D.C.J.; Wang, Y.; Zhu, Z.; Xu, G.; Wang, Q.; Li, S.; Liang, Z.; Xin, H. The ethylene response factor VaERF092 from Amur grape regulates the transcription factor VaWRKY33, improving cold tolerance. Plant J. Cell Mol. Biol. 2019, 99, 988–1002. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bi, B.; Shao, L.; Xu, T.; Du, H.; Li, D. Transcriptomic Analysis Reveals Calcium and Ethylene Signaling Pathway Genes in Response to Cold Stress in Cinnamomum camphora. Horticulturae 2024, 10, 995. https://doi.org/10.3390/horticulturae10090995
Bi B, Shao L, Xu T, Du H, Li D. Transcriptomic Analysis Reveals Calcium and Ethylene Signaling Pathway Genes in Response to Cold Stress in Cinnamomum camphora. Horticulturae. 2024; 10(9):995. https://doi.org/10.3390/horticulturae10090995
Chicago/Turabian StyleBi, Bo, Lingmei Shao, Tong Xu, Hao Du, and Danqing Li. 2024. "Transcriptomic Analysis Reveals Calcium and Ethylene Signaling Pathway Genes in Response to Cold Stress in Cinnamomum camphora" Horticulturae 10, no. 9: 995. https://doi.org/10.3390/horticulturae10090995
APA StyleBi, B., Shao, L., Xu, T., Du, H., & Li, D. (2024). Transcriptomic Analysis Reveals Calcium and Ethylene Signaling Pathway Genes in Response to Cold Stress in Cinnamomum camphora. Horticulturae, 10(9), 995. https://doi.org/10.3390/horticulturae10090995





