Investigating Changes in the Soil Fungal Community Structure, Functions, and Network Stability with Prolonged Grafted Watermelon Cultivation
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site Description and Soil Sample Collection
2.2. Analysis of Physicochemical Properties
2.3. DNA Extraction and Fungal Quantification
2.4. MiSeq Sequencing and Data Processing
2.5. Bioinformatics and Statistical Analysis
3. Results
3.1. Soil Physicochemical Properties
3.2. Soil Fungal Population and Alpha Diversity
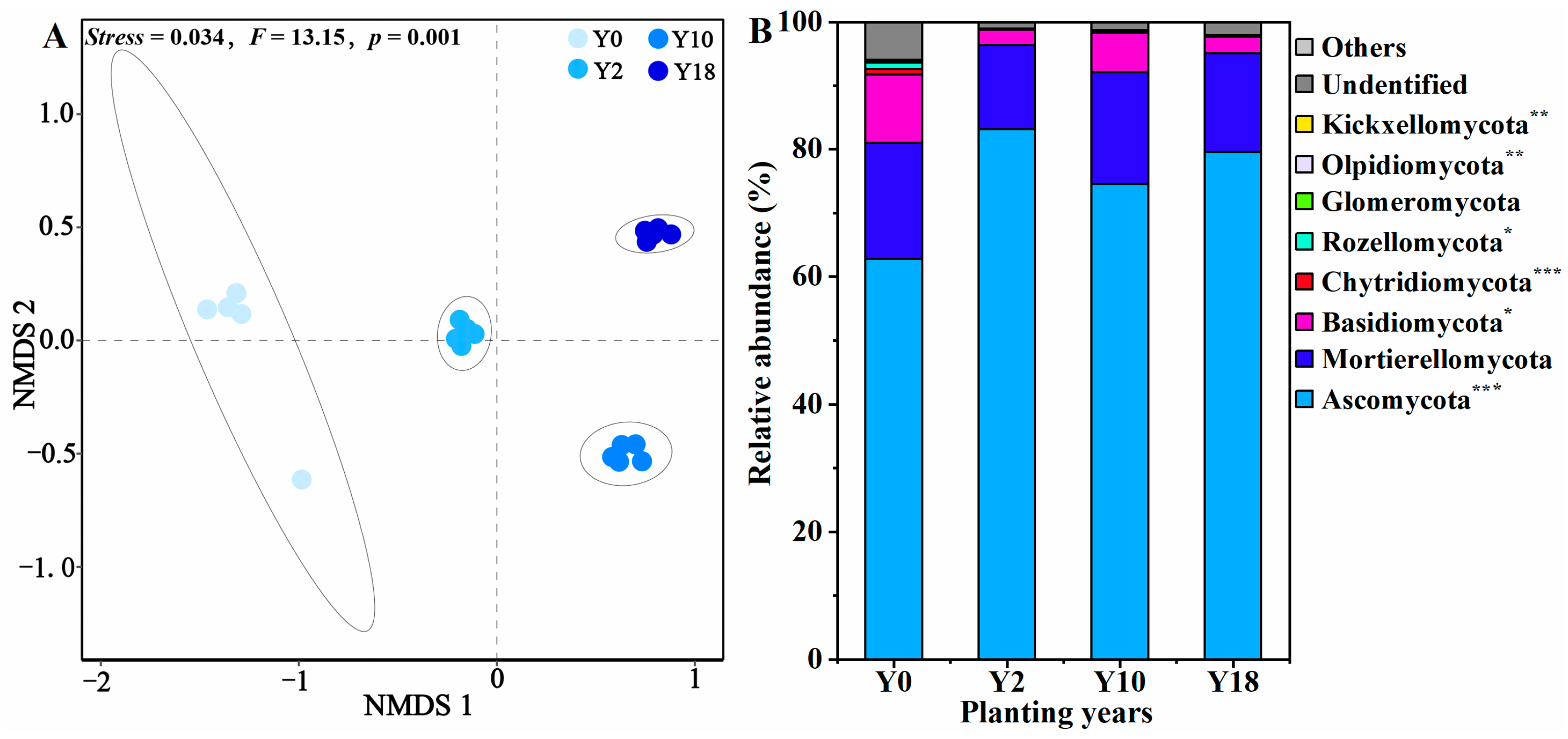
3.3. Soil Fungal Community Structure and Composition
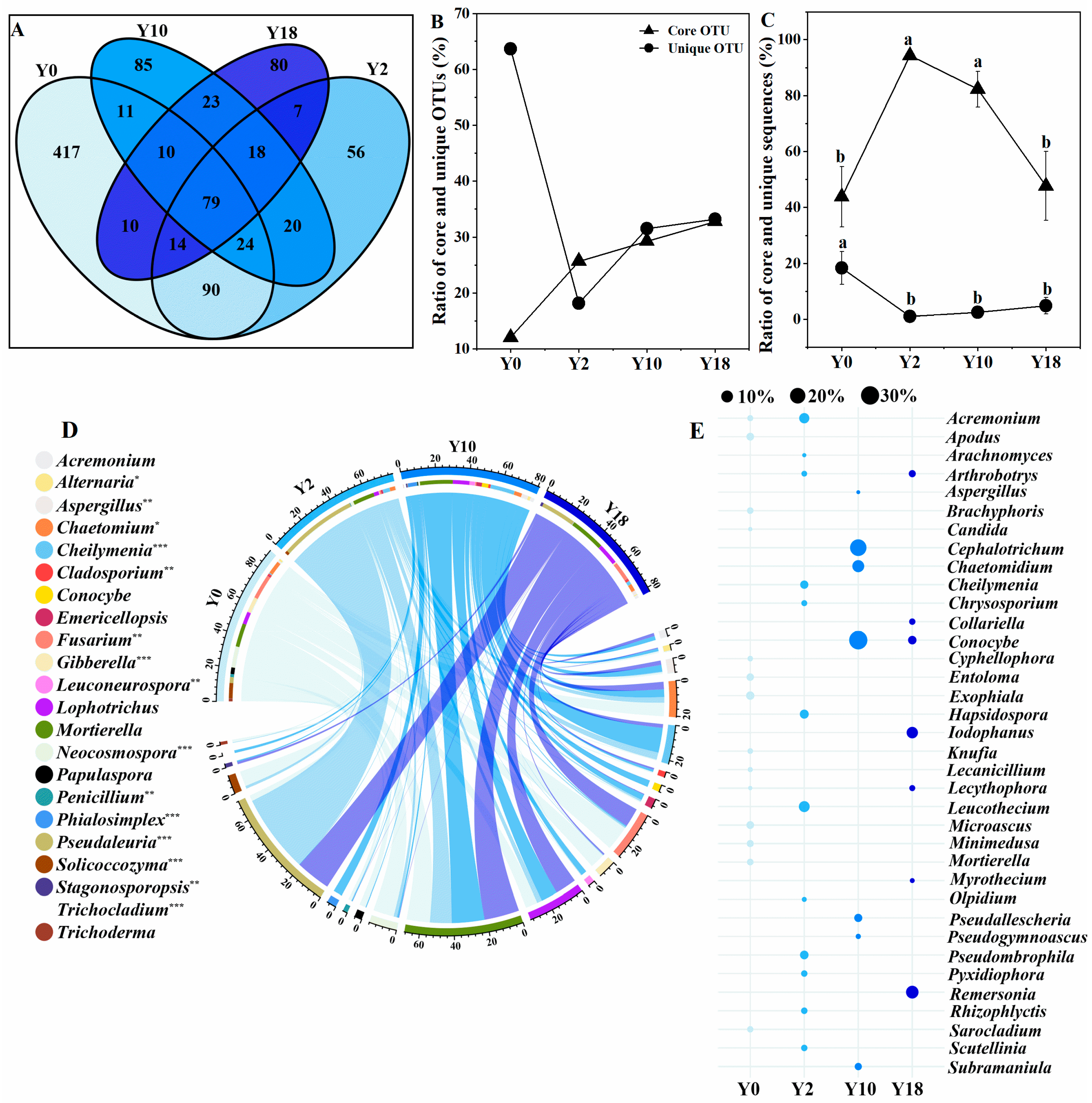
3.4. Compositions of Core and Unique Microbiomes
3.5. Soil Fungal Community Stability
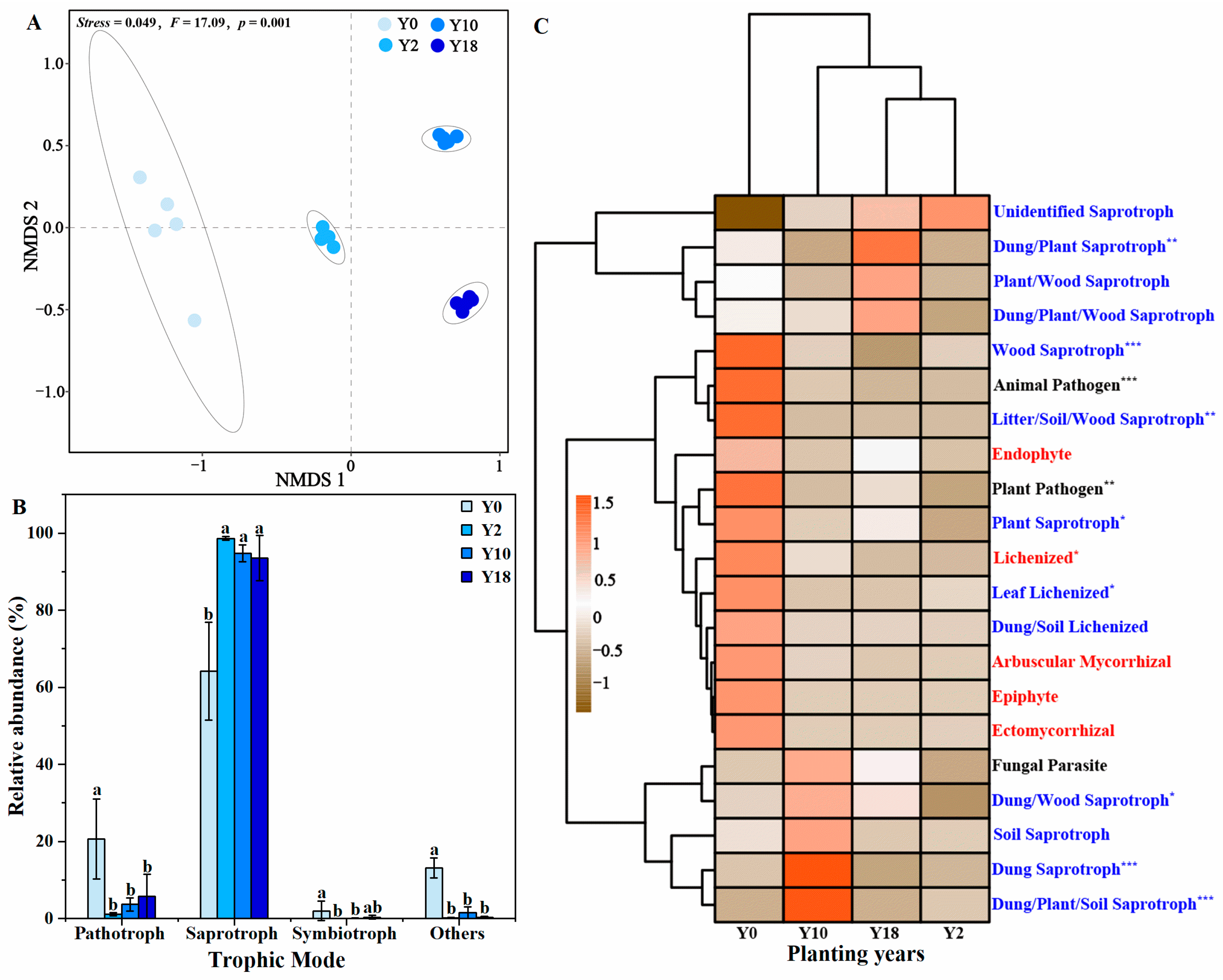
3.6. Functional Composition of Fungal Community
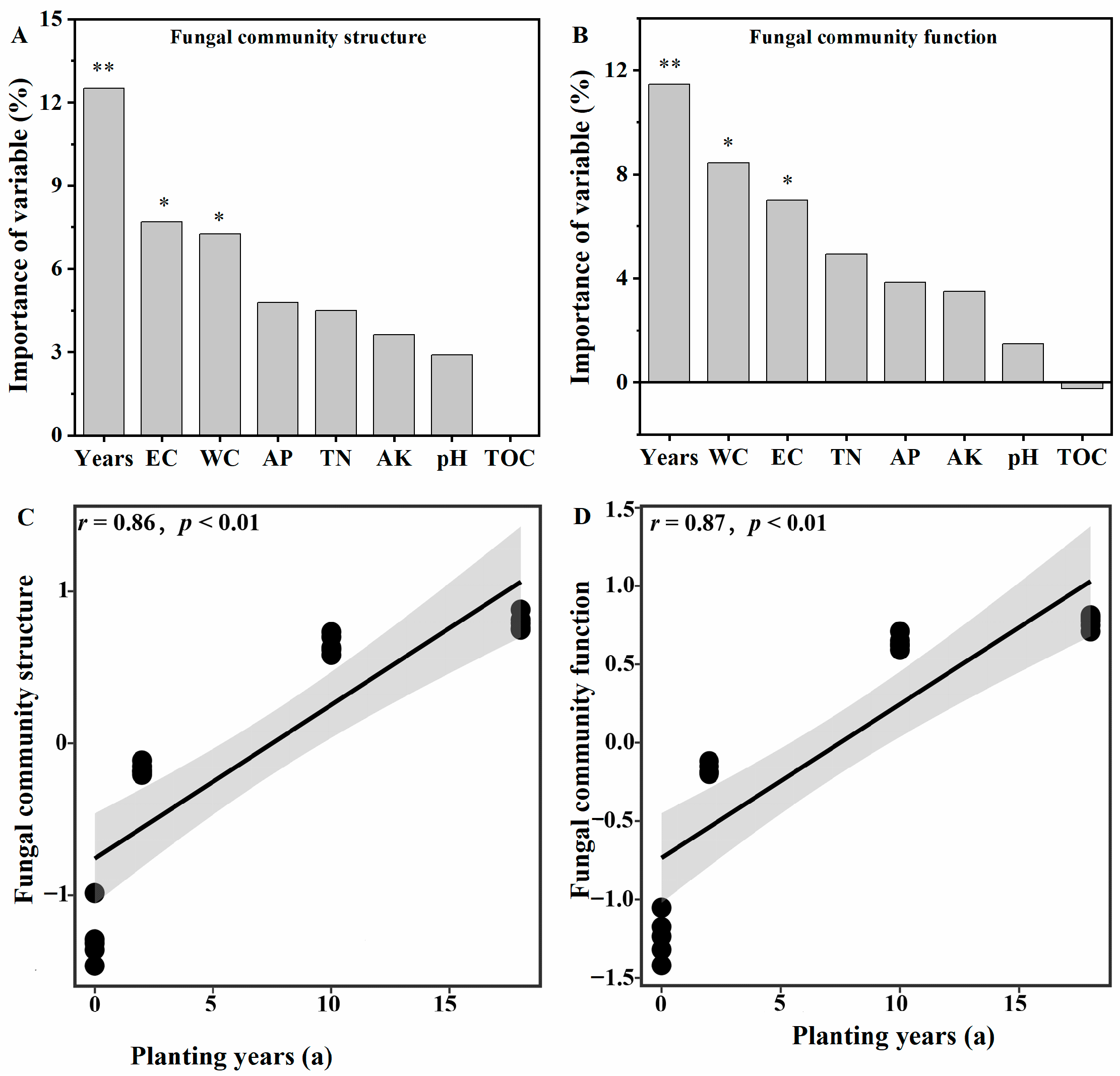
3.7. Driving Factors Affecting the Soil Fungal Community and Functional Composition
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sari, N.; Solmaz, İ. Watermelon Genetic Resources and Diversity. In The Watermelon Genome; Springer: Cham, Switzerland, 2023; pp. 23–36. [Google Scholar]
- Devi, P.; Tymon, L.; Keinath, A.; Miles, C. Progress in grafting watermelon to manage Verticillium wilt. Plant Pathol. 2021, 70, 767–777. [Google Scholar] [CrossRef]
- Ling, N.; Deng, K.Y.; Song, Y.; Wu, Y.C.; Zhao, J.; Raza, W.; Huang, Q.W.; Shen, Q.R. Variation of rhizosphere bacterial community in watermelon continuous mono-cropping soil by long-term application of a novel bioorganic fertilizer. Microbiol. Res. 2014, 169, 570–578. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, D.; Rouphael, Y.; Colla, G.; Venema, J.H. Grafting as a tool to improve tolerance of vegetables to abiotic stresses: Thermal stress, water stress and organic pollutants. Sci. Hortic. 2010, 127, 162–171. [Google Scholar] [CrossRef]
- Feng, M.; Augstein, F.; Kareem, A.; Melnyk, C.W. Plant grafting: Molecular mechanisms and applications. Mol. Plant 2024, 17, 75–91. [Google Scholar] [CrossRef]
- Davis, A.R.; Perkins-Veazie, P.; Hassell, R.; Levi, A.; King, S.R.; Zhang, X.P. Grafting effects on vegetable quality. HortScience 2008, 43, 1670–1672. [Google Scholar] [CrossRef]
- Morais, M.C.; Torres, L.F.; Kuramae, E.E.; de Andrade, S.A.L.; Mazzafera, P. Plant grafting: Maximizing beneficial microbe-plant interactions. Rhizosphere 2024, 29, 100825. [Google Scholar] [CrossRef]
- Thies, J.A.; Ariss, J.J.; Hassell, R.L.; Olson, S.; Kousik, C.S.; Levi, A. Grafting for Management of Southern Root-Knot Nematode, Meloidogyne incognita, in Watermelon. Plant Dis. 2010, 94, 1195–1199. [Google Scholar] [CrossRef]
- Qiao, Y.; Wang, Z.; Sun, H.; Guo, H.; Song, Y.; Zhang, H.; Ruan, Y.; Xu, Q.; Huang, Q.; Shen, Q.; et al. Synthetic community derived from grafted watermelon rhizosphere provides protection for ungrafted watermelon against Fusarium oxysporum via microbial synergistic effects. Microbiome 2024, 12, 101. [Google Scholar] [CrossRef]
- Ruan, Y.; Wang, T.T.; Guo, S.W.; Ling, N.; Shen, Q.R. Plant Grafting Shapes Complexity and Co-occurrence of Rhizobacterial Assemblages. Microb. Ecol. 2020, 80, 643–655. [Google Scholar] [CrossRef]
- Louws, F.J.; Rivard, C.L.; Kubota, C. Grafting fruiting vegetables to manage soilborne pathogens, foliar pathogens, arthropods and weeds. Sci. Hortic. 2010, 127, 127–146. [Google Scholar] [CrossRef]
- Verdejo-Lucas, S.; Cortada, L.; Sorribas, F.J.; Ornat, C. Selection of virulent populations of Meloidogyne javanica by repeated cultivation of Mi resistance gene tomato rootstocks under field conditions. Plant Pathol. 2009, 58, 990–998. [Google Scholar] [CrossRef]
- Hartmann, M.; Six, J. Soil structure and microbiome functions in agroecosystems. Nat. Rev. Earth Environ. 2023, 4, 4–18. [Google Scholar] [CrossRef]
- Zhou, X.; Liu, L.L.; Zhao, J.; Zhang, J.B.; Cai, Z.C.; Huang, X.Q. High carbon resource diversity enhances the certainty of successful plant pathogen and disease control. New Phytol. 2023, 237, 1333–1346. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.L.; Huang, X.Q.; Zhang, J.B.; Cai, Z.C.; Jiang, K.; Chang, Y.Y. Deciphering the relative importance of soil and plant traits on the development of rhizosphere microbial communities. Soil Biol. Biochem. 2020, 148, 107909. [Google Scholar] [CrossRef]
- Song, Y.; Ling, N.; Ma, J.H.; Wang, J.C.; Zhu, C.; Raza, W.; Shen, Y.F.; Huang, Q.W.; Shen, Q.R. Grafting Resulted in a Distinct Proteomic Profile of Watermelon Root Exudates Relative to the Un-Grafted Watermelon and the Rootstock Plant. J. Plant Growth Regul. 2016, 35, 778–791. [Google Scholar] [CrossRef]
- Zhao, J.; Zhou, X.; Jiang, A.Q.; Fan, J.Z.; Lan, T.; Zhang, J.B.; Cai, Z.C. Distinct impacts of reductive soil disinfestation and chemical soil disinfestation on soil fungal communities and memberships. Appl. Microbiol. Biotechnol. 2018, 102, 7623–7634. [Google Scholar] [CrossRef]
- Liu, L.L.; Yan, Y.Y.; Ding, H.X.; Zhao, J.; Cai, Z.C.; Dai, C.C.; Huang, X.Q. The fungal community outperforms the bacterial community in predicting plant health status. Appl. Microbiol. Biotechnol. 2021, 105, 7051. [Google Scholar]
- Liu, L.L.; Yan, Y.Y.; Ali, A.; Zhao, J.; Cai, Z.C.; Dai, C.C.; Huang, X.Q.; Zhou, K.S. Deciphering the Fusarium-wilt control effect and succession driver of microbial communities managed under low-temperature conditions. Appl. Soil Ecol. 2022, 171, 104334. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S.; Michael, W. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Friednly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. R Package 2022, Version 2.5-7. 28 February 2022. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 25 May 2024).
- Steinhauser, D.; Krall, L.; Mussig, C.; Bussis, D.; Usadel, B. Correlation networks. In Analysis of Biological Networks, 2nd ed.; Junker, B.H., Schreiber, F., Eds.; John Wiley & Sons Inc.: Hoboken, NJ, USA, 2007; pp. 305–333. [Google Scholar]
- Bastian, M.; Heymann, S.; Jacomy, M. Gephi: An open source software for exploring and manipulating networks. In Proceedings of the Third International AAAI Conference on Weblogs and Social Media, San Jose, CA, USA, 17–20 May 2009; Volume 8, pp. 361–362. [Google Scholar]
- Yuan, M.M.; Guo, X.; Wu, L.W.; Zhang, Y.; Xiao, N.J.; Ning, D.L.; Shi, Z.; Zhou, X.S.; Wu, L.Y.; Yang, Y.F.; et al. Climate warming enhances microbial network complexity and stability. Nat. Clim. Change 2021, 11, 343–348. [Google Scholar] [CrossRef]
- Jiao, S.; Chen, W.M.; Wei, G.H. Core microbiota drive functional stability of soil microbiome in reforestation ecosystems. Glob. Change Biol. 2022, 28, 1038–1047. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.H.; Song, Z.W.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Yang, T.; Tedersoo, L.; Liu, X.; Gao, G.F.; Dong, K.; Adams, J.M.; Chu, H.Y. Fungi stabilize multi-kingdom community in a high elevation timberline ecosystem. Imeta 2022, 1, e49. [Google Scholar] [CrossRef]
- Bridge, P.; Spooner, B. Soil fungi: Diversity and detection. Plant Soil 2001, 232, 147–154. [Google Scholar] [CrossRef]
- Li, X.G.; Ding, C.F.; Zhang, T.L.; Wang, X.X. Fungal pathogen accumulation at the expense of plant-beneficial fungi as a consequence of consecutive peanut monoculturing. Soil Biol. Biochem. 2014, 72, 11–18. [Google Scholar] [CrossRef]
- Liu, L.; Long, S.; Deng, B.; Kuang, J.; Wen, K.; Li, T.; Bai, Z.; Shao, Q. Effects of Plastic Shed Cultivation System on the Properties of Red Paddy Soil and Its Management by Reductive Soil Disinfestation. Horticulturae 2022, 8, 279. [Google Scholar] [CrossRef]
- Sun, K.; Fu, L.Y.; Song, Y.; Yuan, L.; Zhang, H.R.; Wen, D.; Yang, N.; Wang, X.; Yue, Y.Q.; Li, X.H.; et al. Effects of continuous cucumber cropping on crop quality and soil fungal community. Environ. Monit. Assess. 2021, 193, 436. [Google Scholar] [CrossRef]
- Zhang, C.F.; Shu, D.T.; Wei, G.H. Soybean cropping patterns affect trait-based microbial strategies by changing soil properties. Appl. Soil Ecol. 2021, 167, 104095. [Google Scholar] [CrossRef]
- Chen, L.; Bian, L.S.; Ma, Q.H.; Li, Y.; Wang, X.H.; Liu, Y.P. Defensive alteration of root exudate composition by grafting Prunus sp. onto resistant rootstock contributes to reducing crown gall disease. Hortic. Res. 2024, 11, uhae049. [Google Scholar] [CrossRef]
- Li, Y.; Shi, C.Q.; Wei, D.; Gu, X.J.; Wang, Y.F.; Sun, L.; Cai, S.S.; Hu, Y.; Jin, L.; Wang, W. Soybean continuous cropping affects yield by changing soil chemical properties and microbial community richness. Front. Microbiol. 2022, 13, 1083736. [Google Scholar] [CrossRef] [PubMed]
- Philippot, L.; Chenu, C.; Kappler, A.; Rillig, M.C.; Fierer, N. The interplay between microbial communities and soil properties. Nat. Rev. Microbiol. 2024, 22, 226–239. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.B.; Fu, H.D.; Shi, Q.W.; Shan, X.; Wang, Z.; Sun, Z.P.; Li, T.L. Overfertilization reduces tomato yield under long-term continuous cropping system via regulation of soil microbial community composition. Front. Microbiol. 2022, 13, 952021. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, Q.; Chen, Y.L.; Dai, Q.G.; Hu, J. Mudflat reclamation causes change in the composition of fungal communities under long-term rice cultivation. Can. J. Microbiol. 2019, 65, 530–537. [Google Scholar] [CrossRef] [PubMed]
- Gu, X.; Yang, N.; Zhao, Y.; Liu, W.H.; Li, T.F. Long-term watermelon continuous cropping leads to drastic shifts in soil bacterial and fungal community composition across gravel mulch fields. BMC Microbiol. 2022, 22, 189. [Google Scholar] [CrossRef]
- Talley, S.M.; Coley, P.D.; Kursar, T.A. The effects of weather on fungal abundance and richness among 25 communities in the Intermountain West. BMC Ecol. 2002, 2, 7. [Google Scholar] [CrossRef]
- Li, P.F.; Liu, J.; Saleem, M.; Li, G.L.; Luan, L.; Wu, M.; Li, Z.P. Reduced chemodiversity suppresses rhizosphere microbiome functioning in the mono-cropped agroecosystems. Microbiome 2022, 10, 108. [Google Scholar] [CrossRef]
- Banerjee, S.; Zhao, C.; Garland, G.; Edlinger, A.; García-Palacios, P.; Romdhane, S.; Degrune, F.; Pescador, D.S.; Herzog, C.; Camuy-Velez, L.A.; et al. Biotic homogenization, lower soil fungal diversity and fewer rare taxa in arable soils across Europe. Nat. Commun. 2024, 15, 327. [Google Scholar] [CrossRef]
- Peng, Z.H.; Qian, X.; Liu, Y.; Li, X.M.; Gao, H.; An, Y.N.; Qi, J.J.; Jiang, L.; Zhang, Y.R.; Chen, S.; et al. Land conversion to agriculture induces taxonomic homogenization of soil microbial communities globally. Nat. Commun. 2024, 15, 3624. [Google Scholar] [CrossRef]
- Pang, Q.L.; Alami, M.M.; Yu, W.L.; Ouyang, Z.; Shu, S.H.; Tu, D.Q.; Alami, M.J.; Wang, X.K. A Meta-Analysis in Nine Different Continuous Cropping Fields to Find the Relationship between Plant Species and Rhizosphere Fungal Community. Agronomy 2023, 13, 1827. [Google Scholar] [CrossRef]
- Xu, L.H.; Ravnskov, S.; Larsen, J.; Nilsson, R.H.; Nicolaisen, M. Soil fungal community structure along a soil health gradient in pea fields examined using deep amplicon sequencing. Soil Biol. Biochem. 2012, 46, 26–32. [Google Scholar] [CrossRef]
- Heuchert, B.; Braun, U.; Schubert, K. Morphotaxonomic revision of fungicolous Cladosporium species (hyphomycetes). Schlechtendalia 2005, 13, 1–78. [Google Scholar]
- Derbyshire, M.C.; Raffaele, S. Surface frustration re-patterning underlies the structural landscape and evolvability of fungal orphan candidate effectors. Nat. Commun. 2023, 14, 5244. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Yao, G.; Zhao, H.; Li, X.; Li, G.; Gao, X.; Zhang, Y.; Zhou, H.; Wang, H. First report of Stagonosporopsis pogostemonis causing root rot on Strawberry in China. Plant Dis. 2024, 108, 2565. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Walder, F.; Büchi, L.; Meyer, M.; Held, A.Y.; Gattinger, A.; Keller, T.; Charles, R.; van der Heijden, M.G.A. Agricultural intensification reduces microbial network complexity and the abundance of keystone taxa in roots. ISME J. 2019, 13, 1722–1736. [Google Scholar] [CrossRef]
- Zhang, C.; Lei, S.L.; Wu, H.Y.; Liao, L.R.; Wang, X.T.; Zhang, L.; Liu, G.B.; Wang, G.L.; Fang, L.C.; Song, Z.L. Simplified microbial network reduced microbial structure stability and soil functionality in alpine grassland along a natural aridity gradient. Soil Biol. Biochem. 2024, 191, 109366. [Google Scholar] [CrossRef]
- Gao, M.; Xiong, C.; Gao, C.; Tsui, C.K.M.; Wang, M.M.; Zhou, X.; Zhang, A.M.; Cai, L. Disease-induced changes in plant microbiome assembly and functional adaptation. Microbiome 2021, 9, 187. [Google Scholar] [CrossRef]
- Fernández-González, A.J.; Cardoni, M.; Cabanás, C.G.L.; Valverde-Corredor, A.; Villadas, P.J.; Fernandez-López, M.; Mercado-Blanco, J. Linking belowground microbial network changes to different tolerance level towards Verticillium wilt of olive. Microbiome 2020, 8, 11. [Google Scholar] [CrossRef]
- Mendes, L.W.; Raaijmakers, J.M.; de Hollander, M.; Sepo, E.; Expósito, R.; Chiorato, A.F.; Mendes, R.; Tsai, S.M.; Carrión, V.J. Impact of the fungal pathogen Fusarium oxysporum on the taxonomic and functional diversity of the common bean root microbiome. Environ. Microbiome 2023, 18, 68. [Google Scholar] [CrossRef]
- Yan, Y.; Wu, R.; Li, S.; Su, Z.; Shao, Q.; Cai, Z.; Huang, X.; Liu, L. Reductive Soil Disinfestation Enhances Microbial Network Complexity and Function in Intensively Cropped Greenhouse Soil. Horticulturae 2022, 8, 476. [Google Scholar] [CrossRef]

| Planting Years | Physicochemical Properties | ||||||
|---|---|---|---|---|---|---|---|
| pH | EC (μS·cm−1) | TOC (g·kg−1) | TN (g·kg−1) | AP (mg·kg−1) | AK (mg·kg−1) | WC (%) | |
| Y0 | 6.42 ± 0.31 b | 88.8 ± 54.9 c | 11.01 ± 2.00 ab | 0.82 ± 0.21 c | 106.7 ± 121.6 b | 208.5 ± 21.9 d | 5.06 ± 1.79 b |
| Y2 | 5.86 ± 0.16 c | 314.7 ± 54.9 a | 9.46 ± 0.23 b | 0.99 ± 0.07 c | 137.8 ± 11.6 b | 367.8 ± 31.3 a | 5.76 ± 1.58 b |
| Y10 | 6.53 ± 0.33 ab | 182.1 ± 26.9 b | 12.75 ± 3.59 a | 1.22± 0.11 b | 197.6 ± 63.7 ab | 260.9 ± 44.2 c | 11.35 ± 1.57 a |
| Y18 | 6.86 ± 0.07 a | 211.9 ± 12.9 b | 14.04 ± 1.66 a | 1.68 ± 0.09 a | 259.0 ± 38.6 a | 305.5 ± 28.6 b | 10.41 ± 2.70 a |
| Planting Years | Population of Fungi (lg ITS Copies·g−1) | Richness | Shannon Index | Evenness |
|---|---|---|---|---|
| Y0 | 8.31 ± 0.27 b | 848 ± 160 a | 4.28 ± 0.52 a | 0.63 ± 0.06 a |
| Y2 | 8.75 ± 0.06 a | 366 ± 35 b | 2.30 ± 0.18 c | 0.39 ± 0.02 d |
| Y10 | 8.53 ± 0.07 a | 339 ± 26 b | 3.32 ± 0.10 b | 0.57 ± 0.02 b |
| Y18 | 8.55 ± 0.17 a | 291 ± 27 b | 2.67 ± 0.21 c | 0.47 ± 0.03 c |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, X.; Guo, B.; Zhang, R.; Zhou, L.; Huang, X.; Liu, L. Investigating Changes in the Soil Fungal Community Structure, Functions, and Network Stability with Prolonged Grafted Watermelon Cultivation. Horticulturae 2024, 10, 971. https://doi.org/10.3390/horticulturae10090971
Zhou X, Guo B, Zhang R, Zhou L, Huang X, Liu L. Investigating Changes in the Soil Fungal Community Structure, Functions, and Network Stability with Prolonged Grafted Watermelon Cultivation. Horticulturae. 2024; 10(9):971. https://doi.org/10.3390/horticulturae10090971
Chicago/Turabian StyleZhou, Xing, Bingyu Guo, Ruyi Zhang, Linfei Zhou, Xinqi Huang, and Liangliang Liu. 2024. "Investigating Changes in the Soil Fungal Community Structure, Functions, and Network Stability with Prolonged Grafted Watermelon Cultivation" Horticulturae 10, no. 9: 971. https://doi.org/10.3390/horticulturae10090971
APA StyleZhou, X., Guo, B., Zhang, R., Zhou, L., Huang, X., & Liu, L. (2024). Investigating Changes in the Soil Fungal Community Structure, Functions, and Network Stability with Prolonged Grafted Watermelon Cultivation. Horticulturae, 10(9), 971. https://doi.org/10.3390/horticulturae10090971







