Genomic Organization and Expression Profiling of GOLDEN2-like Transcription Factor Genes in Eggplant and Their Role in Heat Stresses
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Stress Treatments
2.2. Measurement of Relative Electrolyte Leakage and Relative Water Content
2.3. Phenotypic Identification of Heat Tolerance in Eggplant at Seedling Stage
2.4. Identification of GLK Genes in Eggplant Genome
2.5. Phylogenetic Tree Analysis
2.6. Gene Structure Analysis, Motif Analysis, and Cis Regulatory Element (CREs) Analysis of GLK Genes in Eggplant
2.7. Chromosomal Location and Gene Duplication Analysis of SmGLK Genes
2.8. RNA Isolation and Real-Time Quantitative PCR (RT-qPCR) Analysis
3. Results
3.1. Identification of GLK Family Members in Eggplant
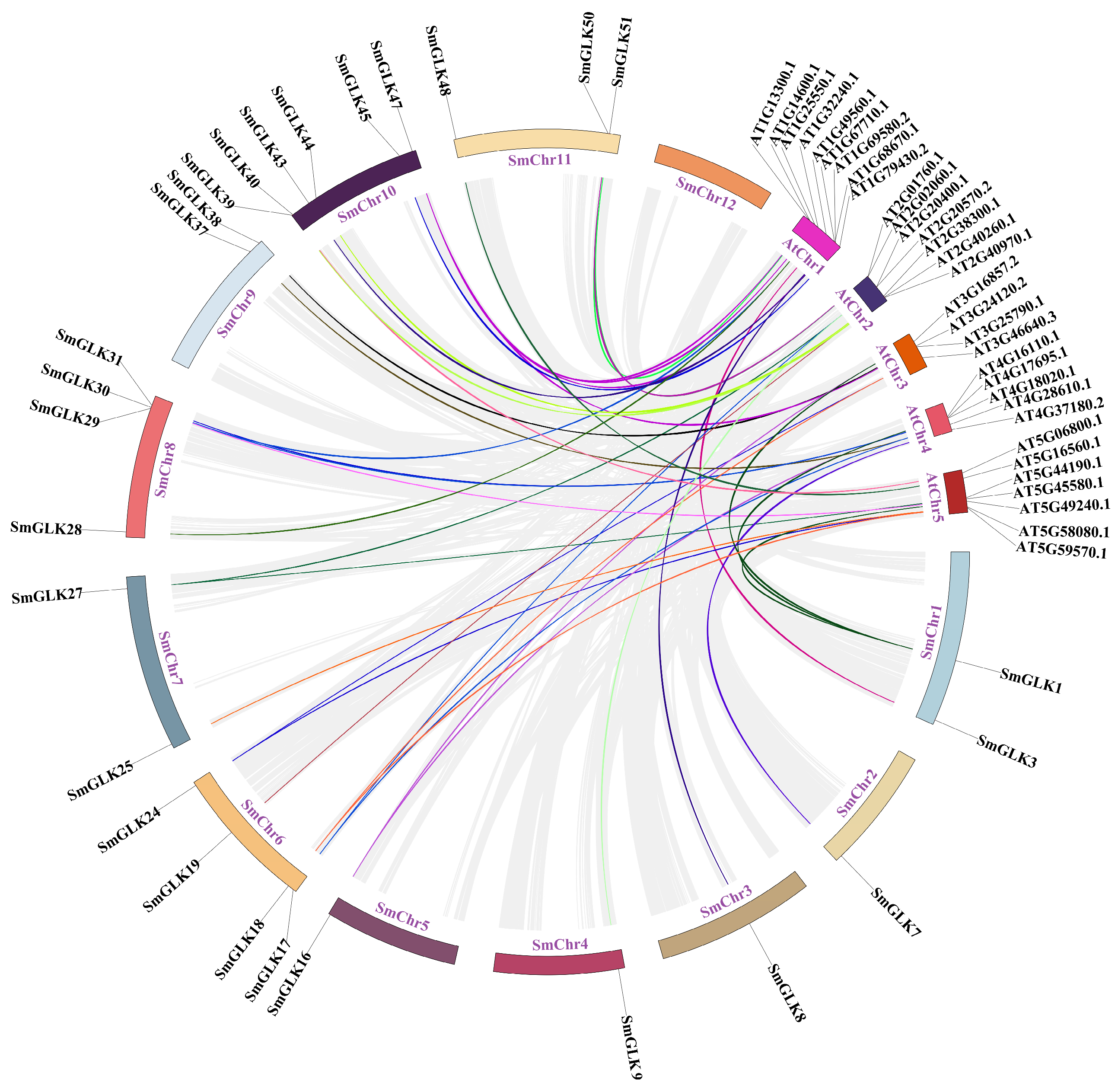
3.2. Evolutionary and Structural Analysis
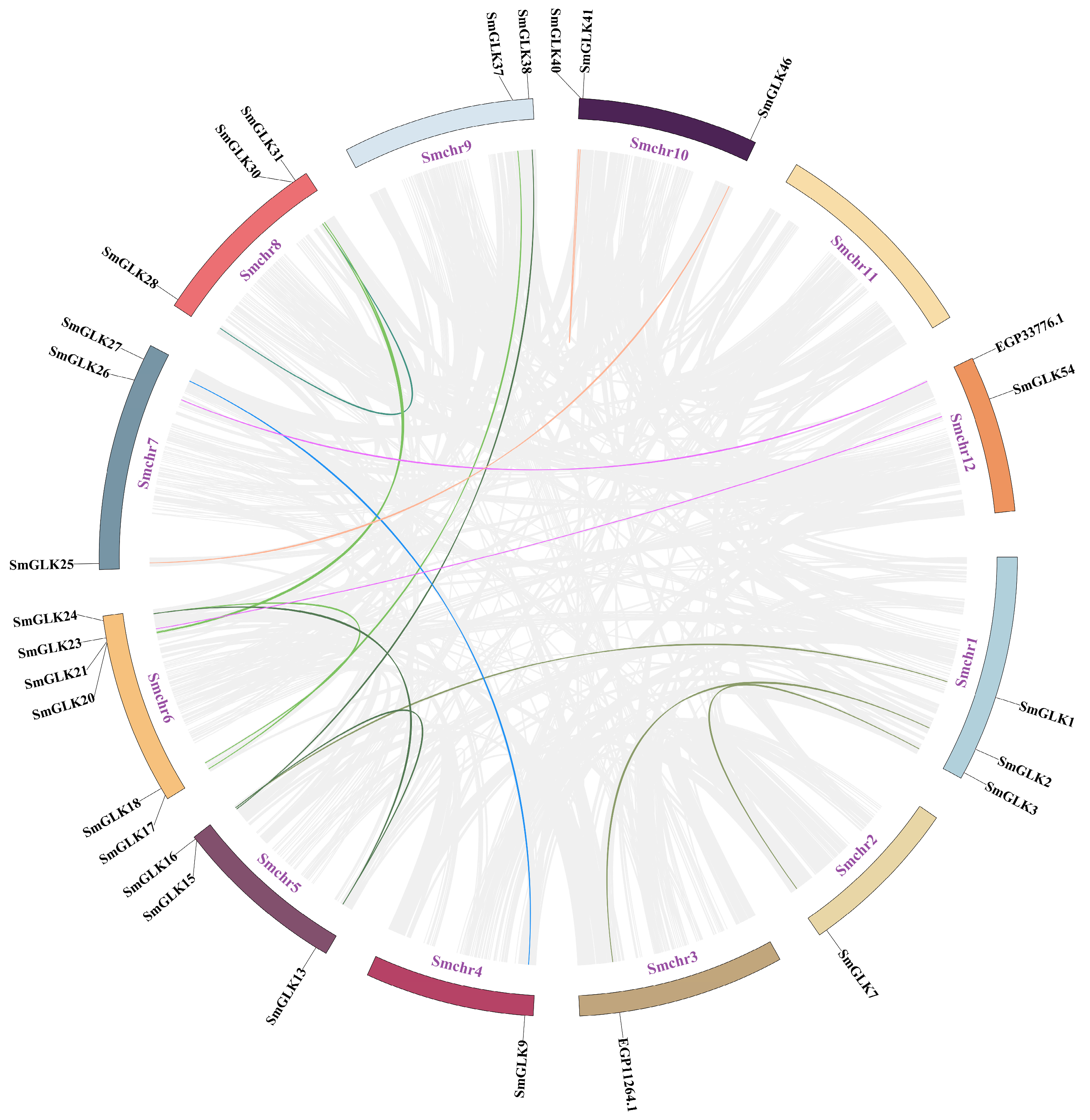
3.3. Chromosome Distribution and Duplication Events of SmGLK Genes
3.4. Promoter Analysis of SmGLK Genes
3.5. Expression Pattern of SmGLK Genes across Different Developmental Stages in Different Eggplant Tissues
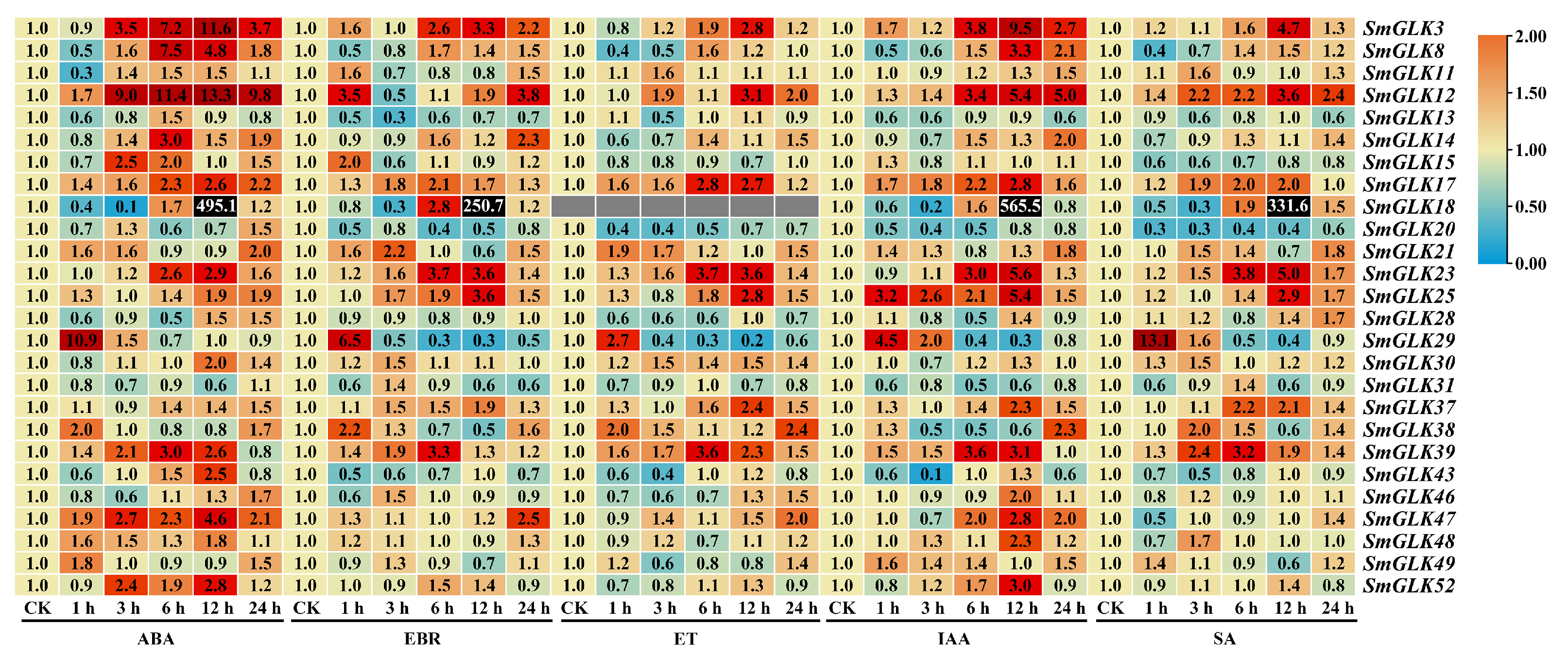
3.6. Expression Patterns of SmGLK Genes in Response to Hormone Treatments
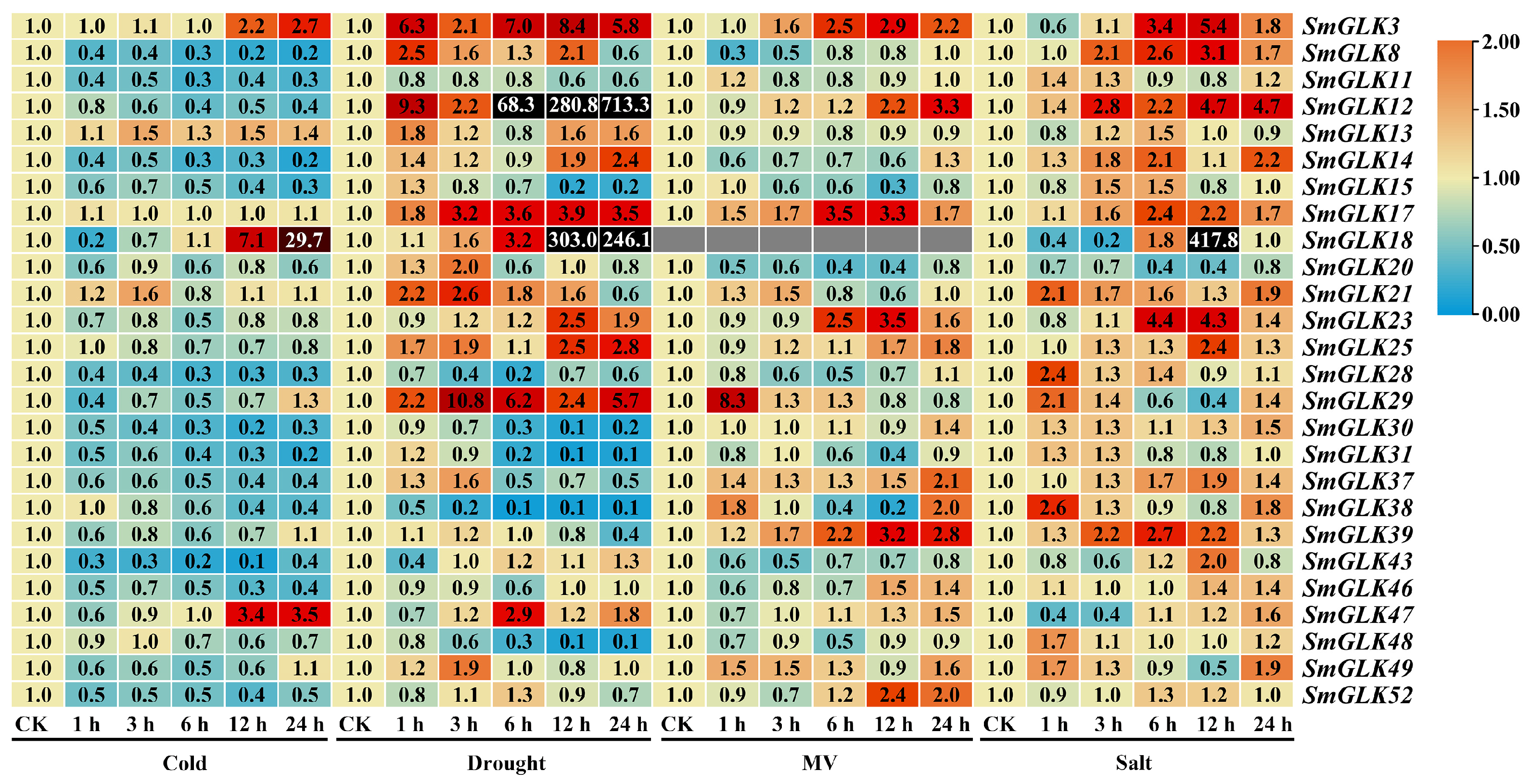
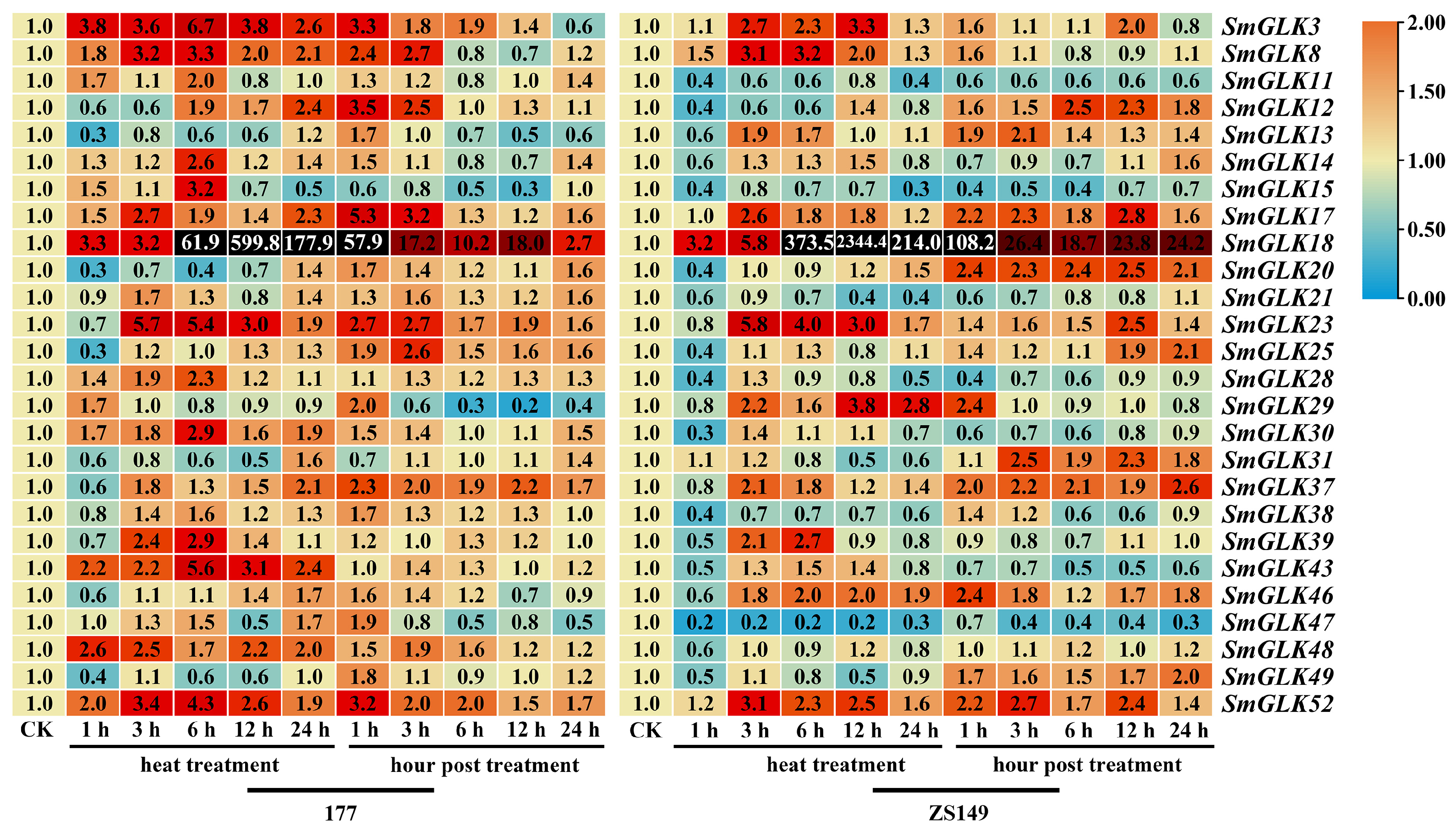
3.7. Expression Patterns of SmGLK Genes in Response to Abiotic Stress Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hernández-Verdeja, T.; Lundgren, M.R. GOLDEN2-LIKE transcription factors: A golden ticket to improve crops? Plants People Planet 2023, 6, 79–93. [Google Scholar] [CrossRef]
- Xiao, Y.; You, S.; Kong, W.; Tang, Q.; Bai, W.; Cai, Y.; Zheng, H.; Wang, C.; Jiang, L.; Wang, C. A GARP transcription factor anther dehiscence defected 1 (OsADD1) regulates rice anther dehiscence. Plant Mol. Biol. 2019, 101, 403–414. [Google Scholar] [CrossRef]
- Jenkins, M.T. A second gene producing golden plant color in maize. Am. Nat. 1926, 60, 484–488. [Google Scholar] [CrossRef]
- Hall, L.N.; Rossini, L.; Cribb, L.; Langdale, J.A. GOLDEN 2: A novel transcriptional regulator of cellular differentiation in the maize leaf. Plant Cell 1998, 10, 925–936. [Google Scholar] [CrossRef]
- Alam, I.; Wu, X.; Yu, Q.; Ge, L. Comprehensive genomic analysis of G2-like transcription factor genes and their role in development and abiotic stresses in Arabidopsis. Diversity 2022, 14, 228. [Google Scholar] [CrossRef]
- Liu, F.; Xu, Y.; Han, G.; Zhou, L.; Ali, A.; Zhu, S.; Li, X. Molecular evolution and genetic variation of G2-like transcription factor genes in maize. PLoS ONE 2016, 11, e0161763. [Google Scholar] [CrossRef] [PubMed]
- Wu, R.; Guo, L.; Wang, R.; Zhang, Q.; Yao, H. Genome-Wide Identification and Characterization of G2-Like Transcription Factor Genes in Moso Bamboo (Phyllostachys edulis). Molecules 2022, 27, 5491. [Google Scholar] [CrossRef] [PubMed]
- Alam, I.; Manghwar, H.; Zhang, H.; Yu, Q.; Ge, L. Identification of GOLDEN2-like transcription factor genes in soybeans and their role in regulating plant development and metal ion stresses. Front. Plant Sci. 2022, 13, 1052659. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Y.; Zhao, S.; Liu, J.F.; Zhao, H.Y.; Sun, X.Y.; Wu, T.R.; Pei, T.; Wang, Y.; Liu, Q.F.; Yang, H.H.; et al. Genome-wide identification of Tomato Golden 2-Like transcription factors and abiotic stress related members screening. BMC Plant Biol. 2022, 22, 82. [Google Scholar] [CrossRef]
- Hu, G.; Li, J.; Wang, X.; Kang, Y.; Li, Y.; Niu, J.; Yin, J. Molecular Evolution and Genetic Variation of G2-Like Transcription Factor Genes in Wheat (Triticum aestivum L.). Genes 2023, 14, 1341. [Google Scholar] [CrossRef]
- Rossini, L.; Cribb, L.; Martin, D.J.; Langdale, J.A. The maize golden2 gene defines a novel class of transcriptional regulators in plants. Plant Cell 2001, 13, 1231–1244. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Shuang, J.; Li, Z.; Xiao, H.; Liu, Y.; Wang, T.; Wei, Y.; Hu, S.; Wan, S.; Peng, R. Identification of the Golden-2-like transcription factors gene family in Gossypium hirsutum. PeerJ 2021, 9, e12484. [Google Scholar] [CrossRef]
- Cackett, L.; Luginbuehl, L.H.; Schreier, T.B.; Lopez-Juez, E.; Hibberd, J.M. Chloroplast development in green plant tissues: The interplay between light, hormone, and transcriptional regulation. New Phytol. 2022, 233, 2000–2016. [Google Scholar] [CrossRef] [PubMed]
- Langdale, J.A.; Kidner, C.A. Bundle sheath defective, a mutation that disrupts cellular differentiation in maize leaves. Development 1994, 120, 673–681. [Google Scholar] [CrossRef]
- Fitter, D.W.; Martin, D.J.; Copley, M.J.; Scotland, R.W.; Langdale, J.A. GLK gene pairs regulate chloroplast development in diverse plant species. Plant J. 2002, 31, 713–727. [Google Scholar] [CrossRef] [PubMed]
- Yasumura, Y.; Moylan, E.C.; Langdale, J.A. A conserved transcription factor mediates nuclear control of organelle biogenesis in anciently diverged land plants. Plant Cell 2005, 17, 1894–1907. [Google Scholar] [CrossRef]
- Chen, M.; Ji, M.; Wen, B.; Liu, L.; Li, S.; Chen, X.; Gao, D.; Li, L. GOLDEN 2-LIKE transcription factors of plants. Front. Plant Sci. 2016, 7, 1509. [Google Scholar] [CrossRef]
- Chang, Y.-M.; Liu, W.-Y.; Shih, A.C.-C.; Shen, M.-N.; Lu, C.-H.; Lu, M.-Y.J.; Yang, H.-W.; Wang, T.-Y.; Chen, S.C.-C.; Chen, S.M. Characterizing regulatory and functional differentiation between maize mesophyll and bundle sheath cells by transcriptomic analysis. Plant Physiol. 2012, 160, 165–177. [Google Scholar] [CrossRef]
- Powell, A.L.; Nguyen, C.V.; Hill, T.; Cheng, K.L.; Figueroa-Balderas, R.; Aktas, H.; Ashrafi, H.; Pons, C.; Fernández-Muñoz, R.; Vicente, A. Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast Development. Science 2012, 336, 1711–1715. [Google Scholar] [CrossRef]
- Cheng, K.L.L. Golden2–like (GLK2) Transcription Factor: Developmental Control of Tomato Fruit Photosynthesis and Its Contribution to Ripe Fruit Characteristics; University of California: Davis, CA, USA, 2013. [Google Scholar]
- Nguyen, C.V.; Vrebalov, J.T.; Gapper, N.E.; Zheng, Y.; Zhong, S.; Fei, Z.; Giovannoni, J.J. Tomato GOLDEN2-LIKE transcription factors reveal molecular gradients that function during fruit development and ripening. Plant Cell 2014, 26, 585–601. [Google Scholar] [CrossRef]
- Brand, A.; Borovsky, Y.; Hill, T.; Rahman, K.A.A.; Bellalou, A.; Van Deynze, A.; Paran, I. CaGLK2 regulates natural variation of chlorophyll content and fruit color in pepper fruit. Theor. Appl. Genet. 2014, 127, 2139–2148. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zhao, D.; Li, X.; Zeng, Y. AtGLK2, an Arabidopsis GOLDEN2-LIKE transcription factor, positively regulates anthocyanin biosynthesis via AtHY5-mediated light signaling. Plant Growth Regul. 2022, 96, 79–90. [Google Scholar] [CrossRef]
- Zhu, X.; Zhang, L.; Kuang, C.; Guo, Y.; Huang, C.; Deng, L.; Sun, X.; Zhan, G.; Hu, Z.; Wang, H. Important photosynthetic contribution of silique wall to seed yield-related traits in Arabidopsis thaliana. Photosynth. Res. 2018, 137, 493–501. [Google Scholar] [CrossRef]
- Zeng, L.; Liu, X.; Zhou, Z.; Li, D.; Zhao, X.; Zhu, L.; Luo, Y.; Hu, S. Identification of a G2-like transcription factor, OsPHL3, functions as a negative regulator of flowering in rice by co-expression and reverse genetic analysis. BMC Plant Biol. 2018, 18, 157. [Google Scholar] [CrossRef] [PubMed]
- Rauf, M.; Arif, M.; Dortay, H.; Matallana-Ramírez, L.P.; Waters, M.T.; Gil Nam, H.; Lim, P.O.; Mueller-Roeber, B.; Balazadeh, S. ORE1 balances leaf senescence against maintenance by antagonizing G2-like-mediated transcription. EMBO Rep. 2013, 14, 382–388. [Google Scholar] [CrossRef] [PubMed]
- Savitch, L.V.; Subramaniam, R.; Allard, G.C.; Singh, J. The GLK1 ‘regulon’encodes disease defense related proteins and confers resistance to Fusarium graminearum in Arabidopsis. Biochem. Biophys. Res. Commun. 2007, 359, 234–238. [Google Scholar] [CrossRef]
- Schreiber, K.J.; Nasmith, C.G.; Allard, G.; Singh, J.; Subramaniam, R.; Desveaux, D. Found in translation: High-throughput chemical screening in Arabidopsis thaliana identifies small molecules that reduce Fusarium head blight disease in wheat. Mol. Plant-Microbe Interact. 2011, 24, 640–648. [Google Scholar] [CrossRef]
- Murmu, J.; Wilton, M.; Allard, G.; Pandeya, R.; Desveaux, D.; Singh, J.; Subramaniam, R. Arabidopsis GOLDEN2-LIKE (GLK) transcription factors activate jasmonic acid (JA)-dependent disease susceptibility to the biotrophic pathogen H yaloperonospora arabidopsidis, as well as JA-independent plant immunity against the necrotrophic pathogen B otrytis cinerea. Mol. Plant Pathol. 2014, 15, 174–184. [Google Scholar]
- Han, X.-Y.; Li, P.-X.; Zou, L.-J.; Tan, W.-r.; Zheng, T.; Zhang, D.-W.; Lin, H.-H. GOLDEN2-LIKE transcription factors coordinate the tolerance to Cucumber mosaic virus in Arabidopsis. Biochem. Biophys. Res. Commun. 2016, 477, 626–632. [Google Scholar] [CrossRef]
- Yuan, Y.; Xu, X.; Gong, Z.; Tang, Y.; Wu, M.; Yan, F.; Zhang, X.; Zhang, Q.; Yang, F.; Hu, X. Auxin response factor 6A regulates photosynthesis, sugar accumulation, and fruit development in tomato. Hortic. Res. 2019, 6, 85. [Google Scholar] [CrossRef]
- Nagatoshi, Y.; Mitsuda, N.; Hayashi, M.; Inoue, S.-i.; Okuma, E.; Kubo, A.; Murata, Y.; Seo, M.; Saji, H.; Kinoshita, T. GOLDEN 2-LIKE transcription factors for chloroplast development affect ozone tolerance through the regulation of stomatal movement. Proc. Natl. Acad. Sci. USA 2016, 113, 4218–4223. [Google Scholar] [CrossRef] [PubMed]
- Lv, L.L.; Feng, X.F.; Li, W.; Li, K. High temperature reduces peel color in eggplant (Solanum melongena) as revealed by RNA-seq analysis. Genome 2019, 62, 503–512. [Google Scholar] [CrossRef] [PubMed]
- Hawkins, E.; Fricker, T.E.; Challinor, A.J.; Ferro, C.A.; Ho, C.K.; Osborne, T.M. Increasing influence of heat stress on French maize yields from the 1960s to the 2030s. Glob. Change Biol. 2013, 19, 937–947. [Google Scholar] [CrossRef] [PubMed]
- Boehlein, S.K.; Liu, P.; Webster, A.; Ribeiro, C.; Suzuki, M.; Wu, S.; Guan, J.C.; Stewart, J.D.; Tracy, W.F.; Settles, A.M. Effects of long-term exposure to elevated temperature on Zea mays endosperm development during grain fill. Plant J. 2019, 99, 23–40. [Google Scholar] [CrossRef]
- Yu, C.; Song, L.; Song, J.; Ouyang, B.; Guo, L.; Shang, L.; Wang, T.; Li, H.; Zhang, J.; Ye, Z. ShCIGT, a Trihelix family gene, mediates cold and drought tolerance by interacting with SnRK1 in tomato. Plant Sci. 2018, 270, 140–149. [Google Scholar] [CrossRef]
- Yamasaki, S.; Dillenburg, L.R. Measurements of leaf relative water content in Araucaria angustifolia. Rev. Bras. Fisiol. Veg. 1999, 11, 69–75. [Google Scholar]
- Liu, R.; Shu, B.; Wang, Y.; Yu, B.; Wang, Y.; Gan, Y.; Liang, Y.; Qiu, Z.; Yang, J.; Yan, S. Transcriptome analysis reveals key genes involved in the eggplant response to high-temperature stress. Environ. Exp. Bot. 2023, 211, 105369. [Google Scholar] [CrossRef]
- Li, D.; Qian, J.; Li, W.; Yu, N.; Gan, G.; Jiang, Y.; Li, W.; Liang, X.; Chen, R.; Mo, Y. A high-quality genome assembly of the eggplant provides insights into the molecular basis of disease resistance and chlorogenic acid synthesis. Mol. Ecol. Resour. 2021, 21, 1274–1286. [Google Scholar] [CrossRef]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y. TBtools-II: A “One for All, All for One” bioinformatics platform for biological big-data mining. Mol. Plant 2023, 16, 1733–1742. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547. [Google Scholar] [CrossRef]
- Xu, D.; Lu, Z.; Jin, K.; Qiu, W.; Qiao, G.; Han, X.; Zhuo, R. SPDE: A multi-functional software for sequence processing and data extraction. Bioinformatics 2021, 37, 3686–3687. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Holub, E.B. The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2001, 2, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Safi, A.; Medici, A.; Szponarski, W.; Ruffel, S.; Lacombe, B.; Krouk, G. The world according to GARP transcription factors. Curr. Opin. Plant Biol. 2017, 39, 159–167. [Google Scholar] [CrossRef]
- Qin, M.; Zhang, B.; Gu, G.; Yuan, J.; Yang, X.; Yang, J.; Xie, X. Genome-wide analysis of the G2-like transcription factor genes and their expression in different senescence stages of tobacco (Nicotiana tabacum L.). Front. Genet. 2021, 12, 626352. [Google Scholar] [CrossRef]
- Chothia, C.; Gough, J.; Vogel, C.; Teichmann, S.A. Evolution of the protein repertoire. Science 2003, 300, 1701–1703. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Birchler, J.A.; Yang, H. The multiple fates of gene duplications: Deletion, hypofunctionalization, subfunctionalization, neofunctionalization, dosage balance constraints, and neutral variation. Plant Cell 2022, 34, 2466–2474. [Google Scholar] [CrossRef]





| Name | Gene ID | Chr ID | Start | End | pI | MW (Da) | CDS Length (bp) | Protein (aa) | Exons | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|---|
| SmGLK1 | EGP04115 | chr1 | 68,643,071 | 68,646,335 | 6.61 | 60,615.06 | 1647 | 548 | 6 | nucl |
| SmGLK2 | EGP05116 | chr1 | 95,471,528 | 95,480,561 | 5.56 | 88,500.67 | 2382 | 793 | 15 | nucl |
| SmGLK3 | EGP06342 | chr1 | 108,107,510 | 108,110,509 | 7.17 | 44,925.73 | 1218 | 405 | 5 | nucl |
| SmGLK4 | EGP29953 | chr2 | 60,788,782 | 60,792,579 | 8.88 | 40,016.61 | 1023 | 340 | 6 | nucl |
| SmGLK5 | EGP30651 | chr2 | 69,219,561 | 69,221,293 | 9.42 | 22,504.46 | 615 | 204 | 6 | nucl |
| SmGLK6 | EGP30658 | chr2 | 69,308,009 | 69,309,947 | 6.45 | 26,915.78 | 714 | 237 | 6 | nucl |
| SmGLK7 | EGP31411 | chr2 | 76,116,607 | 76,119,195 | 7.16 | 40,238.87 | 1077 | 358 | 5 | pero |
| SmGLK8 | EGP12000 | chr3 | 42,770,529 | 42,776,676 | 6.01 | 65,332.49 | 1761 | 586 | 5 | nucl |
| SmGLK9 | EGP26827 | chr4 | 4,028,213 | 4,030,290 | 9.23 | 23,642.87 | 630 | 209 | 3 | nucl |
| SmGLK10 | EGP28551 | chr4 | 79,927,110 | 79,931,297 | 7.02 | 35,226.93 | 942 | 313 | 6 | cyto |
| SmGLK11 | EGP24129 | chr5 | 1,630,169 | 1,637,808 | 7.79 | 44,398.56 | 1197 | 398 | 6 | nucl |
| SmGLK12 | EGP24145 | chr5 | 1,804,572 | 1,808,269 | 7.79 | 33,260.82 | 870 | 289 | 4 | nucl |
| SmGLK13 | EGP24440 | chr5 | 5,613,154 | 5,620,386 | 5.69 | 72,812.13 | 1977 | 658 | 6 | nucl |
| SmGLK14 | EGP24597 | chr5 | 7,563,196 | 7,569,245 | 5.60 | 45,814.18 | 1248 | 415 | 7 | nucl |
| SmGLK15 | EGP26270 | chr5 | 83,294,033 | 83,298,987 | 6.37 | 32,857.76 | 897 | 298 | 6 | nucl |
| SmGLK16 | EGP26358 | chr5 | 84,224,489 | 84,227,940 | 5.00 | 44,165.10 | 1179 | 392 | 7 | nucl |
| SmGLK17 | EGP15888 | chr6 | 2,113,619 | 2,124,522 | 5.13 | 50,749.64 | 1395 | 464 | 7 | nucl |
| SmGLK18 | EGP15695 | chr6 | 5,839,487 | 5,841,423 | 5.91 | 34,094.85 | 921 | 306 | 1 | nucl |
| SmGLK19 | EGP15014 | chr6 | 54,881,706 | 54,883,465 | 7.15 | 40,025.95 | 1047 | 348 | 5 | nucl |
| SmGLK20 | EGP14289 | chr6 | 81,275,437 | 81,280,149 | 6.21 | 61,361.55 | 1656 | 551 | 11 | nucl |
| SmGLK21 | EGP14253 | chr6 | 81,787,799 | 81,793,799 | 9.15 | 43,622.27 | 1167 | 388 | 6 | nucl |
| SmGLK22 | EGP14239 | chr6 | 82,118,616 | 82,120,799 | 8.48 | 28,372.73 | 765 | 254 | 6 | nucl |
| SmGLK23 | EGP14133 | chr6 | 83,555,681 | 83,563,561 | 6.31 | 35,907.49 | 999 | 332 | 6 | nucl |
| SmGLK24 | EGP13249 | chr6 | 91,911,599 | 91,914,246 | 5.47 | 35,110.80 | 939 | 312 | 1 | nucl |
| SmGLK25 | EGP02563 | chr7 | 2,910,686 | 2,916,581 | 6.24 | 75,391.20 | 2064 | 687 | 6 | nucl |
| SmGLK26 | EGP01139 | chr7 | 93,632,358 | 93,637,356 | 6.83 | 25,491.09 | 669 | 222 | 5 | mito |
| SmGLK27 | EGP00675 | chr7 | 104,675,122 | 104,680,177 | 6.08 | 48,777.46 | 1341 | 446 | 6 | nucl |
| SmGLK28 | EGP20881 | chr8 | 3,370,310 | 3,376,379 | 6.93 | 40,606.18 | 1086 | 361 | 6 | nucl |
| SmGLK29 | EGP19297 | chr8 | 82,506,447 | 82,509,595 | 6.98 | 31,661.35 | 852 | 283 | 6 | nucl |
| SmGLK30 | EGP19254 | chr8 | 83,141,169 | 83,147,055 | 9.24 | 32,473.04 | 864 | 287 | 4 | nucl |
| SmGLK31 | EGP19168 | chr8 | 84,417,636 | 84,423,522 | 6.16 | 62,160.30 | 1674 | 557 | 11 | nucl |
| SmGLK32 | EGP18241 | chr9 | 4,886,189 | 4,898,278 | 5.64 | 54,731.15 | 1449 | 482 | 5 | nucl |
| SmGLK33 | EGP18238 | chr9 | 4,957,579 | 4,961,351 | 6.05 | 49,000.02 | 1287 | 428 | 4 | nucl |
| SmGLK34 | EGP18065 | chr9 | 7,912,729 | 7,919,511 | 8.36 | 59,083.64 | 1584 | 527 | 5 | E.R |
| SmGLK35 | EGP17937 | chr9 | 12,229,872 | 12,232,841 | 7.17 | 26,882.06 | 702 | 233 | 6 | nucl |
| SmGLK36 | EGP17858 | chr9 | 16,429,096 | 16,431,149 | 8.36 | 35,911.84 | 969 | 322 | 5 | nucl |
| SmGLK37 | EGP16748 | chr9 | 84,207,659 | 84,216,589 | 5.04 | 48,823.09 | 1338 | 445 | 7 | nucl |
| SmGLK38 | EGP16179 | chr9 | 91,964,731 | 91,969,446 | 5.86 | 31,901.78 | 864 | 287 | 6 | nucl |
| SmGLK39 | EGP21185 | chr10 | 331,005 | 332,779 | 6.48 | 32,238.59 | 882 | 293 | 1 | nucl |
| SmGLK40 | EGP21245 | chr10 | 819,985 | 822,961 | 6.11 | 45,635.78 | 1212 | 403 | 7 | nucl |
| SmGLK41 | EGP21361 | chr10 | 1,736,637 | 1,742,936 | 5.80 | 44,354.21 | 1173 | 390 | 7 | nucl |
| SmGLK42 | EGP21794 | chr10 | 6,478,648 | 6,481,704 | 9.00 | 55,889.28 | 1482 | 493 | 7 | nucl |
| SmGLK43 | EGP22032 | chr10 | 13,141,129 | 13,143,574 | 6.56 | 38,290.57 | 1026 | 341 | 5 | nucl |
| SmGLK44 | EGP22321 | chr10 | 18,625,031 | 18,626,906 | 8.99 | 33,405.03 | 888 | 295 | 5 | nucl |
| SmGLK45 | EGP23367 | chr10 | 77,444,097 | 77,449,402 | 6.93 | 42,472.29 | 1149 | 382 | 8 | nucl |
| SmGLK46 | EGP23752 | chr10 | 85,369,460 | 85,375,848 | 5.91 | 77,539.63 | 2121 | 706 | 6 | nucl |
| SmGLK47 | EGP23800 | chr10 | 85,902,577 | 85,905,785 | 6.84 | 42,768.62 | 1170 | 389 | 5 | nucl |
| SmGLK48 | EGP06828 | chr11 | 1,453,235 | 1,460,575 | 7.90 | 51,124.93 | 1374 | 457 | 6 | nucl |
| SmGLK49 | EGP07802 | chr11 | 29,542,927 | 29,557,840 | 5.82 | 51,410.97 | 1407 | 468 | 7 | nucl |
| SmGLK50 | EGP08969 | chr11 | 97,459,423 | 97,468,863 | 6.30 | 70,201.99 | 1944 | 647 | 5 | nucl |
| SmGLK51 | EGP09012 | chr11 | 98,353,223 | 98,358,334 | 6.57 | 34,346.06 | 897 | 298 | 6 | nucl |
| SmGLK52 | EGP09258 | chr11 | 101,893,426 | 101,898,259 | 6.20 | 34,847.76 | 960 | 319 | 6 | nucl |
| SmGLK53 | EGP33729 | chr12 | 1,379,693 | 1,382,695 | 6.85 | 49,594.35 | 1341 | 446 | 4 | nucl |
| SmGLK54 | EGP33116 | chr12 | 21,405,470 | 21,415,883 | 7.70 | 71,609.34 | 1935 | 644 | 11 | cyto_nucl |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, C.; Xiang, R.; Jiang, Y.; Li, W.; Yang, Q.; Gan, G.; Cai, L.; Wang, P.; Li, W.; Wang, Y. Genomic Organization and Expression Profiling of GOLDEN2-like Transcription Factor Genes in Eggplant and Their Role in Heat Stresses. Horticulturae 2024, 10, 958. https://doi.org/10.3390/horticulturae10090958
Yu C, Xiang R, Jiang Y, Li W, Yang Q, Gan G, Cai L, Wang P, Li W, Wang Y. Genomic Organization and Expression Profiling of GOLDEN2-like Transcription Factor Genes in Eggplant and Their Role in Heat Stresses. Horticulturae. 2024; 10(9):958. https://doi.org/10.3390/horticulturae10090958
Chicago/Turabian StyleYu, Chuying, Rui Xiang, Yaqin Jiang, Weiliu Li, Qihong Yang, Guiyun Gan, Liangyu Cai, Peng Wang, Wenjia Li, and Yikui Wang. 2024. "Genomic Organization and Expression Profiling of GOLDEN2-like Transcription Factor Genes in Eggplant and Their Role in Heat Stresses" Horticulturae 10, no. 9: 958. https://doi.org/10.3390/horticulturae10090958
APA StyleYu, C., Xiang, R., Jiang, Y., Li, W., Yang, Q., Gan, G., Cai, L., Wang, P., Li, W., & Wang, Y. (2024). Genomic Organization and Expression Profiling of GOLDEN2-like Transcription Factor Genes in Eggplant and Their Role in Heat Stresses. Horticulturae, 10(9), 958. https://doi.org/10.3390/horticulturae10090958






