Genome-Wide Identification of DREB Transcription Factor Family and Functional Analysis of PaDREB1D Associated with Low-Temperature Stress in Phalaenopsis aphrodite
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Stress Treatments
2.2. Total RNA Extraction and Real-Time Quantitative PCR (RT-qPCR)
2.3. Screening of DREB Family Members in Phalaenopsis aphrodite
2.4. Phylogenetic Tree Construction and Gene Structure Analysis
2.5. Expression Pattern Analysis of PaDREBs in Different Tissues
2.6. Gene Cloning and Subcellular Localization
2.7. Preparation and Detection of Transgenic Phalaenopsis
2.8. Cold Tolerance Assays of PLBs
2.9. Physiological Measurements
2.10. Statistical Analysis
3. Results
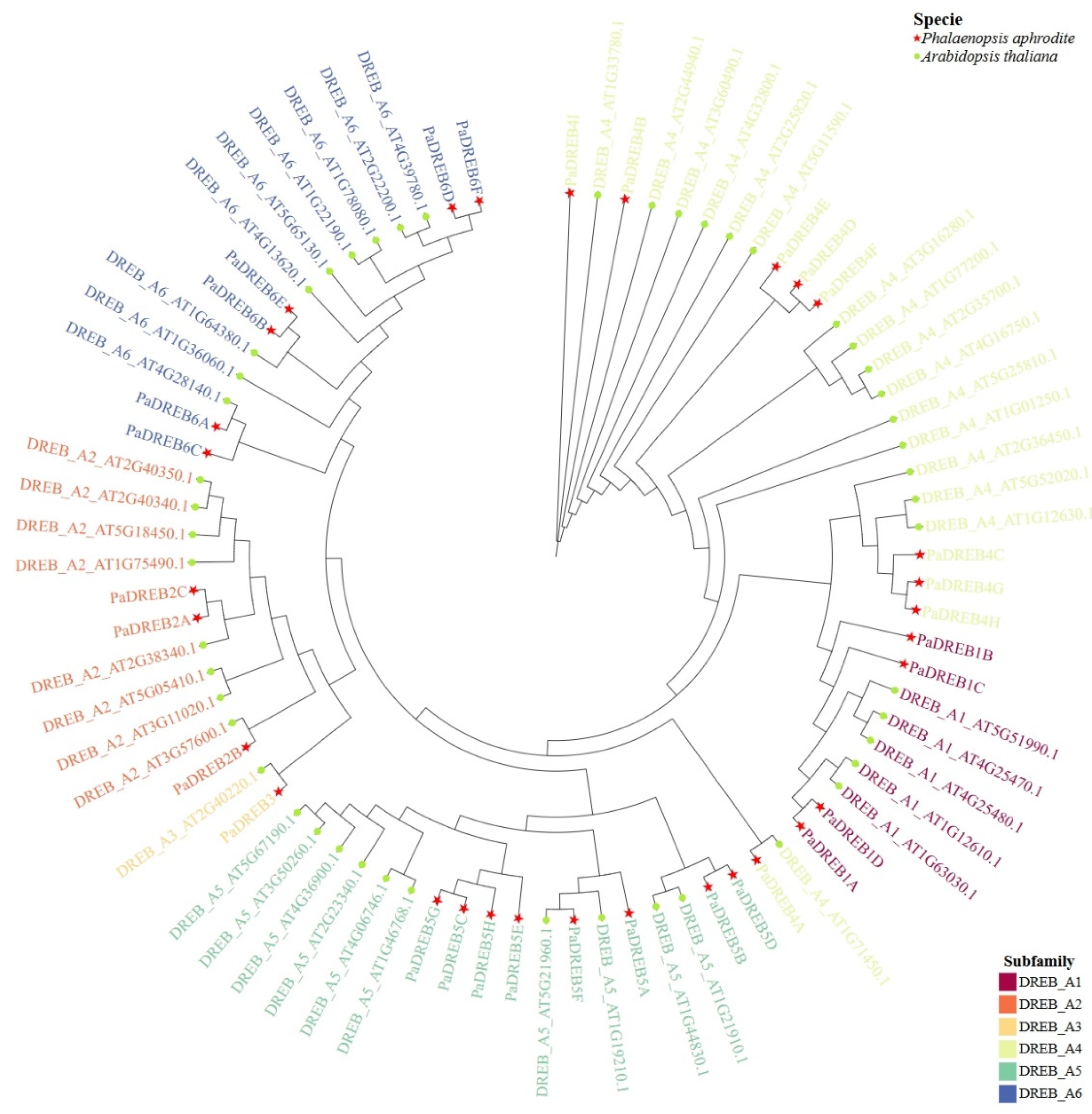
3.1. Identification of DREB Family Members in P. aphrodite and Phylogenetic Tree Construction
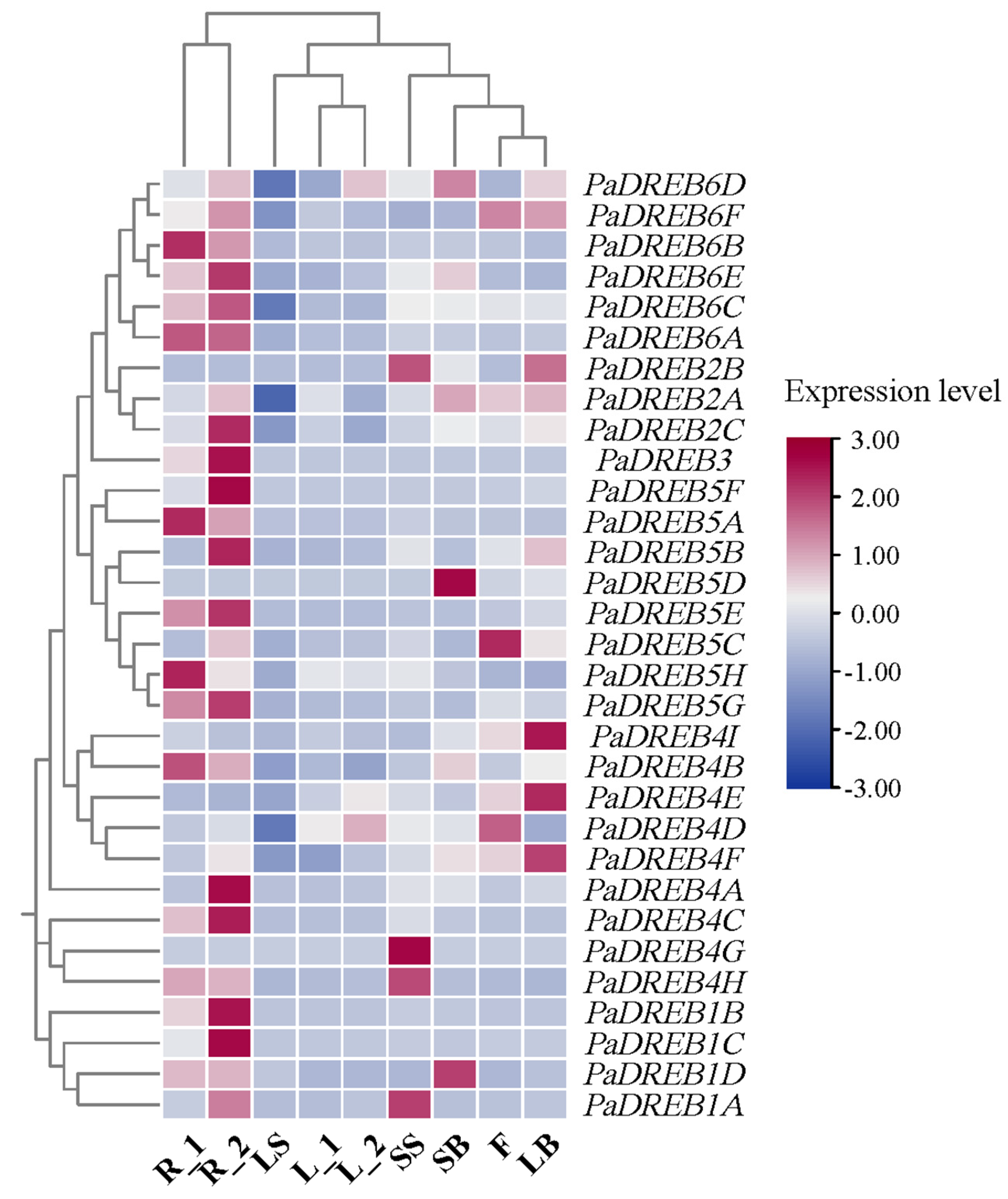
3.2. Analysis of the PaDREB Expression Patterns in Different Tissues
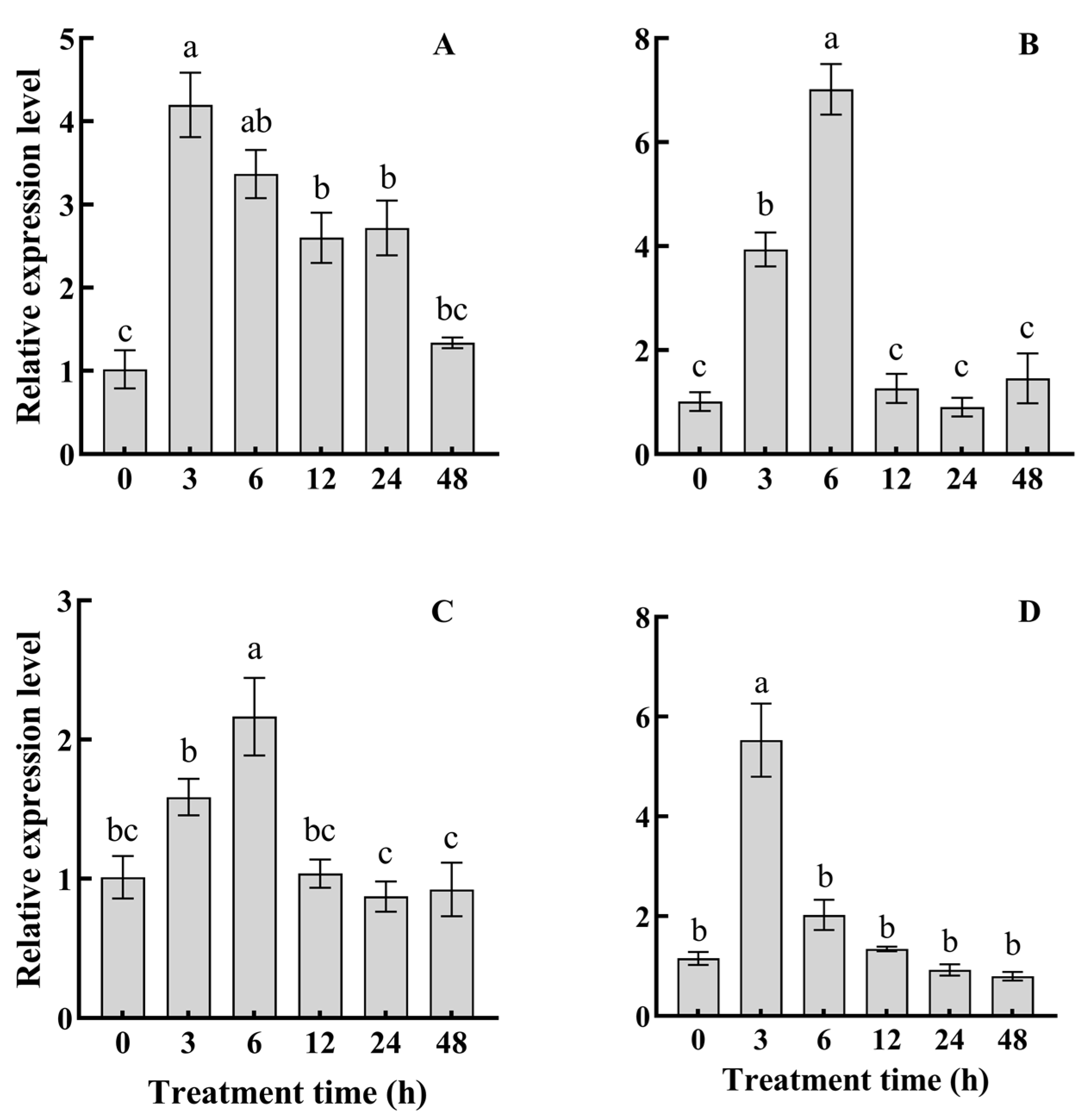
3.3. PaDREB1D Responded to Four Treatments and Was Expressed in Different Tissues
3.4. Gene Isolation and Molecular Characterization Analysis of PaDREB1D
3.5. Subcellular Localization of PaDREB1D
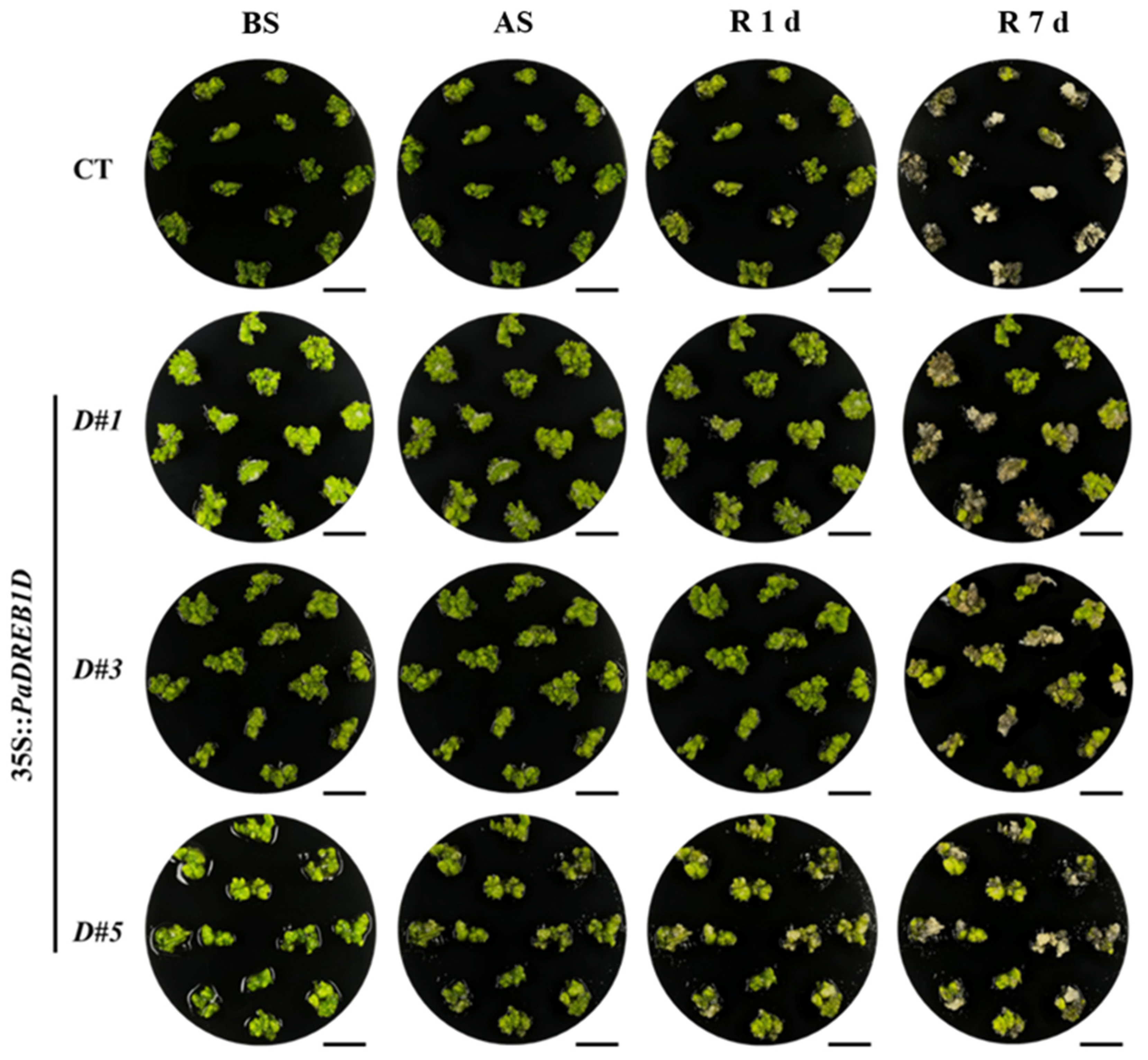
3.6. PaDREB1D Overexpression Enhanced Cold Tolerance in PLBs
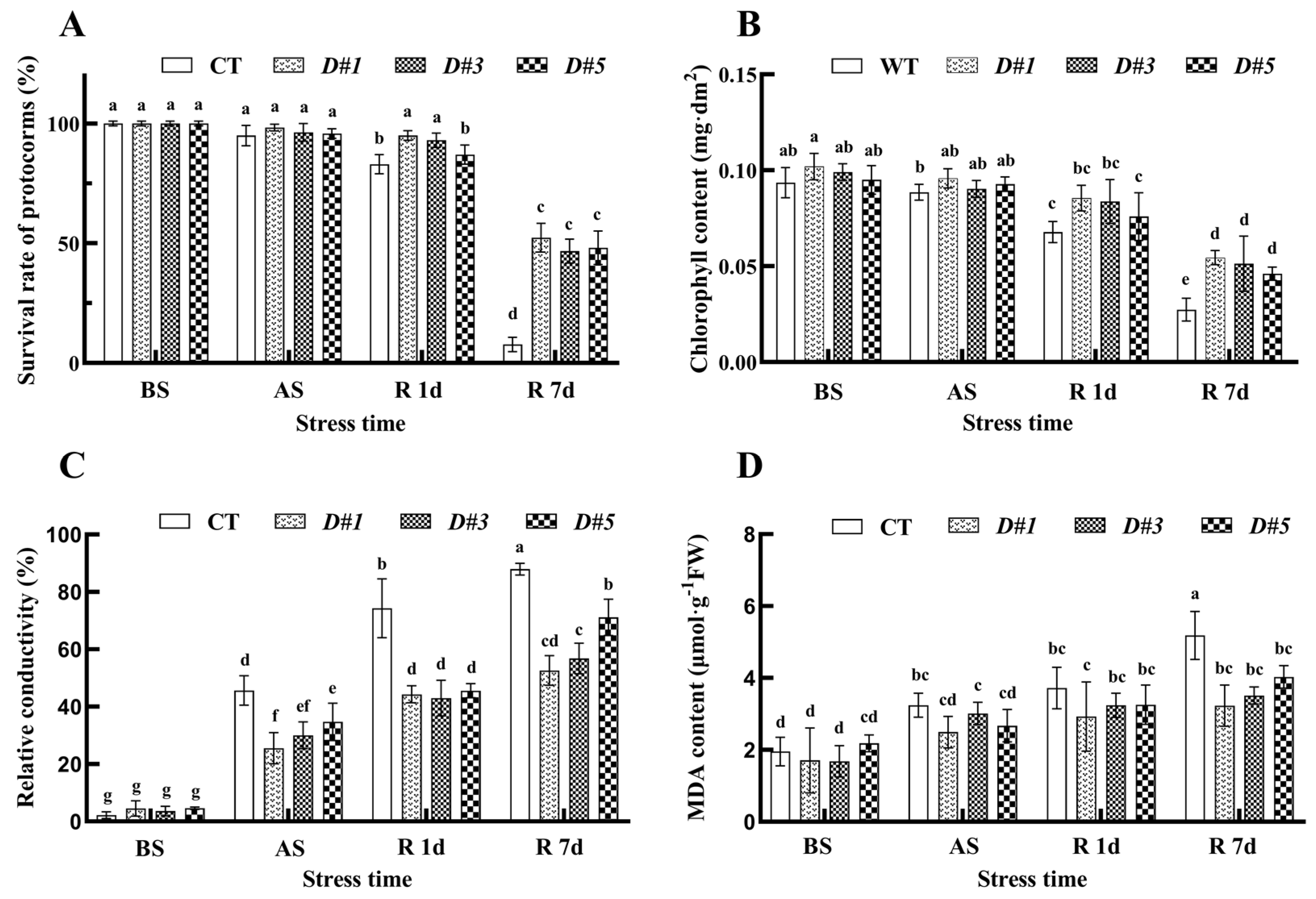
3.7. Overexpression of the PaDREB1D Gene Reduced Cellular Damage to PLBs
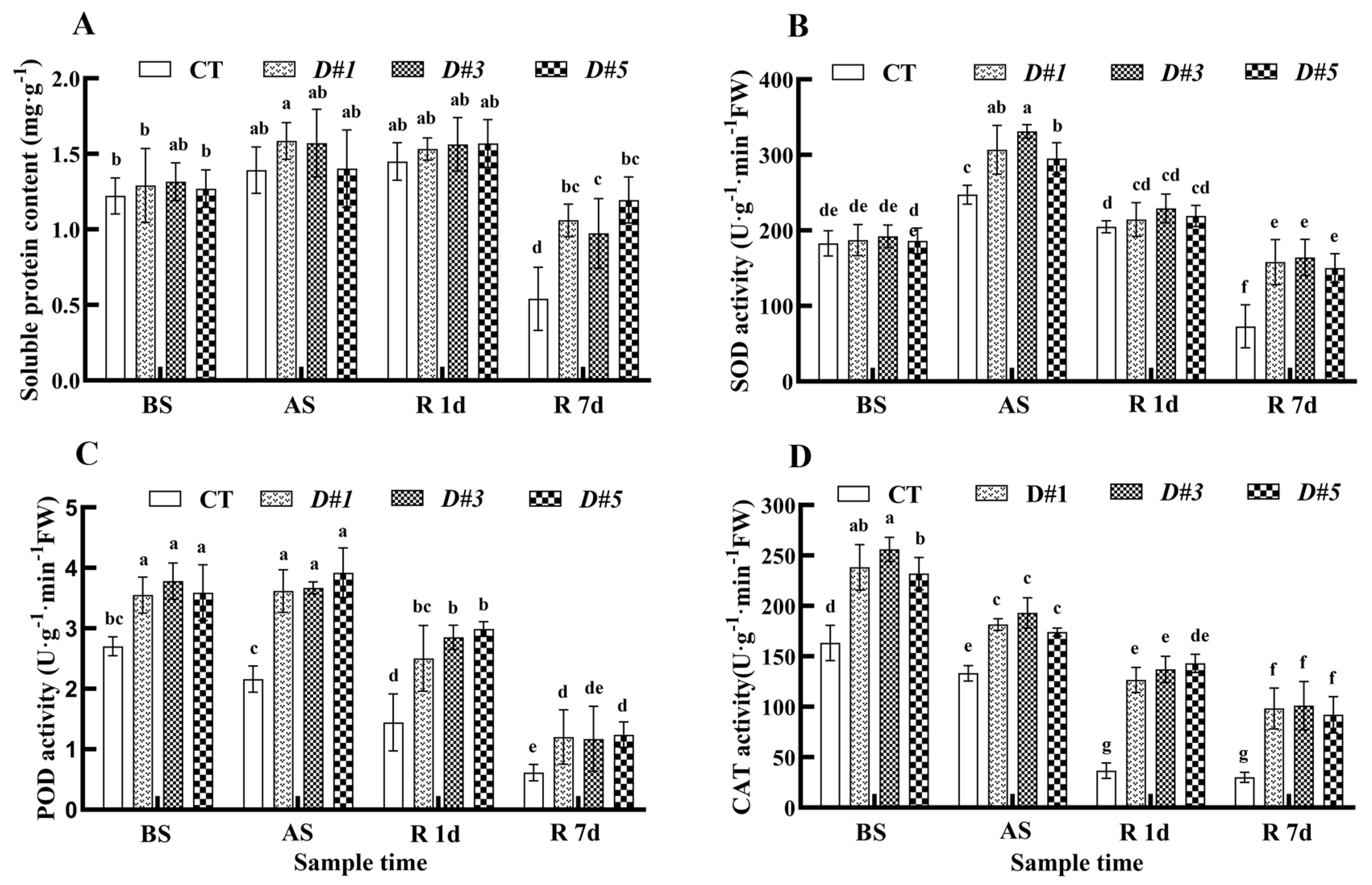
3.8. PaDREB1D Overexpression Enhanced the Antioxidant Enzyme Activities of PLBs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Chen, J.; Chen, C. The effect of temperature on the inflorescence formation model for Phalaenopsis. Plants 2024, 13, 1280. [Google Scholar] [CrossRef]
- Peng, P.H.; Lin, C.H.; Tsai, H.W.; Lin, T.Y. Cold response in Phalaenopsis aphrodite and characterization of PaCBF1 and PaICE1. Plant Cell Physiol. 2014, 55, 1623–1635. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.C.; Chen, Y.J.; Markhart, A.; Lin, T.Y. Temperature effects on systemic endoreduplication in orchid during floral development. Plant Sci. 2007, 172, 588–595. [Google Scholar] [CrossRef]
- Akhtar, M.; Jaiswal, A.; Taj, G.; Jaiswal, J.P.; Qureshi, M.I.; Singh, N.K. DREB1/CBF transcription factors: Their structure, function and role in abiotic stress tolerance in plants. J. Genet. 2012, 91, 385–395. [Google Scholar] [CrossRef]
- Feng, K.; Hou, X.L.; Xing, G.M.; Liu, J.X.; Duan, A.Q.; Xu, Z.S.; Li, M.Y.; Zhuang, J.; Xiong, A.S. Advances in AP2/ERF super-family transcription factors in plant. Crit. Rev. Biotechnol. 2020, 40, 750–776. [Google Scholar] [CrossRef]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef]
- Kim, J.S.; Kidokoro, S.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Regulatory networks in plant responses to drought and cold stress. Plant Physiol. 2024, 195, 170–189. [Google Scholar] [CrossRef]
- Kidokoro, S.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Transcriptional regulatory network of plant cold-stress responses. Trends Plant Sci. 2022, 27, 922–935. [Google Scholar] [CrossRef] [PubMed]
- Jaglo, K.R.; Kleff, S.; Amundsen, K.L.; Zhang, X.; Haake, V.; Zhang, J.Z.; Deits, T.; Thomashow, M.F. Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiol. 2001, 127, 910–917. [Google Scholar] [CrossRef]
- Dubouzet, J.G.; Sakuma, Y.; Ito, Y.; Kasuga, M.; Dubouzet, E.G.; Miura, S.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 2003, 33, 751–763. [Google Scholar] [CrossRef] [PubMed]
- Hwarari, D.; Guan, Y.; Ahmad, B.; Movahedi, A.; Min, T.; Hao, Z.; Lu, Y.; Chen, J.; Yang, L. ICE-CBF-COR signaling cascade and its regulation in plants responding to cold stress. Int. J. Mol. Sci. 2022, 23, 1549. [Google Scholar] [CrossRef] [PubMed]
- Zarka, D.G.; Vogel, J.T.; Cook, D.; Thomashow, M.F. Cold induction of Arabidopsis CBF genes involves multiple ICE (inducer of CBF expression) promoter elements and a cold-regulatory circuit that is desensitized by low temperature. Plant Physiol. 2003, 133, 910–918. [Google Scholar] [CrossRef] [PubMed]
- Haake, V.; Cook, D.; Riechmann, J.L.; Pineda, O.; Thomashow, M.F.; Zhang, J.Z. Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiol. 2002, 130, 639–648. [Google Scholar] [CrossRef]
- Lehti-Shiu, M.D.; Uygun, S.; Moghe, G.D.; Panchy, N.; Fang, L.; Hufnagel, D.E.; Jasicki, H.L.; Feig, M.; Shiu, S.H. Molecular evidence for functional divergence and decay of a transcription factor derived from whole-genome duplication in Arabidopsis thaliana. Plant Physiol. 2015, 168, 1717–1734. [Google Scholar] [CrossRef]
- Xie, Z.; Nolan, T.M.; Jiang, H.; Yin, Y. AP2/ERF transcription factor regulatory networks in hormone and abiotic stress responses in Arabidopsis. Front. Plant Sci. 2019, 10, 228. [Google Scholar] [CrossRef]
- Rubio, S.; Noriega, X.; Pérez, F.J. Abscisic acid (ABA) and low temperatures synergistically increase the expression of CBF/DREB1 transcription factors and cold-hardiness in grapevine dormant buds. Ann. Bot. 2019, 123, 681–689. [Google Scholar] [CrossRef]
- Zhang, S.Y.; Cao, K.F.; Wei, Y.Y.; Jiang, S.; Ye, J.F.; Xu, F.; Chen, Y.; Shao, X.F. PpBZR1, a BES/BZR transcription factor, enhances cold stress tolerance by suppressing sucrose degradation in peach fruit. Plant Physiol. Biochem. 2023, 202, 107972. [Google Scholar] [CrossRef]
- Kazan, K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015, 20, 219–229. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Chao, Y.T.; Chen, W.C.; Chen, C.Y.; Ho, H.Y.; Yeh, C.H.; Kuo, Y.T.; Su, C.L.; Yen, S.H.; Hsueh, H.Y.; Yeh, J.H.; et al. Chromosome-level assembly, genetic and physical mapping of Phalaenopsis aphrodite genome provides new insights into species adaptation and resources for orchid breeding. Plant Biotechnol. J. 2018, 16, 2027–2041. [Google Scholar] [CrossRef] [PubMed]
- Chao, Y.T.; Yen, S.H.; Yeh, J.H.; Chen, W.C.; Shih, M.C. Orchidstra 2.0-A transcriptomics resource for the Orchid family. Plant Cell Physiol. 2017, 58, e9. [Google Scholar] [CrossRef]
- Reiser, L.; Subramaniam, S.; Li, D.; Huala, E. Using the Arabidopsis information resource (TAIR) to find information about Arabidopsis genes. Curr. Protoc. Bioinform. 2017, 60, 1.11.1–1.11.45. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Xie, J.M.; Chen, Y.R.; Cai, G.J.; Cai, R.L.; Hu, Z.; Wang, H. Tree Visualization By One Table (tvBOT): A web application for visualizing, modifying and annotating phylogenetic Trees. Nucleic Acids Res. 2023, 51, W587–W592. [Google Scholar] [CrossRef]
- Gong, X.Q.; Zhang, J.Y.; Hu, J.B.; Wang, W.; Wu, H.; Zhang, Q.H.; Liu, J.H. FcWRKY70, a WRKY protein of Fortunella crassifolia, functions in drought tolerance and modulates putrescine synthesis by regulating arginine decarboxylase gene. Plant Cell Environ. 2015, 38, 2248–2262. [Google Scholar] [CrossRef]
- Dahro, B.; Wang, F.; Peng, T.; Liu, J.H. PtrA/NINV, an alkaline/neutral invertase gene of Poncirus trifoliata, confers enhanced tolerance to multiple abiotic stresses by modulating ROS levels and maintaining photosynthetic efficiency. BMC Plant Biol. 2016, 16, 76. [Google Scholar] [CrossRef]
- Porra, R.J.; Thompson, W.A.; Kriedemann, P.E. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: Verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. BBA-Bioenerg. 1989, 975, 384–394. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Maqsood, H.; Munir, F.; Amir, R.; Gul, A. Genome-wide identification, comprehensive characterization of transcription factors, cis-regulatory elements, protein homology, and protein interaction network of DREB gene family in Solanum Lycopersicum. Front. Plant Sci. 2022, 13, 1031679. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.L.; Cai, M.H.; Zhang, S.Q.; Chai, J.W.; Sun, M.Z.; Wang, Y.W.; Xie, Q.Y.; Chen, Y.H.; Wang, H.Z.; Chen, T. Genome-wide identification of AP2/ERF transcription factor family and functional analysis of DcAP2/ERF#96 associated with abiotic stress in Dendrobium Catenatum. Int. J. Mol. Sci. 2022, 23, 13603. [Google Scholar] [CrossRef]
- Dong, C.; Xi, Y.; Chen, X.L.; Cheng, Z.M. Genome-wide identification of AP2/EREBP in Fragaria Vesca and expression pattern analysis of the FvDREB subfamily under drought stress. BMC Plant Biol. 2021, 21, 295. [Google Scholar] [CrossRef]
- Niu, X.; Luo, T.L.; Zhao, H.Y.; Su, Y.L.; Ji, W.Q.; Li, H.F. Identification of wheat DREB genes and functional characterization of TaDREB3 in response to abiotic stresses. Gene 2020, 740, 144514. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.L.; Wang, Y.; Li, X.T.; Gao, F.; Feng, J.C.; Zhou, Y.J. Characterization of the AP2/ERF transcription factor family and expression profiling of DREB subfamily under cold and osmotic stresses in Ammopiptanthus nanus. Plants 2020, 9, 455. [Google Scholar] [CrossRef] [PubMed]
- Li, J.L.; Li, H.M.; Quan, X.Y.; Shan, Q.; Wang, W.B.; Yin, N.; Wang, S.Q.; Wang, Z.H.; He, W.X. Comprehensive analysis of cucumber C-repeat/dehydration-responsive element binding factor family genes and their potential roles in cold tolerance of Cucumber. BMC Plant Biol. 2022, 22, 270. [Google Scholar] [CrossRef]
- Azzeme, A.M.; Abdullah, S.N.A.; Aziz, M.A.; Wahab, P.E.M. Oil palm drought inducible DREB1 induced expression of DRE/CRT- and non-DRE/CRT-containing genes in lowland transgenic tomato under cold and PEG treatments. Plant Physiol. Biochem. 2017, 112, 129–151. [Google Scholar] [CrossRef]
- Yang, W.; Liu, X.D.; Chi, X.J.; Wu, C.A.; Li, Y.Z.; Song, L.L.; Liu, X.M.; Wang, Y.F.; Wang, F.W.; Zhang, C.; et al. Dwarf apple MbDREB1 enhances plant tolerance to low temperature, drought, and salt stress via both ABA-dependent and ABA-independent pathways. Planta 2011, 233, 219–229. [Google Scholar] [CrossRef]
- Bolaños-Villegas, P.; Chen, F.C. Advances and perspectives for polyploidy breeding in Orchids. Plants 2022, 11, 1421. [Google Scholar] [CrossRef]
- Teixeira da Silva, J.A.; Jin, X.; Dobránszki, J.; Lu, J.; Wang, H.; Zotz, G.; Cardoso, J.C.; Zeng, S. Advances in Dendrobium molecular research: Applications in genetic variation, identification and breeding. Mol. Phylogenet. Evol. 2016, 95, 196–216. [Google Scholar] [CrossRef]
- Popova, E.; Kim, H.H.; Saxena, P.K.; Engelmann, F.; Pritchard, H.W. Frozen beauty: The cryobiotechnology of orchid diversity. Biotechnol. Adv. 2016, 34, 380–403. [Google Scholar] [CrossRef] [PubMed]
- Iiyama, C.M.; Vilcherrez-Atoche, J.A.; Germanà, M.A.; Vendrame, W.A.; Cardoso, J.C. Breeding of ornamental orchids with focus on Phalaenopsis: Current approaches, tools, and challenges for this century. Heredity 2024, 132, 163–178. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, J.C.; Zanello, C.A.; Chen, J.T. An overview of orchid protocorm-like bodies: Mass propagation, biotechnology, molecular aspects, and breeding. Int. J. Mol. Sci. 2020, 21, 985. [Google Scholar] [CrossRef]
- Lou, Y.X.; Zhang, Q.Y.; Xu, Q.Y.; Xu, X.Y.; Wang, W.X.; Gai, R.N.; Ming, F. PhCHS5 and PhF3’5’H Genes over-expression in Petunia (Petunia hybrida) and Phalaenopsis (Phalaenopsis aphrodite) regulate flower color and branch number. Plants 2023, 12, 2204. [Google Scholar] [CrossRef]
- Xu, Q.Y.; Xia, M.; He, G.R.; Zhang, Q.Y.; Meng, Y.; Ming, F. New insights into the influence of NHX-type Cation/H+ antiporter on flower color in Phalaenopsis orchids. J. Plant Physiol. 2022, 279, 153857. [Google Scholar] [CrossRef]
- Chen, W.H.; Jiang, Z.Y.; Hsu, H.F.; Yang, C.H. Silencing of FOREVER YOUNG FLOWER-like genes from Phalaenopsis orchids promotes flower senescence and abscission. Plant Cell Physiol. 2021, 62, 111–124. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.Y.; Rengasamy, K.P.; Huang, L.H.; Hsu, C.C.; Jeng, M.F.; Chen, W.H.; Chen, H.W. Assessment of violet-blue color formation in Phalaenopsis orchids. BMC Plant Biol. 2020, 20, 212. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, Z.; Wang, S.; Wang, Y.; Li, J.; Luo, P.; Xin, J.; Cui, Y. Genome-Wide Identification of DREB Transcription Factor Family and Functional Analysis of PaDREB1D Associated with Low-Temperature Stress in Phalaenopsis aphrodite. Horticulturae 2024, 10, 933. https://doi.org/10.3390/horticulturae10090933
Hu Z, Wang S, Wang Y, Li J, Luo P, Xin J, Cui Y. Genome-Wide Identification of DREB Transcription Factor Family and Functional Analysis of PaDREB1D Associated with Low-Temperature Stress in Phalaenopsis aphrodite. Horticulturae. 2024; 10(9):933. https://doi.org/10.3390/horticulturae10090933
Chicago/Turabian StyleHu, Ziang, Shuang Wang, Yaoling Wang, Jiaming Li, Ping Luo, Jingjing Xin, and Yongyi Cui. 2024. "Genome-Wide Identification of DREB Transcription Factor Family and Functional Analysis of PaDREB1D Associated with Low-Temperature Stress in Phalaenopsis aphrodite" Horticulturae 10, no. 9: 933. https://doi.org/10.3390/horticulturae10090933
APA StyleHu, Z., Wang, S., Wang, Y., Li, J., Luo, P., Xin, J., & Cui, Y. (2024). Genome-Wide Identification of DREB Transcription Factor Family and Functional Analysis of PaDREB1D Associated with Low-Temperature Stress in Phalaenopsis aphrodite. Horticulturae, 10(9), 933. https://doi.org/10.3390/horticulturae10090933



