Abstract
PLCPs (papain-like cysteine proteases) are one of the most abundant groups of cysteine proteases and play vital roles in multiple processes. The pepper (Capsicum annuum) is an important Solanaceae vegetable crop; its commercial hybrid seeds are widely used in production. Male sterility is a valuable trait for hybrid seed production. However, the function of PLCPs and the underlying mechanisms of male sterility in peppers remain unclear. In this study, we comprehensively identified the PLCP gene family in peppers, identifying 31 CaPLCPs. A phylogenetic analysis classified 31 members into eight clades. These CaPLCPs were unevenly distributed across eight chromosomes, and five segmental duplicated pairs were observed. The promoter cis-acting element analysis indicated that CaPLCP promoters contained abundant hormone-responsive and stress-responsive cis-elements, suggesting that CaPLCPs may play important roles in responding to abiotic stress, such as drought and low temperatures, as well as in plant immunity. The qRT-PCR analysis demonstrated that the expression levels of CaPLCP1, CaPLCP5, CaPLCP11, CaPLCP12, CaPLCP13, CaPLCP17, CaPLCP19, and CaPLCP21 were significantly reduced in the flowers of MS (male sterile pepper) at least at one stage, indicating their potential roles as regulatory factors in pepper male sterility. These findings provide important insights into the functional analysis of the PLCP gene family in peppers and other species, laying a crucial foundation for understanding the mechanisms of male sterility in peppers.
1. Introduction
Proteases play a pivotal role in protein hydrolysis and act as key regulators of a variety of biological processes, encompassing multiple families, such as cysteine proteases, aspartic proteases, serine proteases, and metallo proteases [1]. Notably, papain-like cysteine proteases (PLCPs) are among the most abundant cysteine proteases, which exist in animals, plants, and microorganisms, and are extensively involved in diverse life activities [2].
In plants, PLCPs are classified into nine subfamilies: RD21, CEP, XCP, XBCP3, THI, SAG12, RD19, ALP, and CTB [3]. Structurally, PLCPs are characterized by a typical papain-like fold domain, comprising an α-helix and a β-sheet domain. These two domains are linked to each other and form the catalytic triplet Cys-His-Asn at the two-domain interface [4]. PLCPs contain an N-terminal signal peptide, an auto-inhibitory pro-domain, and the mature protease domain. Some subfamily members have the C-terminal granulin domain [2].
PLCPs exhibit diverse functions and play important roles in multiple processes, including programmed cell death (PCD), senescence, the abiotic stress response, and plant immunity. In Arabidopsis, AtXCP1 and AtXCP2 redundantly regulate the PCD of tracheary elements with other regulators [5]. The exact timing of tapetum PCD is crucial for plant reproductive capacity [6]. AtCEP1 is a key regulator of tapetum PCD, and its functional deficiency mutants cep1 exhibited impaired tapetum PCD, essential for pollen fertility [7]. PLCPs are also integral to the proteolysis process during senescence, where SAG12 is widely used as a senescence marker gene and involved in nitrogen distribution during seed production [8,9]. In rice, the SAG12 homologs OsSAG12-1 and OsSAG12-2 negatively regulate senescence-associated cell death [10,11]. Several PLCPs are induced in response to abiotic stress. For example, AtRD21A and AtRD19A, two dehydration response marker genes in Arabidopsis, are induced by drought and salt stress [12]. TaCP in wheat was triggered by PEG, salt, and cold stress, and the Arabidopsis plants overexpressing TaCP displayed enhanced drought resistance [13]. In peppers, CaCP is associated with leaf senescence, and its silencing enhanced the resistance to salt and osmotic stress [14]. Additionally, PLCPs are vital for plant immunity [15], with mutants like Arabidopsis rd19 and rd21 [16,17], tomato Pip1 [18], and cotton GhRD21-7 [19] showing increased sensitivity to various pathogens.
Although PLCPs have been identified in Arabidopsis [3], upland cotton [19], rice [20], soybeans [21], grapes [22], and other species, no comprehensive study of PLCP genes has been reported in peppers. The pepper (Capsicum annuum) is a vital vegetable crop extensively cultivated globally, with commercial hybrid seeds widely used in production. The hybrid seeds typically display heterosis and play a significant role in improving yield, resistance, and fruit quality. Male sterile lines are valuable tools for hybrid seed production [23]. However, the mechanism of male sterility in peppers is still unclear. To investigate whether PLCPs are involved in regulating male infertility in peppers, the PLCP gene family of peppers was comprehensively identified, and CaPLCP1, CaPLCP5, CaPLCP11, CaPLCP12, CaPLCP13, CaPLCP17, CaPLCP19, and CaPLCP21 were found to be potentially related to male sterility in peppers. This study provides an important foundation for the study of PLCPs in other species and the application of male sterile lines in pepper hybrid seed production.
2. Materials and Methods
2.1. Identification of PLCP Family Members
The pepper genome data (Capsicum annuum, zunla_v2) were downloaded from Solanaceae Genomics Network. To identify the pepper PLCP gene family, the Hidden Markov Model (HMM) of the peptidase_C1 domain of PLCP genes was obtained from the Pfam database (PF00112) [24]. HMMER SEARCH3.0, with set E value ≤ 0.01, was used for preliminary retrieval of pepper PLCP genes. Candidate members were further confirmed by blasting against the SMART database [25] and the NCBI-Conserved Domain Database [26]. The theoretical isoelectric point (pI) and the molecular weight (Mv) of CaPLCP members were calculated using the ExPASY online website [27], and their subcellular localizations were predicted using Plant mPLoc in Cell-PLoc 2.0 [28].
2.2. Multiple Sequence Alignment and Phylogenetic Analysis
Multiple sequence alignment was performed using Muscle in MEGA7 [29], and the maximum-likelihood tree was constructed with 1000 bootstrap replications. The phylogenetic tree was visualized using iTOL (v5.7) [30].
2.3. Duplication Analysis
Using the One Step MCScanX plugin in TBtools (v1.120), duplication analysis among CaPLCP genes was carried out, and the results were visualized using the Advanced Circos plugin [31].
2.4. Conserved Motifs and Gene Structure Analysis
The MEME website was used to identify the conserved motifs of CaPLCPs with the following parameters: the number of motifs was set to 15, and the optimum width of each motif was between 6 and 50 residues [32]. Gene structure analysis was performed based on the reference genome annotation (Capsicum annuum, zunla_v2). The results of conserved motifs and gene structure were visualized using the Gene Structure View (Advanced) plugin in TBtools (v1.120) [31].
2.5. Prediction of Cis-Acting Elements in the Promoters of CaPLCPs
The 2000 bp sequence upstream of the gene coding region of CaPLCPs was obtained, and the cis-acting elements in the promoters were predicted using PlantCARE [33]. The prediction results were visualized using the Gene Structure View (Advanced) plugin in TBtools (v1.120) [31].
2.6. Expression Data Analysis
The transcriptome data of various tissues of CaPLCP genes were obtained from PepperHub [34]. Using TBtools’ (v1.120) HeatMap plugin, the tissue expression profile of CaPLCP genes was drawn [31]. The FPKM (Fragments Per Kilobase Million) values of CaPLCP genes are listed in Supplementary Table S1.
2.7. GO Enrichment Analysis
GO (Gene Ontology) enrichment analyses were performed using TBtools (v1.120) [31], and significantly enriched GO terms were selected based on the threshold p ≤ 0.05.
2.8. Quantitative Real-Time PCR
Floral buds of MS (male fertile pepper) and ms (male sterile pepper) were sampled at three stages based on their length (S1: <5 mm; S2: 5–10 mm; S3: >10 mm). Total RNA was extracted using TRIzol reagent (Invitrogen Biotechnology Co. Ltd., Carlsbad, CA, USA) and reverse transcribed to cDNA using HiScript II 1st Strand cDNA Synthesis Kit (Vazyme Biotechnology Co. Ltd., Nanjing, China). The qRT-PCR was conducted according to published article [35]. Pepper UBI3 served as the internal control [36]. Supplementary Table S2 shows the primer list. All experiments in this study were conducted at the Wuhan Academy of Agricultural Sciences.
3. Results
3.1. The PLCP Family Members in Pepper
Following a comprehensive search and verification, 31 PLCP family members were identified in the pepper genome and designated as CaPLCP1 to CaPLCP31 (Table S3). Detailed information for each PLCP, including the gene name, Capana ID, chromosome location, genome location, gene length, protein theoretical isoelectric point (pI), molecular weight (Mv), and prediction of subcellular localization, are presented in Table S3. The characteristic analysis of the CaPLCP gene family revealed that this gene family is predominantly distributed on chromosome 2 in the pepper genome. The members of the CaPLCP gene family exhibited considerable variation in gene length, which results in differences in the length, pI, and Mv of their encoded proteins. The gene lengths of CaPLCPs ranged from 913 bp (CaPLCP7) to 8886 bp (CaPLCP2), and the CDS lengths ranged from 678 bp to 1953 bp, encoding proteins from 225 (CaPLCP15) to 650 (CaPLCP23) amino acid residues. The 31 CaPLCP proteins had a pI ranging from 4.40 (CaPLCP15) to 9.32 (CaPLCP30) and Mv ranging from 24.13 (CaPLCP15) to 69.95 (CaPLCP15) kDa. Subcellular localization predictions indicated that CaPLCP proteins were mainly localized in the vacuole, and CaPLCP15 was localized in both the Golgi apparatus and vacuole. Additionally, some members were localized to the endoplasmic reticulum (CaPLCP11, CaPLCP7, CaPLCP5, and CaPLCP4), while CaPLCP23 was shown to be localized to the nucleus.
3.2. Phylogenetic Analysis of the CaPLCP Family
To identify the phylogenetic relationship of CaPLCP proteins, a phylogenetic tree was constructed. The 31 CaPLCP family members were divided into eight clades (Figure 1). To further elucidate the subfamily clustering of CaPLCPs, a phylogenetic analysis of CaPLCPs and AtPLCPs [3] was performed. CaPLCP21, CaPLCP24, and CaPLCP25 clustered with the Arabidopsis RD19 subfamily; CaPLCP4 and CaPLCP5 clustered with the Arabidopsis CEP subfamily; CaPLCP8 and CaPLCP9 clustered with the Arabidopsis XCP subfamily; CaPLCP17 and the Arabidopsis ALP subfamily clustered together; CaPLCP27, CaPLCP28, CaPLCP29, CaPLCP30, and CaPLCP31 clustered with the Arabidopsis CTB subfamily; CaPLCP12 showed a closed relation to the THI subfamily; CaPLCP10 and CaPLCP23 were most closely related to the XBCP subfamily; and CaPLCP1, CaPLCP2, CaPLCP3, CaPLCP16, and the RD21 subfamily clustered together. The remaining CaPLCPs clustered with the SAG12 subfamily. It is notable that in the evolutionary tree constructed from CaPLCPs and AtPLCPs, the SAG12 and THI subfamilies and the XBCP and RDL subfamilies could not be clearly divided (Figure S1).
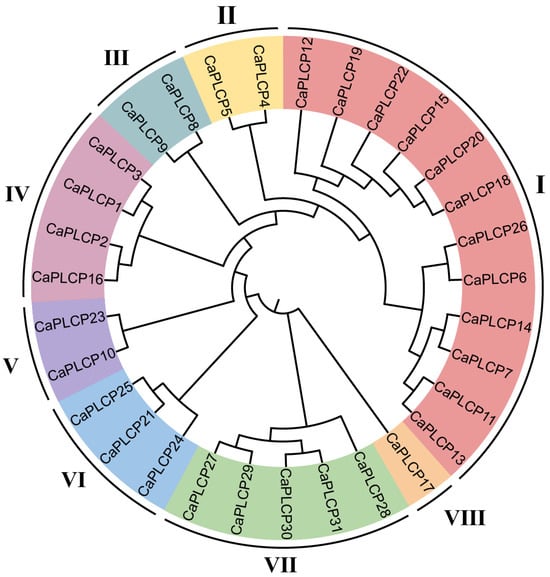
Figure 1.
The phylogenetic tree of PLCP family members from peppers. The phylogenetic tree was drawn using MEGA with the maximum-likelihood method, and the tree was visualized using the iTOL (v5.7) web server. Roman numerals I to VIII represent clades 1 to 8, respectively. Different colors represent different clades.
3.3. Chromosomal Localization and Gene Segmental Duplication of CaPLCP Genes
According to the physical positional information annotated in the zunla genome, the 31 CaPLCP genes were distributed across all chromosomes of peppers except chromosomes 1, 5, 6, and 9. Among them, the largest number of CaPLCP genes was found on chromosome 2, with 14 genes, followed by chromosome 4, containing 5 CaPLCP genes, and chromosome 12, containing 3, while chromosomes 7, 8, and 10 each contained 2 CaPLCP genes. Chromosomes 3 and 11 contained only one CaPLCP gene. In peppers, five segmental duplicated gene pairs of CaPLCP genes were identified: CaPLCP1-CaPLCP3, CaPLCP2-CaPLCP3, CaPLCP8-CaPLCP9, CaPLCP11-CaPLCP14, and CaPLCP21-CaPLCP25. No tandem duplicates were detected among CaPLCP genes (Figure 2).
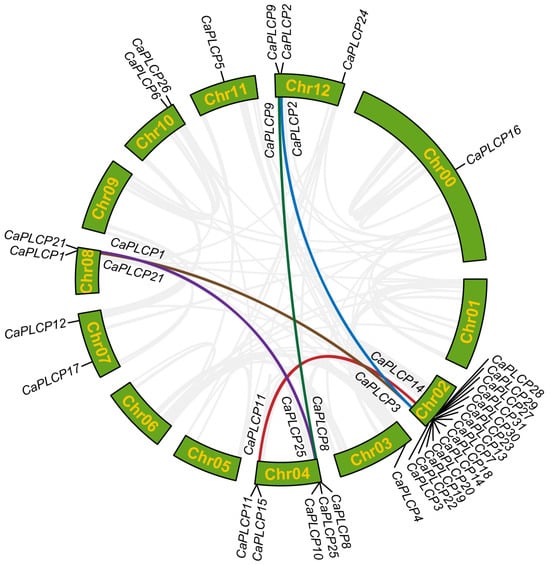
Figure 2.
The chromosomal localization and duplication relationship among the CaPLCP genes. The positions of CaPLCPs in the genome are labeled on chromosomes. The colored lines indicate segmental duplicated gene pairs. Chr refers to chromosomes.
3.4. Distribution of Conserved Motifs and Gene Structure of CaPLCPs
The conservative motifs and gene structure of CaPLCP members were analyzed. Using the MEME website, 15 conservative motifs were predicted and designated motifs 1–15 (Figure 3A and Figure S2). Motif5 and motif7 were characterized as the Inhibitor_I29 domain (I29, PF08246). Motif1, 2, 3, 4, 6, 8, 9, 10, and 13 were characterized as the Petidase_C1 domain (PF00112), while motif12 was characterized as the granulin-like domain (PF00396). Motif15 was peculiar to the CTB subfamily members. The CaPLCP11 and CaPLCP15 of the CTB subfamily and SAG12 subfamily members did not contain the Inhibitor-I29 domain. Members of the same CaPLCP subfamily contained similar conserved motifs.
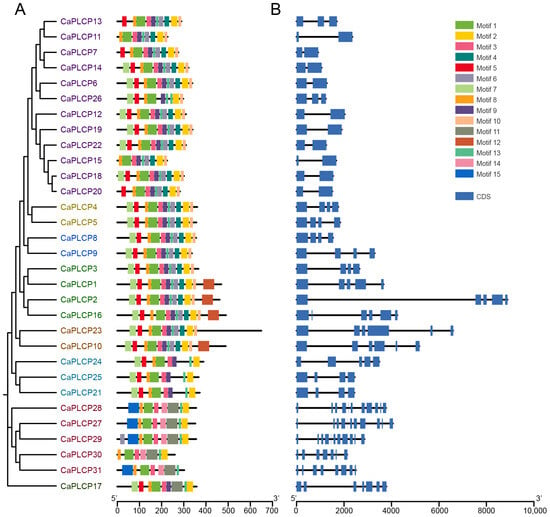
Figure 3.
Conserved motifs and gene structure of CaPLCP family members. (A) The distribution of conserved motifs of PLCP family members in peppers. Fifteen conserved motifs are represented by blocks with different colors. (B) The gene structure of PLCP family members in peppers. Blue blocks and black lines represent exons and introns, respectively.
The gene lengths varied significantly among different CaPLCP members, and the number of exons ranged from 2 to 11. However, the exon number of genes within the same subfamily was similar (Figure 3B). Subfamily I members contained 2 to 3 exons, the number of exons in subfamily II and III was 4, subfamily IV members contained 4 to 6 exons, subfamily V members all contained 6 exons, subfamily VI members all contained 5 exons, and subfamily VII members have the largest number of exons, ranging from 7 to 11. These results also confirm the accuracy of the clustering of CaPLCP members.
3.5. Cis-Acting Elements in the Promoters of CaPLCP Genes
To further understand the transcriptional regulation and gene function of CaPLCPs, we analyzed the cis-acting elements in the 2 kb promoters of CaPLCP genes (Figure 4). Six typical light-responsive elements were identified in the promoters of CaPLCPs, including Box4, G-box1, G-box2, GT1-motif, MRE, and TCT-motif. Each member promoter contained at least one type of light-responsive element. Additionally, nine hormone response-related elements were identified in the promoters of 31 CaPLCP members, including auxin-responsive elements (AuxRR-core and TGA-element), salicylic acid and auxin-responsive element (as-1), abscisic acid-responsive element (ABRE), gibberellin-responsive elements (GARE-motif, P-box, and TATC-box), and jasmonic acid-responsive element (GCTCA-motif). Furthermore, many CaPLCP promoters contained cis-acting elements related to stress response, such as low-temperature responsiveness element (LTR), MYB binding site involved in drought inducibility (MBS), anaerobic induction-related elements (ARE and GC-motif), and wound response elements (W-box, WRE3, and WUN-motif). These results suggest that CaPLCPs may have diverse roles in response to different hormones and stress.

Figure 4.
The distribution of cis-acting elements in the promoters of CaPLCP family genes. The inverted triangles with different colors represent different cis-acting elements, with their names show on the right side. The 2000 bp upstream sequences of CaPLCPs were used for cis-acting element prediction through PlantCARE. The functions of cis-acting elements are listed in Supplementary Table S4.
3.6. Tissue Expression Profile of CaPLCP Genes
To deeply understand the tissue expression patterns of CaPLCP family members, expression data of 31 CaPLCP genes were downloaded from PepperHub and used to draw the tissue expression profile (Figure 5). CaPLCP2, CaPLCP10, CaPLCP16, CaPLCP17, CaPLCP25, CaPLCP27, and CaPLCP29 were expressed in almost all the tested tissues. CaPLCP4 was expressed in roots, stems, and developing flowers and seeds. CaPLCP1 and CaPLCP21 were mainly expressed in the developing flowers and seeds. CaPLCP15, CaPLCP18, CaPLCP19, and CaPLCP22 exhibited higher expression in stems and developing leaves. CaPLCP3, CaPLCP5, and CaPLCP24 were mainly expressed during seed development. CaPLCP11, CaPLCP12, and CaPLCP13 were primarily expressed in developing flowers, with CaPLCP11 highly expressed during late flower development and CaPLCP12 during middle flower development. While CaPLCP6, CaPLCP7, CaPLCP14, CaPLCP20, CaPLCP26, CaPLCP30, and CaPLCP31 were almost unexpressed or lowly expressed in all tissues. The different tissue expression patterns of CaPLCP family genes indicate that they may perform distinct biological functions at different stages of pepper growth and development.
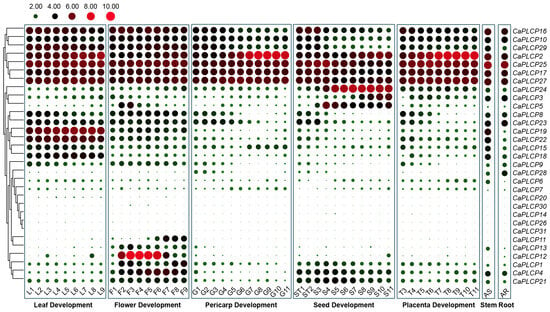
Figure 5.
The expression profiles of CaPLCP family genes in different tissues and developmental stages. RNA-seq data were downloaded from PepperHub. Log2 and row scale normalization were applied to the FPKM values of RNA-seq data. Red and green circles indicate high and low expression levels for each gene, respectively. The specific development stages of the samples refer to Liu et al. (2017) [34]. The FPKM values of CaPLCP genes are listed in Supplementary Table S1.
3.7. GO Enrichment of CaPLCP Genes
The GO enrichment analysis of the 31 CaPLCP genes revealed significant enrichment in several biological processes, including anther wall tapetum development, the macromolecule catabolic process, programmed cell death involved in cell development, proteolysis, and the protein metabolic process. In terms of cell components, CaPLCP members were mainly enriched in the lysosomes, vacuoles, extracellular regions, and cell walls. The GO molecular function terms included catalytic activity acting on a protein and cysteine-type endopeptidase activity (Figure 6). Notably, CaPLCP members were significantly enriched in biological processes related to male fertility development, such as anther wall tapetum development, anther development, and pollen development.
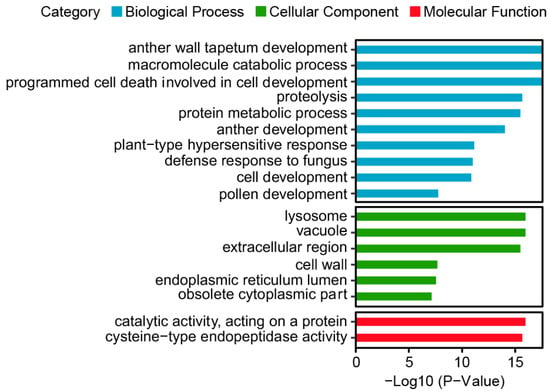
Figure 6.
The GO enrichment analysis of CaPLCP family genes. The blue, green, and red bars represent biological processes, cellular components, and molecular functions, respectively. The Y-axis represents GO terms, and the X-axis represents -Log10 (p-value).
3.8. Expression Analysis of CaPLCP Genes in Male Sterile Materials
To further explore the function of CaPLCPs in male reproductive development and identify the CaPLCP genes involved in the regulation of the male sterility of peppers, CaPLCP genes with an FPKM value greater than 15 at any stage of flower development (F1–F9) were selected based on tissue expression data from PepperHub (the FPKM values of CaPLCP genes are listed in Supplementary Table S1). A total of 19 candidate CaPLCP genes were screened. Floral buds were divided into three stages according to their length: S1, S2, and S3. The floral buds of MF (male fertile pepper material) and MS (male sterile pepper material) were collected at these three stages, and the relative expression levels of the 19 candidate CaPLCPs were detected, respectively. The results showed that the expression of CaPLCP1, CaPLCP5, CaPLCP13, CaPLCP17, and CaPLCP19 was significantly decreased in MS at the S1 stage (Figure 7A,D,I,K,L). At the S2 stage, the expressions of CaPLCP2, CaPLCP11, CaPLCP13, and CaPLCP21 in the floral buds of MS were significantly decreased (Figure 7B,G,I,M). At the S3 stage, the expression of CaPLCP1, CaPLCP5, CaPLCP11, CaPLCP12, CaPLCP13, and CaPLCP21 were significantly reduced in the floral buds of MS (Figure 7A,D,G–I,M). The expression of CaPLCP1 in MS was significantly lower than in MF at all three stages (Figure 7A). The expression of CaPLCP11 and CaPLCP21 was significantly decreased in MS at two consecutive stages (S2 and S3) (Figure 7G,M), while the expression of CaPLCP13 was significantly lower at two consecutive stages (S1 and S2) (Figure 7I). In addition, the expression of some CaPLCP genes, such as CaPLCP22, CaPLCP23, and CaPLCP29, was significantly increased in the floral buds of MS compared to MF (Figure 7N,O,S).
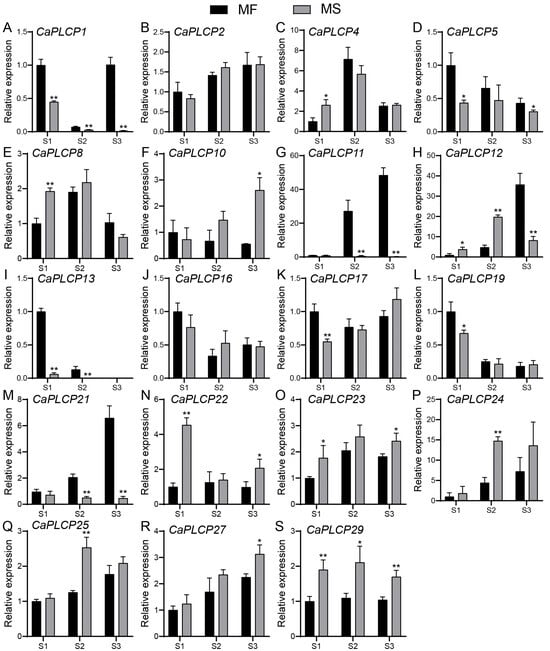
Figure 7.
The expression of CaPLCP members in MF and MS floral buds at three stages. The stages are divided according to the length of the floral buds: S1 (F1–F3, <5 mm), S2 (F4–F6, 5–10 mm), and S3 (F7–F10, >10 mm). S1, S2, and S3 represent stage 1, stage 2, and stage 3, respectively. MF and MS represent male fertile and male sterile pepper materials, respectively. Error bars indicate the standard deviations (SDs). ** p < 0.01, * p < 0.05, Student’s t-test.
4. Discussion
4.1. Characterization of the CaPLCPs
Papain-like cysteine proteases (PLCPs) are major peptidases found across organisms [2]. The number of PLCP members varies significantly across different species. For instance, the soybean has the highest number of PLCP genes identified to date, with 97 members [21]. Upland cotton contains 78 members [19], while Arabidopsis has 31 members [3], rice has 33 members [20], grapes have 23 members [22], papaya has 33 members [37], and rubber has 43 members [38]. In this study, we identified 31 CaPLCP genes within the pepper genome (Table S3). The variation in the number of members may be attributed to whole-genome duplication, tandem duplication, and large-scale segmental duplication [19,21,37].
The phylogenetic analysis within the species revealed that the 31 CaPLCPs were divided into eight clades, with members of the same clades containing similar conserved motifs and exon numbers (Figure 1 and Figure 3). Previous studies have generally divided PLCP members into nine subfamilies: RD21, CEP, XCP, XBCP3, THI, SAG12, RD19, ALP, and CTB [3,19,22]. In the phylogenetic tree constructed using both CaPLCPs and AtPLCPs, we found that SAG12 and THI subfamilies as well as XBCP and RD21 subfamilies could not be clearly distinguished (Figure S1). We speculate that it is due to the incomplete differentiation of CaPLCPs during the evolution of peppers.
4.2. Cis-Acting Elements and the Potential Functions of CaPLCPs
To date, there are very limited reports on the function of PLCP genes in peppers. Cis-acting elements are crucial molecular switches that regulate dynamic gene activity networks and control various biological processes [39]. The prediction of cis-acting elements can offer valuable insights into the expression and function of CaPLCP genes. Abundant cis-acting elements associated with hormone and stress responses were identified in the CaPLCP promoters, suggesting that CaPLCPs are involved in a complex regulatory network in hormone and stress responses during plant development (Figure 4). The as-1 element was identified in the promoters of several salicylic acid (SA)-inducible glutathione S-transferase (GST) genes, activated by SA through oxidizing substances [40,41]. SA is an important plant hormone that plays a role in responding to abiotic stresses, biotic stresses, and pathogenic mechanisms [42]. The as-1 and the TCA-element (another salicylic acid-responsive element) were widely identified in the promoters of multiple CaPLCP members. For instance, the promoter of CaPLCP8 contained five as-1 and one TCA-element, CaPLCP27 contained four as-1, CaPLCP26 contained three as-1, CaPLCP29 contained two each of TCA-element and as-1, and CaPLCP1 contained three TCA-element and one as-1. Jasmonic acid (JA) is considered a stress hormone implicated in plant responses to biotic and abiotic stress [43,44]. The GCTCA-motif, related to JA response, was also detected in the promoters of several CaPLCP genes. For example, the promoter of CaPLCP8 contained five, CaPLCP27 contained four, and CaPLCP26 contained three. Moreover, eight consecutive wun-motifs, associated with wound stress responses [45,46], were detected in the promoter of CaPLCP4. The promoter of CaPLCP26 contained five MBS elements, and the MYB transcription factors can bind to the MBS cis-element, which is involved in drought inducibility [47,48]. These findings indicate that CaPLCP members may also play significant roles in peppers’ response to abiotic stress and plant immunity.
4.3. Potential Role of CaPLCPs in the Regulation of Male Sterility in Pepper
The pepper is an important vegetable crop with obvious heterosis [49]. The utilization of male sterile lines is an effective method to reduce the cost of hybrid production and ensure high breed purity. Male gamete development in plants is a finely tuned process, and any variation that interferes with anther or pollen development may result in male sterility [50]. The tapetum is the innermost layer among the four somatic layers of anther lobes and undergoes apoptosis mediated by programmed cell death (PCD) at the tetrad stage, leading to pollen grain maturation [6]. The exact timing of tapetal PCD is critical for plant reproductive ability [6,51]. Proteases, including cysteine proteases and aspartic proteases, play essential roles in regulating PCD in many plants, particularly in tapetum PCD [52]. Currently, research on male sterile genes in peppers mainly focuses on several transcription factors, including the R2R3-MYB transcription factor Capana10g000198 [53], the PHD-finger transcription factor Msc-2 [54], and the bHLH transcription factor Msc-1 [55]. From the GO enrichment analysis of CaPLCP members, we observed significant enrichment in biological processes related to male fertility development, including anther wall tapetum development, anther development, and pollen development (Figure 6). Moreover, qRT-PCR results revealed that the expression of CaPLCP1 was significantly reduced in the flowers of MS compared to MF at all three stages. The expression of CaPLCP11, CaPLCP13, and CaPLCP21 was significantly decreased during two consecutive stages in the flowers of MS. Additionally, the expression of CaPLCP5, CaPLCP12, CaPLCP17, and CaPLCP19 decreased significantly in the flowers of MS only at a certain stage (Figure 7). Our phylogenetic analysis revealed that CaPLCP5 and Arabidopsis CEP1 were in the same clade. CEP1 is an irreplaceable executor during tapetal cell PCD, and the loss of CPE1 function resulted in abnormal PCD and reduced pollen fertility [7]. Similarly, CaPLCP12 is in the same clade as Arabidopsis THI1 (AtCP51), which is expressed during anther development. Silencing AtCP51 using RNAi caused premature tapetum degeneration and male sterility [56]. OsCP1 and NtCP56, the homologous genes of THI1 in rice and tobacco, have also been confirmed to be involved in male reproductive development [57,58]. CaPLCP19 shares a clade with Arabidopsis PAP2, whose homolog in Brassica napus (BnaC.CP20.1) is required for tapetum degradation and the formation of pollen wall [59]. These findings suggest that CaPLCP5, CaPLCP12, and CaPLCP19 may also be involved in regulating tapetal degradation and pollen fertility in peppers. However, the specific functions of CaPLCP5, CaPLCP12, and CaPLCP19 in the male reproductive development of peppers and whether other CaPLCP genes with significantly reduced expression in MS are also involved in pollen fertility regulation still need further investigation using virus-induced gene silencing (VIGS) or other technologies. Given the material specificity of MS, we cannot exclude the possibility that other CaPLCP members are involved in regulating male sterility in peppers. Our results provide valuable insights into the functions of CaPLCP family members and will accelerate the analysis of the mechanism underlying male sterility in peppers.
5. Conclusions
Thirty-one CaPLCP members were identified in the whole genome of peppers, and these were categorized into eight clades. Comprehensive analyses were conducted on gene segmental duplication, gene structure, conserved motifs, cis-acting elements, and expression patterns. The prediction of cis-acting elements indicated that CaPLCP members may play a crucial role in responding to abiotic stress and plant immunity in peppers. Additionally, the expression levels of CaPLCP1, CaPLCP5, CaPLCP11, CaPLCP12, CaPLCP13, CaPLCP17, CaPLCP19, and CaPLCP21 were significantly reduced in MS, suggesting their potential functions in the male reproductive development of peppers. Our research provides a significant foundation for the further analysis of the function of CaPLCP members and the mechanism underlying the male sterility of peppers.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/horticulturae10080892/s1, Figure S1: Phylogenetic tree of PLCP family members from peppers and Arabidopsis; Figure S2: The sequence logo of different conserved motifs identified in the CaPLCPs.; Table S1: FPKM value of CaPLCP genes from PepperHub; Table S2: Primer list; Table S3: Detailed information of CaPLCPs; Table S4: A list of cis-acting element functions in the prompters of CaPLCP genes.
Author Contributions
R.C., B.W. and M.Z.: conceptualization. R.C. and S.H.: methodology and writing-original draft. X.C., J.T., H.Z. and J.W.: software, investigation, and formal analysis. B.W. and M.Z.: funding acquisition. R.C. and M.Z.: writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the development project of the Hubei Province Department of Science and Technology (2022BBA0074, 2022BBA0060) and the Hubei Provincial Funding Program to Support High-Quality Development of the Seed Industry (HBZY2023B004).
Data Availability Statement
The data presented in this study are available in PepperHub at [https://pubmed.ncbi.nlm.nih.gov/28343897/, accessed on 22 November 2022], reference number [34].
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Van Der Hoorn, R.A. Plant proteases: From phenotypes to molecular mechanisms. Annu. Rev. Plant Biol. 2008, 59, 191–223. [Google Scholar] [CrossRef]
- Liu, H.; Hu, M.; Wang, Q.; Cheng, L.; Zhang, Z. Role of papain-like cysteine proteases in plant development. Front. Plant Sci. 2018, 9, 1717. [Google Scholar] [CrossRef] [PubMed]
- Richau, K.H.; Kaschani, F.; Verdoes, M.; Pansuriya, T.C.; Niessen, S.; Stüber, K.; Colby, T.; Overkleeft, H.S.; Bogyo, M.; Van der Hoorn, R.A. Subclassification and biochemical analysis of plant papain-like cysteine proteases display subfamily-specific characteristics. Plant Physiol. 2012, 158, 1583–1599. [Google Scholar] [CrossRef] [PubMed]
- Turk, V.; Turk, B.; Turk, D. Lysosomal cysteine proteases: Facts and opportunities. EMBO J. 2001, 20, 4629–4633. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Johnson, B.J.; Kositsup, B.; Beers, E.P. Exploiting secondary growth in Arabidopsis. Construction of xylem and bark cDNA libraries and cloning of three xylem endopeptidases. Plant Physiol. 2000, 123, 1185–1196. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Lou, Y.; Xu, X.; Yang, Z.N. A genetic pathway for tapetum development and function in Arabidopsis. J. Integr. Plant Biol. 2011, 53, 892–900. [Google Scholar] [CrossRef]
- Zhang, D.; Liu, D.; Lv, X.; Wang, Y.; Xun, Z.; Liu, Z.; Li, F.; Lu, H. The cysteine protease CEP1, a key executor involved in tapetal programmed cell death, regulates pollen development in Arabidopsis. Plant Cell 2014, 26, 2939–2961. [Google Scholar] [CrossRef] [PubMed]
- Lohman, K.N.; Gan, S.; John, M.C.; Amasino, R.M. Molecular analysis of natural leaf senescence in Arabidopsis thaliana. Physiol. Plant 1994, 92, 322–328. [Google Scholar] [CrossRef]
- James, M.; Poret, M.; Masclaux-Daubresse, C.; Marmagne, A.; Coquet, L.; Jouenne, T.; Chan, P.; Trouverie, J.; Etienne, P. SAG12, a major cysteine protease involved in nitrogen allocation during senescence for seed production in Arabidopsis thaliana. Plant Cell Physiol. 2018, 59, 2052–2063. [Google Scholar] [CrossRef]
- Singh, S.; Giri, M.K.; Singh, P.K.; Siddiqui, A.; Nandi, A.K. Down-regulation of OsSAG12-1 results in enhanced senescence and pathogen-induced cell death in transgenic rice plants. J. Biosci. 2013, 38, 583–592. [Google Scholar] [CrossRef]
- Singh, S.; Singh, A.; Nandi, A.K. The rice OsSAG12-2 gene codes for a functional protease that negatively regulates stress-induced cell death. J. Biosci. 2016, 41, 445–453. [Google Scholar] [CrossRef]
- Koizumi, M.; Yamaguchi-Shinozaki, K.; Tsuji, H.; Shinozaki, K. Structure and expression of two genes that encode distinct drought-inducible cysteine proteinases in Arabidopsis thaliana. Gene 1993, 129, 175–182. [Google Scholar] [CrossRef]
- Zang, Q.W.; Wang, C.X.; Li, X.Y.; Guo, Z.A.; Jing, R.L.; Zhao, J.; Chang, X.P. Isolation and characterization of a gene encoding a polyethylene glycol-induced cysteine protease in common wheat. J. Biosci. 2010, 35, 379–388. [Google Scholar] [CrossRef] [PubMed]
- Xiao, H.J.; Yin, Y.X.; Chai, W.G.; Gong, Z.H. Silencing of the CaCP gene delays salt-and osmotic-induced leaf senescence in Capsicum annuum L. Int. J. Mol. Sci. 2014, 15, 8316–8334. [Google Scholar] [CrossRef] [PubMed]
- Misas-Villamil, J.C.; van der Hoorn, R.A.; Doehlemann, G. Papain-like cysteine proteases as hubs in plant immunity. New Phytol. 2016, 212, 902–907. [Google Scholar] [CrossRef]
- Bernoux, M.; Timmers, T.; Jauneau, A.; Briere, C.; de Wit, P.J.; Marco, Y.; Deslandes, L. RD19, an Arabidopsis cysteine protease required for RRS1-R–mediated resistance, is relocalized to the nucleus by the Ralstonia solanacearum PopP2 effector. Plant Cell 2008, 20, 2252–2264. [Google Scholar] [CrossRef]
- Shindo, T.; Misas-Villamil, J.C.; Hörger, A.C.; Song, J.; van der Hoorn, R.A. A role in immunity for Arabidopsis cysteine protease RD21, the ortholog of the tomato immune protease C14. PLoS ONE 2012, 7, e29317. [Google Scholar] [CrossRef] [PubMed]
- Ilyas, M.; Hörger, A.C.; Bozkurt, T.O.; Van Den Burg, H.A.; Kaschani, F.; Kaiser, M.; Belhaj, K.; Smoker, M.; Joosten, M.H.; Kamoun, S. Functional divergence of two secreted immune proteases of tomato. Curr. Biol. 2015, 25, 2300–2306. [Google Scholar] [CrossRef]
- Zhang, S.; Xu, Z.; Sun, H.; Sun, L.; Shaban, M.; Yang, X.; Zhu, L. Genome-wide identification of papain-like cysteine proteases in Gossypium hirsutum and functional characterization in response to Verticillium dahliae. Front. Plant Sci. 2019, 10, 134. [Google Scholar] [CrossRef] [PubMed]
- Niño, M.C.; Kang, K.K.; Cho, Y.G. Genome-wide transcriptional response of papain-like cysteine protease-mediated resistance against Xanthomonas oryzae pv. oryzae in rice. Plant Cell Rep. 2020, 39, 457–472. [Google Scholar] [CrossRef]
- Yuan, S.; Ke, D.; Li, R.; Li, X.; Wang, L.; Chen, H.; Zhang, C.; Huang, Y.; Chen, L.; Hao, Q. Genome-wide survey of soybean papain-like cysteine proteases and their expression analysis in root nodule symbiosis. BMC Plant Biol. 2020, 20, 517. [Google Scholar] [CrossRef]
- Kang, J.; Gong, P.; Ge, M.; Sadeghnezhad, E.; Liu, Z.; Zhang, M.; Shangguan, L.; Fang, J. The PLCP gene family of grapevine (Vitis vinifera L.): Characterization and differential expression in response to Plasmopara Viticola. BMC Plant Biol. 2021, 21, 499. [Google Scholar]
- Chen, L.; Liu, Y.G. Male sterility and fertility restoration in crops. Annu. Rev. Plant Biol. 2014, 65, 579–606. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.; Tosatto, S.C.; Paladin, L.; Raj, S.; Richardson, L.J. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Gonzales, N.R.; Gwadz, M.; Lu, S.; Marchler, G.H.; Song, J.S.; Thanki, N.; Yamashita, R.A. The conserved domain database in 2023. Nucleic Acids Res. 2023, 51, D384–D388. [Google Scholar] [CrossRef] [PubMed]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; De Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Yu, H.; Deng, Y.; Zheng, J.; Liu, M.; Ou, L.; Yang, B.; Dai, X.; Ma, Y.; Feng, S. PepperHub, an informatics hub for the chili pepper research community. Mol. Plant 2017, 10, 1129–1132. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Yang, C.; Gao, H.; Shi, C.; Zhang, Z.; Lu, G.; Shen, X.; Tang, Y.; Li, F.; Lu, Y. Induced mutation in ELONGATED HYPOCOTYL5 abolishes anthocyanin accumulation in the hypocotyl of pepper. Theor. Appl. Genet. 2022, 135, 3455–3468. [Google Scholar] [CrossRef]
- Wan, H.; Yuan, W.; Ruan, M.; Ye, Q.; Wang, R.; Li, Z.; Zhou, G.; Yao, Z.; Zhao, J.; Liu, S. Identification of reference genes for reverse transcription quantitative real-time PCR normalization in pepper (Capsicum annuum L.). Biochem. Biophys. Res. Commun. 2011, 416, 24–30. [Google Scholar] [CrossRef]
- Liu, J.; Sharma, A.; Niewiara, M.J.; Singh, R.; Ming, R.; Yu, Q. Papain-like cysteine proteases in Carica papaya: Lineage-specific gene duplication and expansion. BMC Genom. 2018, 19, 26. [Google Scholar] [CrossRef]
- Zou, Z.; Xie, G.; Yang, L. Papain-like cysteine protease encoding genes in rubber (Hevea brasiliensis): Comparative genomics, phylogenetic, and transcriptional profiling analysis. Planta 2017, 246, 999–1018. [Google Scholar] [CrossRef]
- Yamaguchi-Shinozaki, K.; Shinozaki, K. Organization of cis-acting regulatory elements in osmotic-and cold-stress-responsive promoters. Trends Plant Sci. 2005, 10, 88–94. [Google Scholar] [CrossRef]
- Xiang, C.; Miao, Z.H.; Lam, E. Coordinated activation of as-1-type elements and a tobacco glutathione S-transferase gene by auxins, salicylic acid, methyl-jasmonate and hydrogen peroxide. Plant Mol. Biol. 1996, 32, 415–426. [Google Scholar] [CrossRef] [PubMed]
- Garretón, V.; Carpinelli, J.; Jordana, X.; Holuigue, L. The as-1 promoter element is an oxidative stress-responsive element and salicylic acid activates it via oxidative species. Plant Physiol. 2002, 130, 1516–1526. [Google Scholar] [CrossRef] [PubMed]
- Hara, M.; Furukawa, J.; Sato, A.; Mizoguchi, T.; Miura, K. Abiotic stress and role of salicylic acid in plants. In Abiotic Stress Responses in Plants: Metabolism, Productivity and Sustainability; Springer: New York, NY, USA, 2012; pp. 235–251. [Google Scholar]
- Ghorbel, M.; Brini, F.; Sharma, A.; Landi, M. Role of jasmonic acid in plants: The molecular point of view. Plant Cell Rep. 2021, 40, 1471–1494. [Google Scholar] [CrossRef] [PubMed]
- Ruan, J.; Zhou, Y.; Zhou, M.; Yan, J.; Khurshid, M.; Weng, W.; Cheng, J.; Zhang, K. Jasmonic acid signaling pathway in plants. Int. J. Mol. Sci. 2019, 20, 2479. [Google Scholar] [CrossRef] [PubMed]
- Moin, M.; Bommineni, P.R.; Tyagi, W. Exploration of the pearl millet phospholipase gene family to identify potential candidates for grain quality traits. BMC Genom. 2024, 25, 581. [Google Scholar] [CrossRef]
- Siebertz, B.; Logemann, J.; Willmitzer, L.; Schell, J. cis-analysis of the wound-inducible promoter wun1 in transgenic tobacco plants and histochemical localization of its expression. Plant Cell 1989, 1, 961–968. [Google Scholar]
- Abe, H.; Yamaguchi-Shinozaki, K.; Urao, T.; Iwasaki, T.; Hosokawa, D.; Shinozaki, K. Role of Arabidopsis MYC and MYB homologs in drought-and abscisic acid-regulated gene expression. Plant Cell 1997, 9, 1859–1868. [Google Scholar]
- Su, L.T.; Li, J.W.; Liu, D.Q.; Zhai, Y.; Zhang, H.J.; Li, X.W.; Zhang, Q.L.; Wang, Y.; Wang, Q.Y. A novel MYB transcription factor, GmMYBJ1, from soybean confers drought and cold tolerance in Arabidopsis thaliana. Gene 2014, 538, 46–55. [Google Scholar] [CrossRef]
- Shu, H.; Zhou, H.; Mu, H.; Wu, S.; Jiang, Y.; Yang, Z.; Hao, Y.; Zhu, J.; Bao, W.; Cheng, S.; et al. Integrated analysis of mRNA and non-coding RNA transcriptome in pepper (Capsicum chinense) hybrid at seedling and flowering stages. Front. Genet. 2021, 12, 685788. [Google Scholar] [CrossRef] [PubMed]
- An, X.; Ma, B.; Duan, M.; Dong, Z.; Liu, R.; Yuan, D.; Hou, Q.; Wu, S.; Zhang, D.; Liu, D. Molecular regulation of ZmMs7 required for maize male fertility and development of a dominant male-sterility system in multiple species. Proc. Natl. Acad. Sci. USA 2020, 117, 23499–23509. [Google Scholar] [CrossRef]
- Varnier, A.L.; Mazeyrat-Gourbeyre, F.; Sangwan, R.S.; Clément, C. Programmed cell death progressively models the development of anther sporophytic tissues from the tapetum and is triggered in pollen grains during maturation. J. Struct. Biol. 2005, 152, 118–128. [Google Scholar] [CrossRef]
- Buono, R.A.; Hudecek, R.; Nowack, M.K. Plant proteases during developmental programmed cell death. J. Exp. Bot. 2019, 70, 2097–2112. [Google Scholar] [CrossRef] [PubMed]
- Dong, J.; Hu, F.; Guan, W.; Yuan, F.; Lai, Z.; Zhong, J.; Liu, J.; Wu, Z.; Cheng, J.; Hu, K. A 163-bp insertion in the Capana10g000198 encoding a MYB transcription factor causes male sterility in pepper (Capsicum annuum L.). Plant J. 2023, 113, 521–535. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Li, T.; Ai, Y.; Lu, Q.; Wang, Y.; Wu, L.; Liu, J.; Sun, L.; Shen, H. Phenotypic, genetic, and molecular function of msc-2, a genic male sterile mutant in pepper (Capsicum annuum L.). Theor. Appl. Genet. 2020, 133, 843–855. [Google Scholar] [CrossRef]
- Cheng, Q.; Wang, P.; Liu, J.; Wu, L.; Zhang, Z.; Li, T.; Gao, W.; Yang, W.; Sun, L.; Shen, H. Identification of candidate genes underlying genic male-sterile msc-1 locus via genome resequencing in Capsicum annuum L. Theor. Appl. Genet. 2018, 131, 1861–1872. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Dong, C.; Yu, J.; Shi, L.; Tong, C.; Li, Z.; Huang, J.; Liu, S. Cysteine Protease 51 (CP51), an anther-specific cysteine protease gene, is essential for pollen exine formation in Arabidopsis. Plant Cell Tissue Organ Cult. 2014, 119, 383–397. [Google Scholar] [CrossRef]
- Lee, S.; Jung, K.H.; An, G.; Chung, Y.Y. Isolation and characterization of a rice cysteine protease gene, OsCP1, using T-DNA gene-trap system. Plant Mol. Biol. 2004, 54, 755–765. [Google Scholar] [CrossRef]
- Zhang, X.M.; Wang, Y.; Lv, X.M.; Li, H.; Sun, P.; Lu, H.; Li, F.L. NtCP56, a new cysteine protease in Nicotiana tabacum L., involved in pollen grain development. J. Exp. Bot. 2009, 60, 1569–1577. [Google Scholar] [CrossRef]
- Song, L.; Zhou, Z.; Tang, S.; Zhang, Z.; Xia, S.; Qin, M.; Li, B.; Wen, J.; Yi, B.; Shen, J. Ectopic expression of BnaC.CP20. 1 results in premature tapetal programmed cell death in Arabidopsis. Plant Cell Physiol. 2016, 57, 1972–1984. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).