Comparative Analysis of Transcriptomes to Identify Genes during Bud Dormancy of Pyrus pyrifolia ‘Huanghua’
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Determination of Flower Bud Break Percentage and Respiration Intensity
2.3. RNA Extraction and Library Preparation for RNA Sequencing
2.4. Differential Expression Gene Analysis
2.5. GO and KEGG Enrichment Analysis of Differentially Expressed Genes
2.6. Real-Time Fluorescence Quantitative PCR
2.7. Statistical Analysis
3. Results
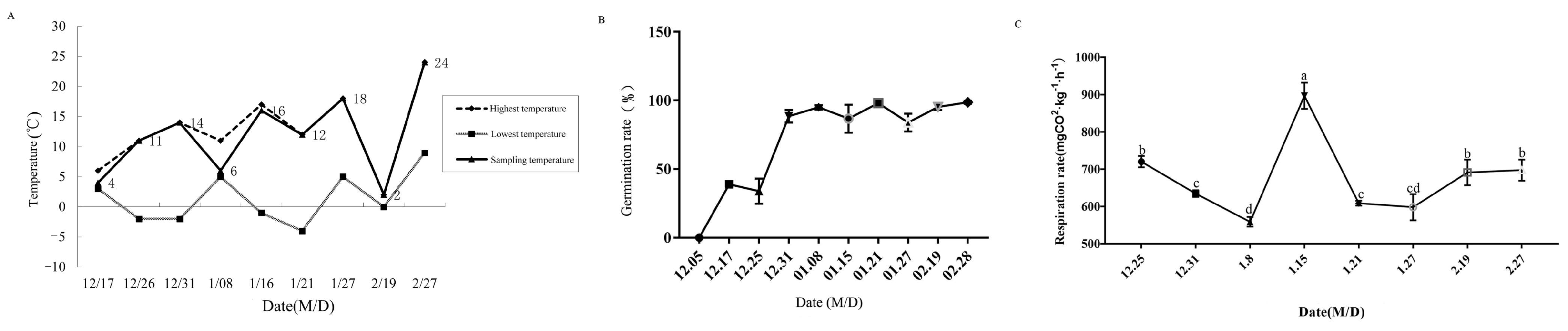
3.1. Physiological Indexes of Pyrus pyrifolia ‘Huanghua’ during Dormancy
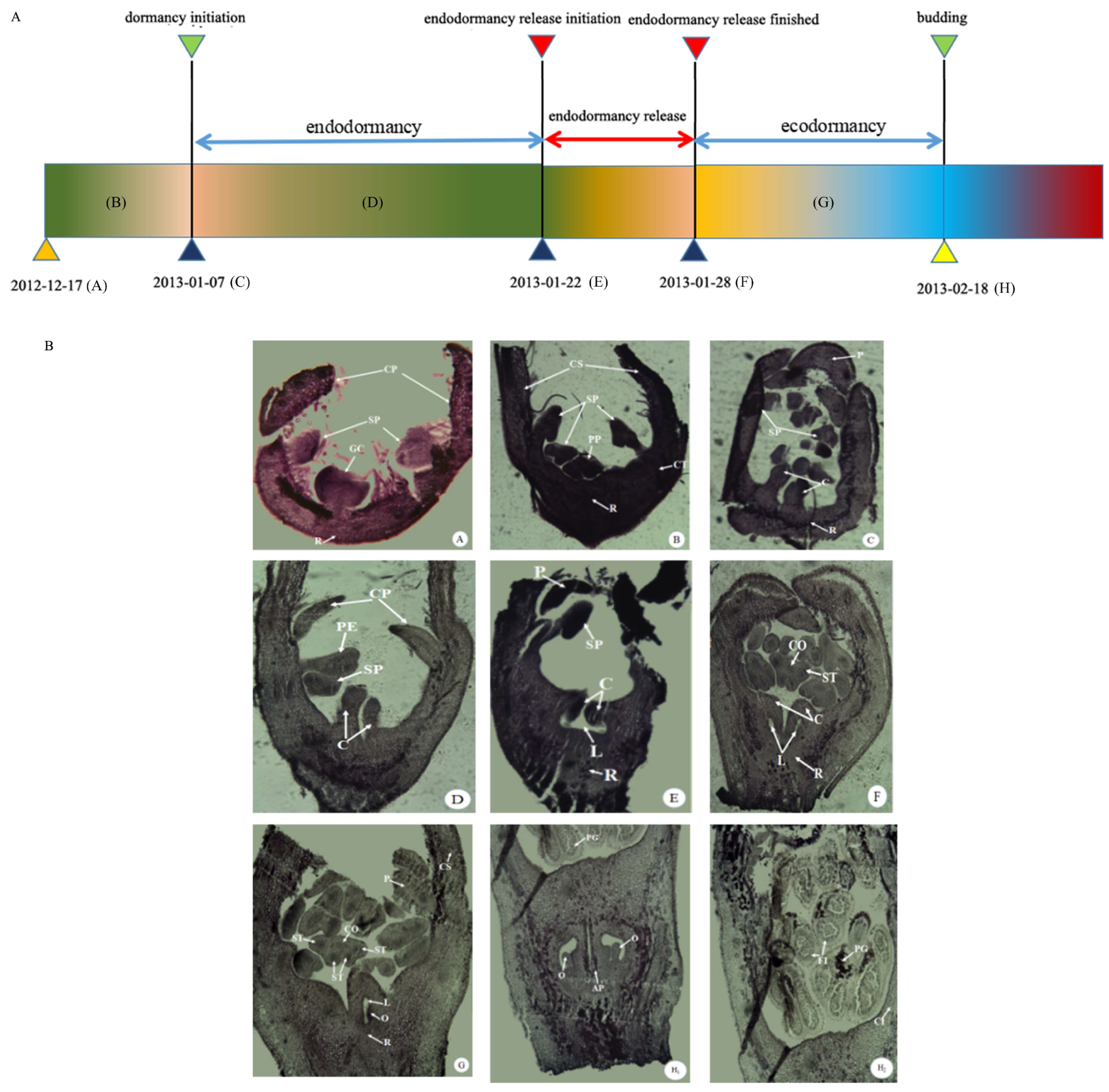
3.2. Histomorphological Analysis of Pear Buds during the Dormancy Process in 2012/2013 Year
3.3. Differentially Expressed Gene Analysis in Pyrus pyrifolia ‘Huanghua’
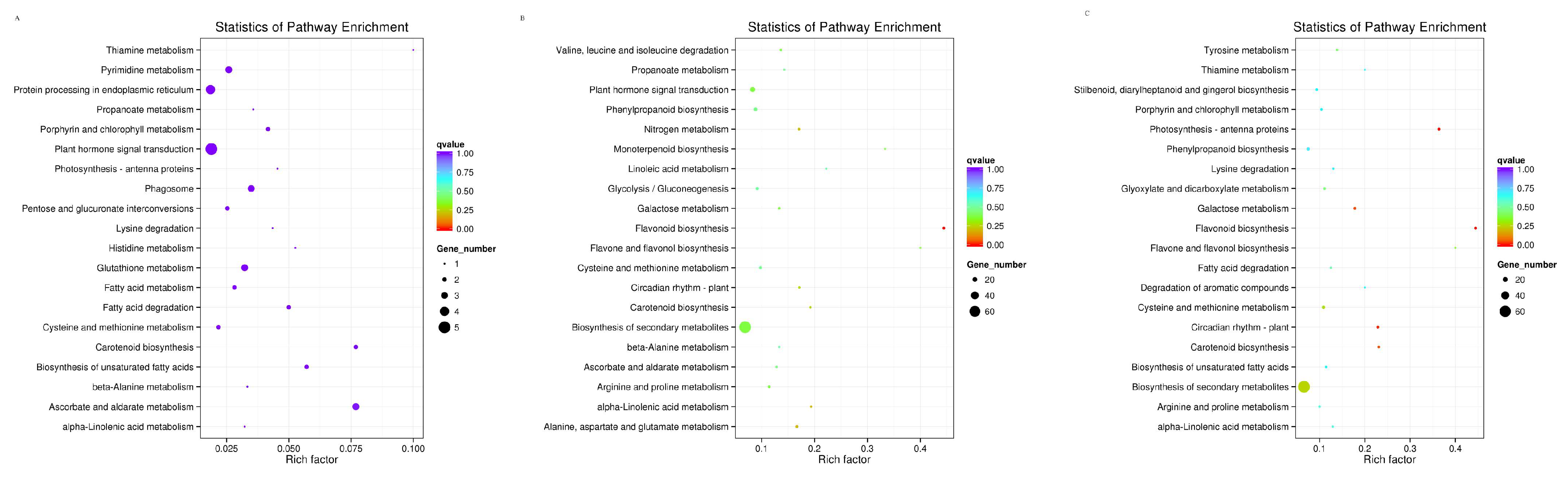
3.4. KEGG Functional Enrichment Analysis of Differentially Expressed Genes in Pyrus pyrifolia ‘Huanghua’
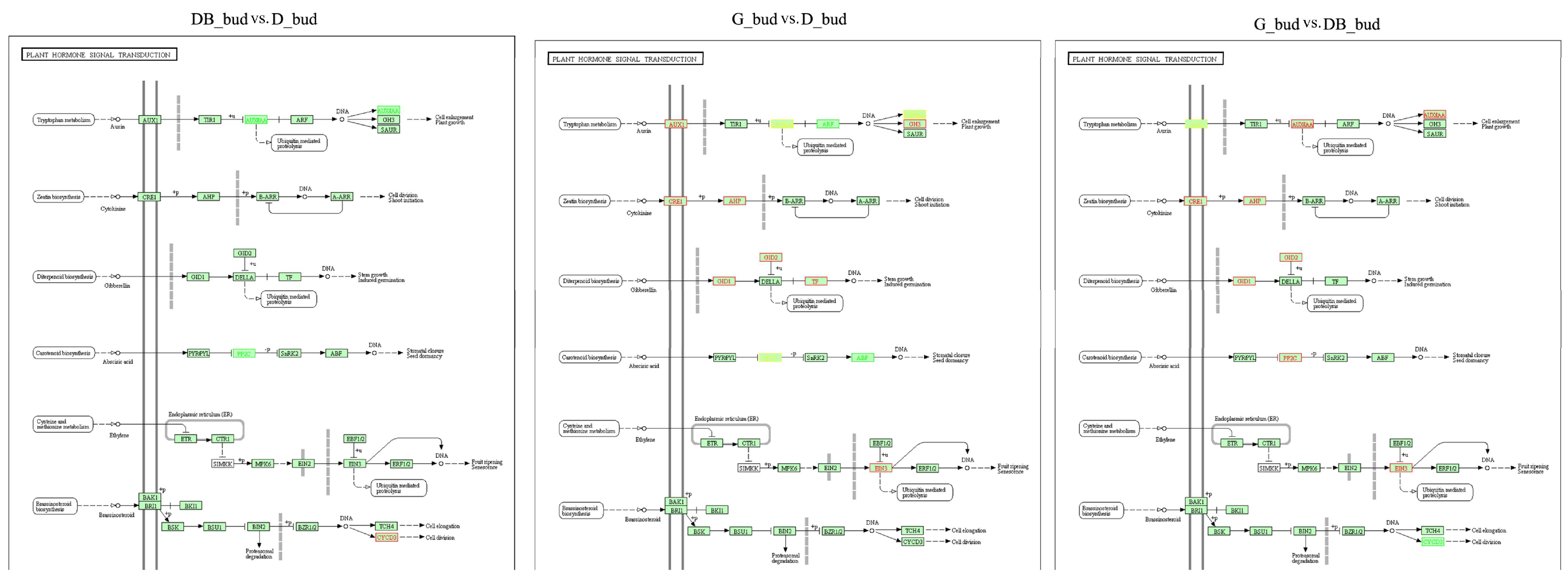
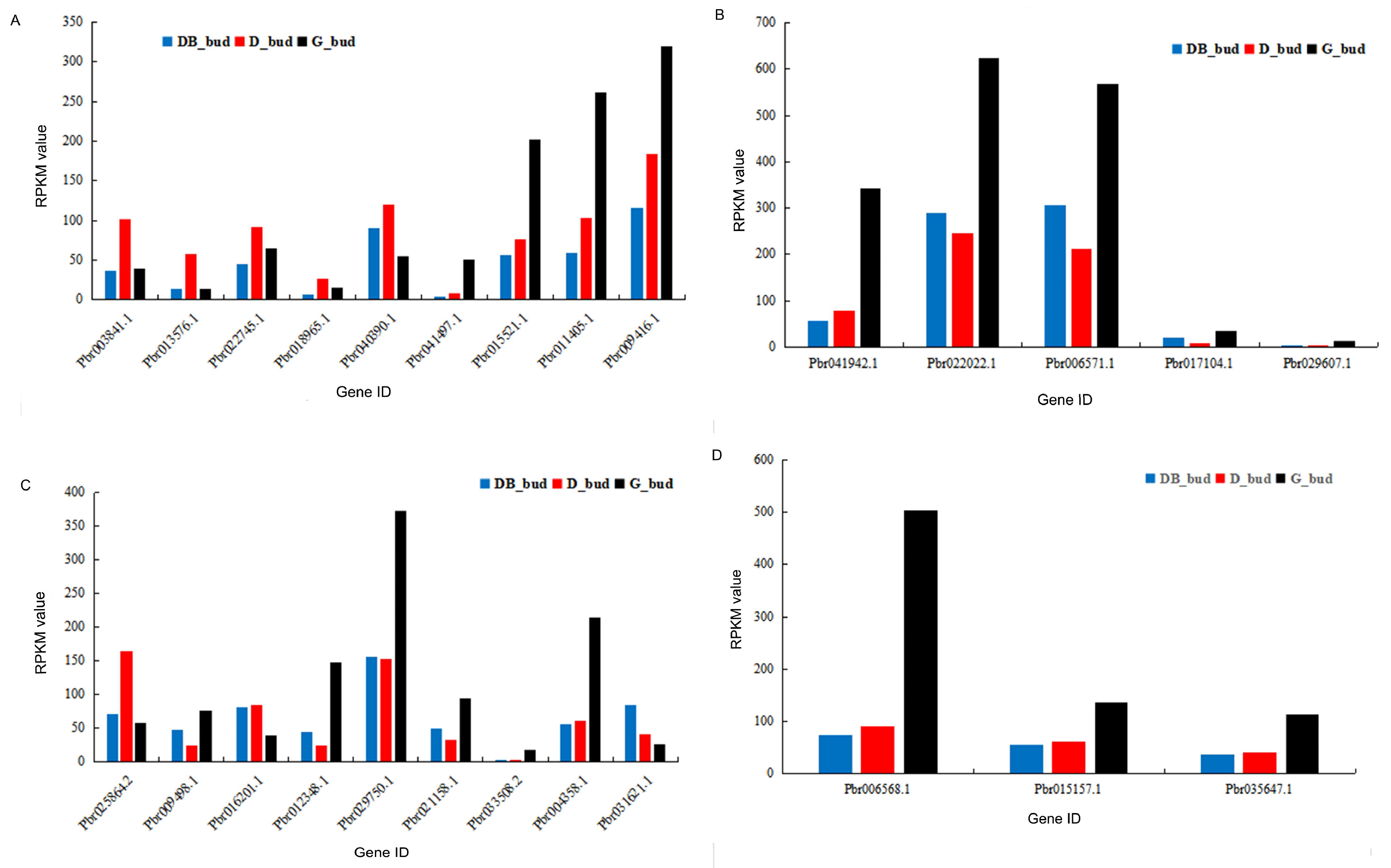
3.5. Expression Analysis of Differential Genes Related to Plant Hormone Metabolism in Pyrus pyrifolia ‘Huanghua’
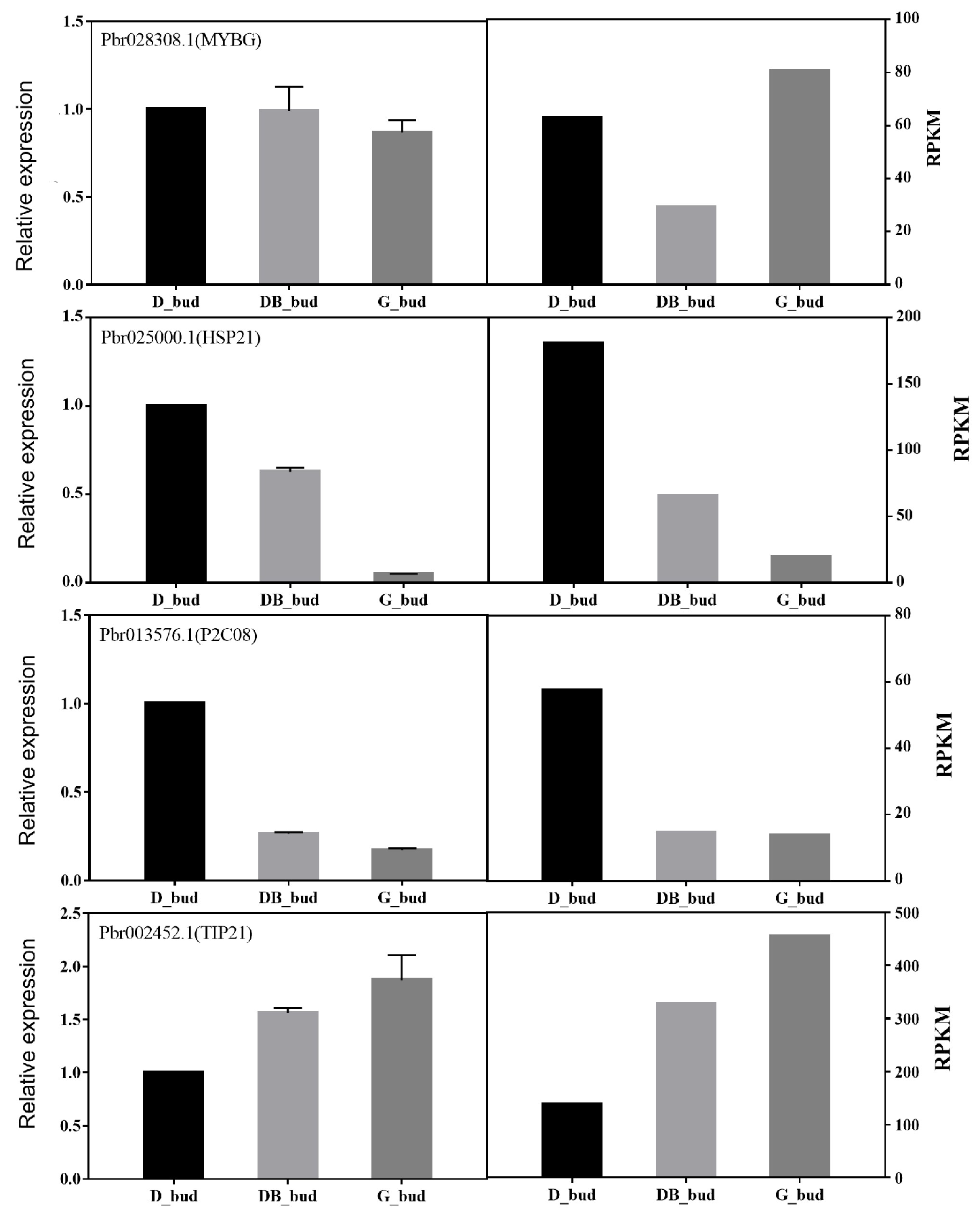
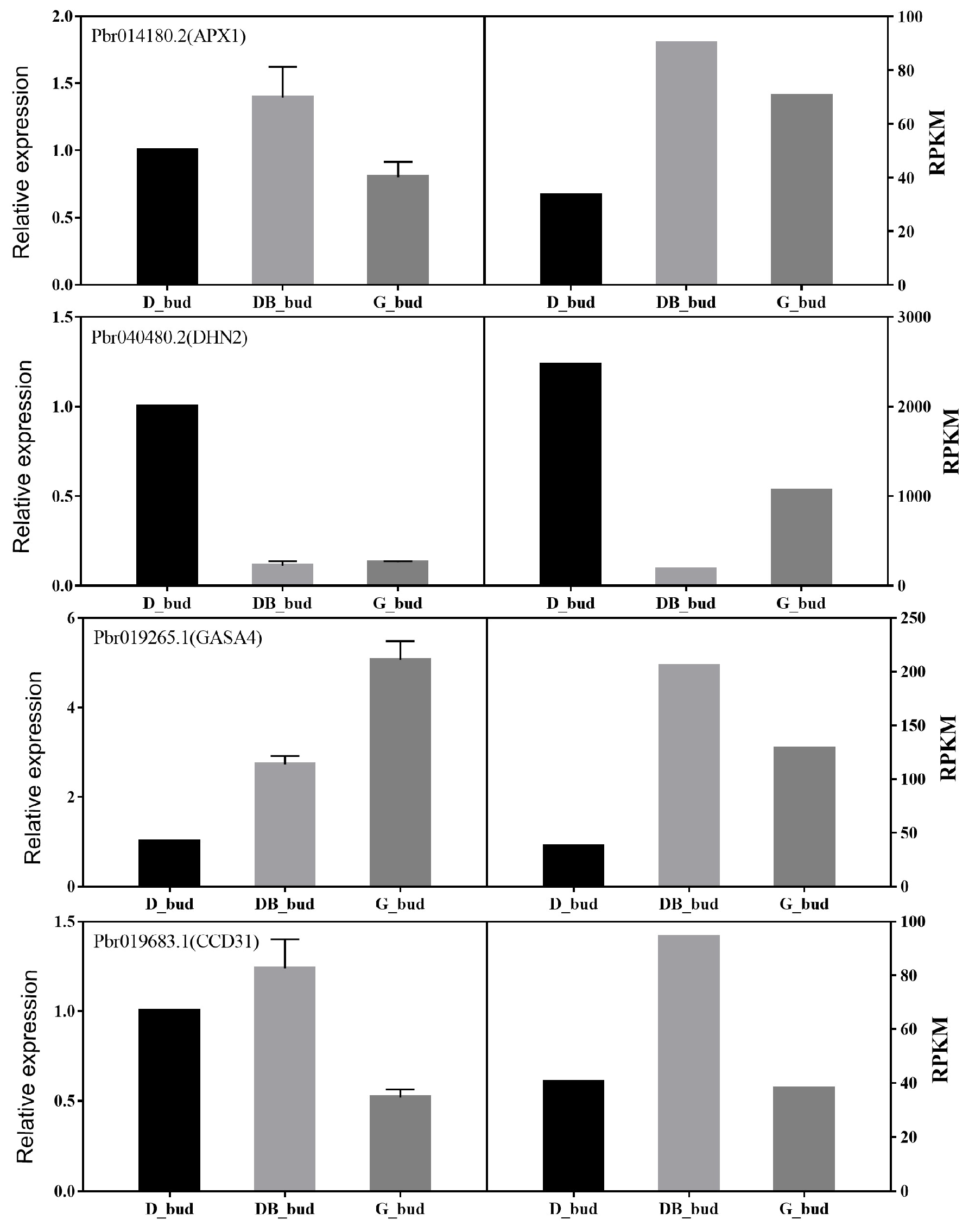
3.6. Expression Analysis of DEG in Pyrus pyrifolia ‘Huanghua’ by Real-Time PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wu, J.; Wang, Z.W.; Shi, Z.B.; Zhang, S.; Ming, R.; Zhu, S.L.; Khan, M.A.; Tao, S.T.; Korban, S.S.; Wang, H.; et al. The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res. 2013, 23, 396–408. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, J.Y.; Gao, W.Y.; Wang, Y.; Wang, H.Y.; Cao, J.G.; Huang, L.Q. Chemical Composition and Anti-inflammatory and Antioxidant Activities of Eight Pear Cultivars. J. Agric. Food Chem. 2012, 60, 8738–8744. [Google Scholar] [CrossRef] [PubMed]
- Fan, G.J.; Wang, S.Y.; Shi, W.J.; Gong, Z.F.; Gao, M. Simulation Parameter Calibration and Test of Typical Pear Varieties Based on Discrete Element Method. Agronomy 2022, 12, 1720. [Google Scholar] [CrossRef]
- Jia, X.; Wang, W.; Fu, J.; Du, Y.; Wang, Y.; Zhou, R.; Liu, B. Biological Characteristics and Host Range of Athelia bombacina Causing Postharvest Fruit Rot on Pear. Acta Hortic. Sin. 2020, 47, 1253–1263. [Google Scholar]
- Petri, J.L.; Herter, F. Nashi pear (Pyrus pyrifolia) dormancy under mild temperate climate conditions. Acta Hortic. 2002, 587, 353–361. [Google Scholar] [CrossRef]
- Lin, H.T.; Xi, Y.F.; Chen, S.J. Postharvest Softening Physiological Mechanism of Huanghua Pear Fruit. Sci. Agric. Sin. 2003, 36, 349–352. [Google Scholar]
- Faust, M.; Erez, A.; Rowland, L.J.; Wang, S.Y.; Norman, H.A. Bud dormancy in perennial fruit trees: Physiological basis for dormancy induction, maintenance, and release. Hortscience 1997, 32, 623–629. [Google Scholar] [CrossRef]
- Lang, G.A. Dormancy—A New Universal Terminology. Hortscience 1987, 22, 817–820. [Google Scholar] [CrossRef]
- Canton, M.; Forestan, C.; Bonghi, C.; Varotto, S. Meta-analysis of RNA-Seq studies reveals genes with dominant functions during flower bud endo- to eco-dormancy transition in Prunus species. Sci. Rep. 2021, 11, 13173. [Google Scholar] [CrossRef]
- Prudencio, A.S.; Hoeberichts, F.A.; Dicenta, F.; Martínez-Gómez, P.; Sánchez-Pérez, R. Identification of early and late flowering time candidate genes in endodormant and ecodormant almond flower buds. Tree Physiol. 2021, 41, 589–605. [Google Scholar] [CrossRef]
- Saure, M.C. Dormancy release in deciduous fruit trees. Hortic. Rev. 1985, 7, 239. [Google Scholar]
- Sugiura, T.; Kuroda, H.; Sugiura, H.J. Influence of the Current State of Global Warming on Fruit Tree Growth in Japan. Hortic. Res. (Jpn.) 2007, 6, 257–263. [Google Scholar] [CrossRef]
- Houghton, J. Climate Change 2013—The Physical Science Basis: Working Group I Contribution to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change; Cambridge University Press: Cambridge, UK, 2014. [Google Scholar]
- Cai, P.M.; Song, Y.Z.; Meng, L.T.; Liu, R.J.; Lin, J.; Zhao, M.T.; Nie, C.P.; Li, Y.Y.; Ji, Q.G. Climate warming affects phenology of Bactrocera dorsalis: A case study of Fujian and Guangxi, China. Bull. Insectol. 2023, 76, 73–81. [Google Scholar]
- Salama, A.M.; Ezzat, A.; El-Ramady, H.; Alam-Eldein, S.M.; Holb, I.J. Temperate Fruit Trees under Climate Change: Challenges for Dormancy and Chilling Requirements in Warm Winter Regions. Horticulturae 2021, 7, 86. [Google Scholar] [CrossRef]
- Anderson, J.V.; Horvath, D.P.; Chao, W.S.; Foley, M.E. Bud Dormancy in Perennial Plants: A Mechanism for Survival. Dormancy Resist. Harsh Environ. 2010, 21, 69–90. [Google Scholar]
- Nishitani, C.; Saito, T.; Ubi, B.E.; Shimizu, T.; Itai, A.; Saito, T.; Yamamoto, T.; Moriguchi, T. Transcriptome analysis of Pyrus pyrifolia leaf buds during transition from endodormancy to ecodormancy. Sci. Hortic. 2012, 147, 49–55. [Google Scholar] [CrossRef]
- Khalil-Ur-Rehman, M.; Sun, L.; Li, C.X.; Faheem, M.; Wang, W.; Tao, J.M. Comparative RNA-seq based transcriptomic analysis of bud dormancy in grape. BMC Plant Biol. 2017, 17, 18. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Gómez, P.; Dicenta, F. Mechanisms of dormancy in seeds of peach (Prunus persica (L.) Batsch) cv. GF305. Sci. Hortic. 2001, 91, 51–58. [Google Scholar] [CrossRef]
- Martínez, J.J.; Gardea, A.A.; Sagnelli, S.; Olivas, J. Sweet cherry and adaptation to mild winters. Fruit Var. J. 1999, 53, 181–183. [Google Scholar]
- Cook, N.C.; Calitz, F.J.; Aliderman, L.A.; Steyn, W.J.; Louw, E.D. Diverse patterns in dormancy progression of apple buds under variable winter conditions. Sci. Hortic. 2017, 226, 307–315. [Google Scholar] [CrossRef]
- Finch-Savage, W.E.; Leubner-Metzger, G. Seed dormancy and the control of germination. New Phytol. 2006, 171, 501–523. [Google Scholar] [CrossRef] [PubMed]
- Leubner-Metzger, G. Functions and regulation of β-1,3-glucanases during seed germination, dormancy release and after-ripening. Seed Sci. Res. 2003, 13, 17–34. [Google Scholar] [CrossRef]
- Nambara, E.; Marion-Poll, A. ABA action and interactions in seeds. Trends Plant Sci. 2003, 8, 213–217. [Google Scholar] [CrossRef] [PubMed]
- Belin, C.; Megies, C.; Hauserová, E.; Lopez-Molina, L. Abscisic Acid Represses Growth of the Arabidopsis Embryonic Axis after Germination by Enhancing Auxin Signaling. Plant Cell 2009, 21, 2253–2268. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.L.; Gao, Z.Z.; Du, P.Y.; Xiao, W.; Tan, Q.P.; Chen, X.D.; Li, L.; Gao, D.S. Expression of ABA Metabolism-Related Genes Suggests Similarities and Differences Between Seed Dormancy and Bud Dormancy of Peach (Prunus persica). Front. Plant Sci. 2016, 6, 01248. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, K.; Uemachi, A.; Nonaka, M.; Yazawa, S. Effect of endogenous gibberellins in the early stages of fruit growth and development of the ‘Severianin’ tomato. J. Hortic. Sci. Biotechnol. 2004, 79, 54–58. [Google Scholar] [CrossRef]
- Bazhenov, M.S.; Chernook, A.G.; Goncharov, N.P.; Chikida, N.N.; Belousova, M.K.; Karlov, G.I.; Divashuk, M.G. The Allelic Diversity of the Gibberellin Signaling Pathway Genes in Aegilops tauschii Coss. Plants 2020, 9, 1696. [Google Scholar] [CrossRef] [PubMed]
- Takemura, Y.; Kuroki, K.; Shida, Y.; Araki, S.; Takeuchi, Y.; Tanaka, K.; Ishige, T.; Yajima, S.; Tamura, F. Comparative Transcriptome Analysis of the Less-Dormant Taiwanese Pear and the Dormant Japanese Pear during Winter Season. PLoS ONE 2015, 10, 0139595. [Google Scholar] [CrossRef]
- Sugiura, T. Influence of the Current State of Global Warming on Agricultural Production in Japan; The Society of Agricultural Meteorology of Japan: Tsukuba, Japan, 2008; p. 136. [Google Scholar]
- Cai, F.F.; Jin, X.; Tian, Y.X.; Huang, Z.M.; Wang, X.L.; Zhang, Y.P.; Sun, Y.Q.; Shao, C.S. Molecular regulation of bud dormancy in perennial plants. Plant Growth Regul. 2023, 102, 1–11. [Google Scholar] [CrossRef]
- Zhang, L.; Jia, F.; Wang, J.S. The mechanism of bud dormancy in plants. Adv. Mater. Res. 2012, 518–523, 5376–5380. [Google Scholar] [CrossRef]
- Herrick, G.I.; Fox, G.A. Assessing fitness of dormancy from reproductive values of dormant plants. Evol. Ecol. Res. 2011, 13, 779–795. [Google Scholar]
- Yue, C.; Cao, H.L.; Hao, X.Y.; Zeng, J.M.; Qian, W.J.; Guo, Y.Q.; Ye, N.X.; Yang, Y.J.; Wang, X.C. Differential expression of gibberellin- and abscisic acid-related genes implies their roles in the bud activity-dormancy transition of tea plants. Plant Cell Rep. 2018, 37, 425–441. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Ran, J.Z.; Li, X.W.; Wang, Z.Q.; Chen, R.F.; Wu, F.; Ye, M.; Jia, F.; Niklas, K.J.; Deng, J.M. A General Model for Seed and Seedling Respiratory Metabolism. Am. Nat. 2020, 195, 534–546. [Google Scholar] [CrossRef] [PubMed]
- Yi, X.K.; Xu, Y.L.; Gao, Z.H.; Zhang, J.Y.; Pan, H.F.; Qi, Y.J.; Qin, G.H.; Li, B. Warm winter temperature induced changes in the dormant buds of ‘Dangshansuli’ pear (Pyrus bretschneideri Rehd.). Indian J. Hortic. 2020, 77, 273–278. [Google Scholar] [CrossRef]
- Yamane, H. Regulation of Bud Dormancy and Bud Break in Japanese Apricot (Prunus mume Siebold & Zucc.) and Peach [Prunus persica (L.) Batsch]: A Summary of Recent Studies. J. Jpn. Soc. Hortic. Sci. 2014, 83, 187–202. [Google Scholar] [CrossRef]
- Sudhakaran, S.; Thakral, V.; Padalkar, G.; Rajora, N.; Dhiman, P.; Raturi, G.; Sharma, Y.; Tripathi, D.K.; Deshmukh, R.; Sharma, T.R.; et al. Significance of solute specificity, expression, and gating mechanism of tonoplast intrinsic protein during development and stress response in plants. Physiol. Plant. 2021, 172, 258–274. [Google Scholar] [CrossRef] [PubMed]
- Fouquet, R.; Léon, C.; Ollat, N.; Barrieu, F. Identification of grapevine aquaporins and expression analysis in developing berries. Plant Cell Rep. 2008, 27, 1541–1550. [Google Scholar] [CrossRef]
- Sugaya, S.; Gemma, H.; Iwahori, S. Isolation and expression analysis of a gene encoding a vacuolar-type water channel protein in peach fruit. J. Jpn. Soc. Hortic. Sci. 2001, 70, 716–718. [Google Scholar] [CrossRef]
- Santamaría, M.E.; Rodríguez, R.; Cañal, M.J.; Toorop, P.E. Transcriptome analysis of chestnut (Castanea sativa) tree buds suggests a putative role for epigenetic control of bud dormancy. Ann. Bot. 2011, 108, 485–498. [Google Scholar] [CrossRef]
- Mazzitelli, L.; Hancock, R.D.; Haupt, S.; Walker, P.G.; Pont, S.D.A.; McNicol, J.; Cardle, L.; Morris, J.; Viola, R.; Brennan, R.; et al. Co-ordinated gene expression during phases of dormancy release in raspberry (Rubus idaeus L.) buds. J. Exp. Bot. 2007, 58, 1035–1045. [Google Scholar] [CrossRef]
- Hussain, S.; Liu, G.; Liu, D.; Ahmed, M.; Hussain, N.; Teng, Y. Study on the expression of dehydrin genes and activities of antioxidative enzymes in floral buds of two sand pear (Pyrus pyrifolia Nakai) cultivars requiring different chilling hours for bud break. Turk. J. Agric. For. 2015, 39, 9. [Google Scholar] [CrossRef]
- Yu, M.; Chen, H.; Liu, Q.; Huang, J.; Semagn, K.; Liu, D.; Li, Y.C.; Yang, B.; He, Y.L.; Sui, C.; et al. Analysis of unigenes involved in lateral root development in Bupleurum chinense and B. scorzonerifolium. Planta 2021, 253, 128. [Google Scholar] [CrossRef] [PubMed]
- Ramaih, S.; Guedira, M.; Paulsen, G.M. Relationship of indoleacetic acid and tryptophan to dormancy and preharvest sprouting of wheat. Funct. Plant Biol. 2003, 30, 939–945. [Google Scholar] [CrossRef] [PubMed]
- Nagar, P.K.; Sood, S. Changes in endogenous auxins during winter dormancy in tea (Camellia sinensis L.) O. Kuntze. Acta Physiol. Plant. 2006, 28, 165–169. [Google Scholar] [CrossRef]
- Ueguchi-Tanaka, M.; Nakajima, M.; Katoh, E.; Ohmiya, H.; Asano, K.; Saji, S.; Xiang, H.Y.; Ashikari, M.; Kitano, H.; Yamaguchi, I.; et al. Molecular interactions of a soluble gibberellin receptor, GID1, with a rice DELLA protein, SLR1, and gibberellin. Plant Cell 2007, 19, 2140–2155. [Google Scholar] [CrossRef]
- Voegele, A.; Linkies, A.; Müller, K.; Leubner-Metzger, G. Members of the gibberellin receptor gene family GID1 (GIBBERELLIN INSENSITIVE DWARF1) play distinct roles during Lepidium sativum and Arabidopsis thaliana seed germination. J. Exp. Bot. 2011, 62, 5131–5147. [Google Scholar] [CrossRef]
- Hauvermale, A.L.; Tuttle, K.M.; Takebayashi, Y.; Seo, M.; Steber, C.M. Loss of Arabidopsis thaliana Seed Dormancy is Associated with Increased Accumulation of the GID1 GA Hormone Receptors. Plant Cell Physiol. 2015, 56, 1773–1785. [Google Scholar] [CrossRef]
- Falavigna, V.D.S.; Porto, D.D.; Buffon, V.; Margis-Pinheiro, M.; Pasquali, G.; Revers, L.F. Differential Transcriptional Profiles of Dormancy-Related Genes in Apple Buds. Plant Mol. Biol. Rep. 2014, 32, 796–813. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Liu, C.; Ye, Q.; Shen, Y.; Wu, S.; Lin, L. Comparative Analysis of Transcriptomes to Identify Genes during Bud Dormancy of Pyrus pyrifolia ‘Huanghua’. Horticulturae 2024, 10, 619. https://doi.org/10.3390/horticulturae10060619
Wang H, Liu C, Ye Q, Shen Y, Wu S, Lin L. Comparative Analysis of Transcriptomes to Identify Genes during Bud Dormancy of Pyrus pyrifolia ‘Huanghua’. Horticulturae. 2024; 10(6):619. https://doi.org/10.3390/horticulturae10060619
Chicago/Turabian StyleWang, Huiquan, Chunying Liu, Qinghua Ye, Yunyu Shen, Shaohua Wu, and Lizhong Lin. 2024. "Comparative Analysis of Transcriptomes to Identify Genes during Bud Dormancy of Pyrus pyrifolia ‘Huanghua’" Horticulturae 10, no. 6: 619. https://doi.org/10.3390/horticulturae10060619
APA StyleWang, H., Liu, C., Ye, Q., Shen, Y., Wu, S., & Lin, L. (2024). Comparative Analysis of Transcriptomes to Identify Genes during Bud Dormancy of Pyrus pyrifolia ‘Huanghua’. Horticulturae, 10(6), 619. https://doi.org/10.3390/horticulturae10060619




