Whole-Genome Analysis of ZF-HD Genes among Three Dendrobium Species and Expression Patterns in Dendrobium chrysotoxum
Abstract
1. Introduction
2. Results
2.1. Identification and Analysis of ZF-HD Proteins
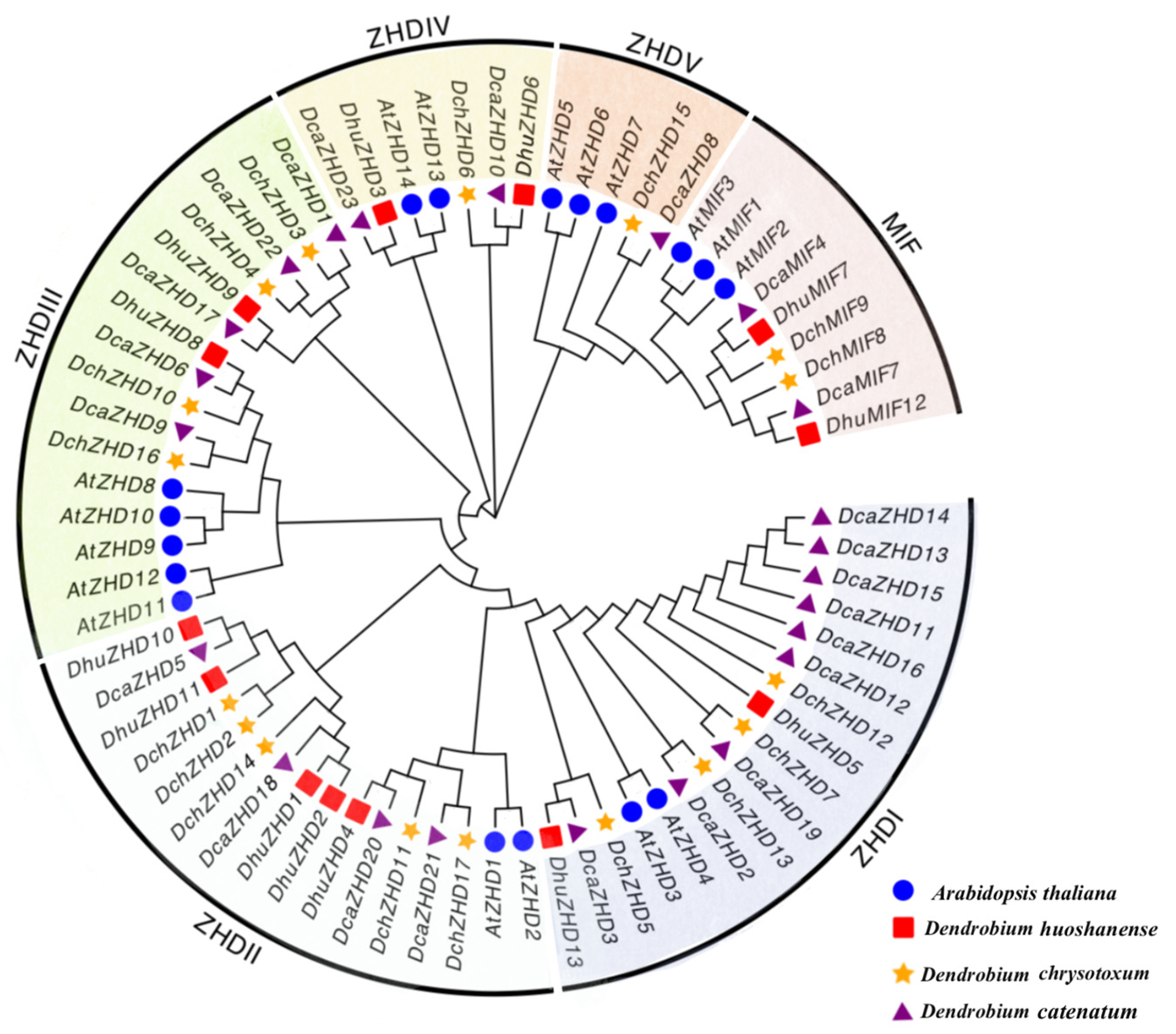
2.2. Phylogeny and Classification of ZF-HDs
2.3. ZF-HD Structure and Motif Analysis
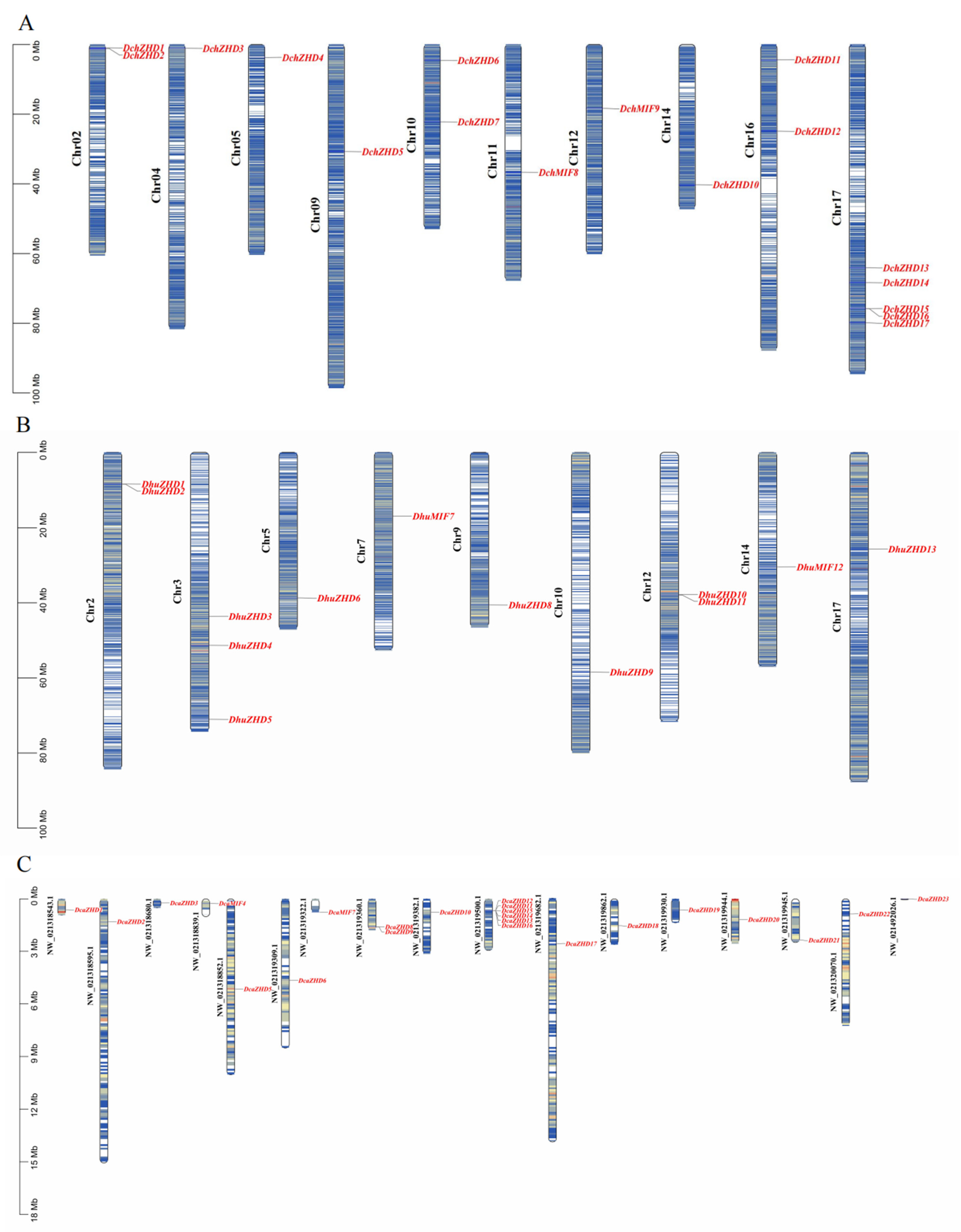
2.4. Chromosomal Localization, Collinearity Analysis, and Promoter Analysis of ZF-HDs
2.5. Prediction of Protein Structure
2.6. Expression Pattern of ZF-HDs in D. chrysotoxum
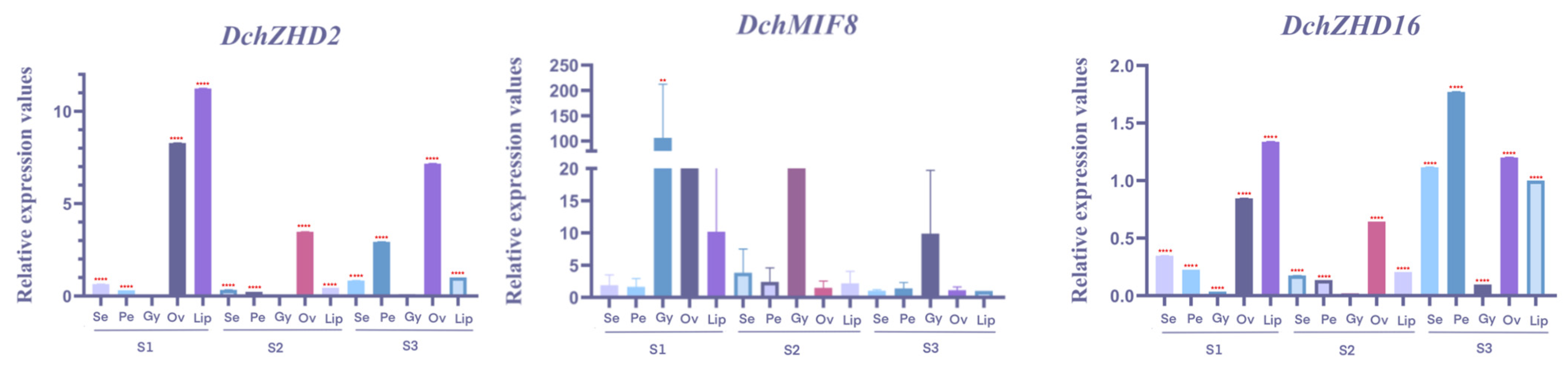
2.7. qRT-PCR Analysis of ZF-HDs in D. chrysotoxum
3. Discussion
4. Material and Methods
4.1. Experimental Materials and Data Sources
4.2. Identification and Physicochemical Properties of ZF-HD Genes
4.3. Phylogenetic Analysis of ZF-HDs
4.4. Gene Structures and Conserved Motif Analysis
4.5. Localization and Collinearity Analysis of ZF-HDs on Chromosomes
4.6. Analysis of ZF-HD Promoter
4.7. Protein Structure Prediction
4.8. Analysis of Expression and RT-qPCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Windhövel, A.; Hein, I.; Dabrowa, R.; Stockhaus, J. Characterization of a novel class of plant homeodomain proteins that bind to the C4 phosphoenolpyruvate carboxylase gene of Flaveria trinervia. Plant Mol. Biol. 2001, 45, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; de Pamphilis, C.W.; Ma, H. Phylogenetic analysis of the plant-specific Zinc Finger-Homeobox and Mini Zinc Finger gene families. J. Integr. Plant Biol. 2008, 50, 1031–1045. [Google Scholar] [CrossRef] [PubMed]
- Vision, T.J.; Brown, D.G.; Tanksley, S.D. The origins of genomic duplications in Arabidopsis. Science 2000, 290, 2114–2117. [Google Scholar] [CrossRef] [PubMed]
- Schoof, H.; Zaccaria, P.; Gundlach, H.; Lemcke, K.; Rudd, S.; Kolesov, G.; Arnold, R.; Mewes, H.W.; Mayer, K.F.X. MIPS Arabidopsis thaliana Database (MAtDB): An integrated biological knowledge resource based on the first complete plant genome. Nucleic Acids Res. 2002, 30, 91–93. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Ma, H. Characterization of a novel putative zinc finger gene MIF1: Involvement in multiple hormonal regulation of Arabidopsis development. Plant J. 2006, 45, 399–422. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Ma, H.; Zhou, J.; Li, Z.; Peng, Z.; Guo, F.; Zhang, J. TsHD1 and TsNAC1 cooperatively play roles in plant growth and abiotic stress resistance of Thellungiella halophile. Plant J. Cell Mol. Biol. 2019, 99, 81–97. [Google Scholar] [CrossRef]
- Khatun, K.; Nath, U.K.; Robin, A.H.K.; Park, J.I.; Lee, D.J.; Kim, M.B.; Kim, C.K.; Lim, K.B.; Nou, I.S.; Chung, M.Y. Genome-wide analysis and expression profiling of zinc finger homeodomain (ZHD) family genes reveal likely roles in organ development and stress responses in tomato. BMC Genom. 2017, 18, 695. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Li, C.; Zhang, Z.; Zhan, L.; Cong, C.; Zhang, D.; Cai, H. Genome-wide investigation of the ZF-HD gene family in two varieties of alfalfa (Medicago sativa L.) and its expression pattern under alkaline stress. BMC Genom. 2022, 23, 150. [Google Scholar] [CrossRef] [PubMed]
- Jain, M.; Tyagi, A.K.; Khurana, J.P. Genome-wide identification, classification, evolutionary expansion and expression analyses of homeobox genes in rice. FEBS J. 2008, 275, 2845–2861. [Google Scholar] [CrossRef]
- Wang, W.; Wu, P.; Li, Y.; Hou, X. Genome-wide analysis and expression patterns of ZF-HD transcription factors under different developmental tissues and abiotic stresses in Chinese cabbage. Mol. Genet. Genom. 2016, 291, 1451–1464, Erratum in Mol. Genet. Genom. 2016, 291, 1465. [Google Scholar] [CrossRef]
- Wang, H.; Yin, X.; Li, X.; Wang, L.; Zheng, Y.; Xu, X.; Zhang, Y.; Wang, X. Genome-wide identification, evolution and expression analysis of the grape (Vitis vinifera L.) zinc finger-homeodomain gene family. Int. J. Mol. Sci. 2014, 15, 5730–5748. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Wang, Y.; Bowers, C.; Ma, H. Isolation, sequence analysis, and expression studies of florally expressed cDNAs in Arabidopsis. Plant Mol. Biol. 2003, 53, 545–563. [Google Scholar] [CrossRef] [PubMed]
- Tan, Q.K.G.; Irish, V.F. The Arabidopsis zinc finger-homeodomain genes encode proteins with unique biochemical properties that are coordinately expressed during floral development. Plant Physiol. 2006, 140, 1095–1108. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Zhu, C.; Xie, S.; Weng, J.; Lin, Y.; Lai, Z.; Guo, Y. Genome-wide analysis of zinc finger motif-associated homeodomain (ZF-HD) family genes and their expression profiles under abiotic stresses and phytohormones stimuli in tea plants (Camellia sinensis). Sci. Hortic. 2021, 281, 109976. [Google Scholar] [CrossRef]
- Xu, Y.; Wang, Y.; Long, Q.; Huang, J.; Wang, Y.; Zhou, K.; Zheng, M.; Sun, J.; Chen, H.; Chen, S.; et al. Overexpression of OsZHD1, a zinc finger homeodomain class homeobox transcription factor, induces abaxially curled and drooping leaf in rice. Planta 2014, 239, 803–816. [Google Scholar] [CrossRef] [PubMed]
- Figueiredo, D.D.; Barros, P.M.; Cordeiro, A.M.; Serra, T.S.; Lourenço, T.; Chander, S.; Oliveira, M.M.; Saibo, N.J.M. Seven zinc-finger transcription factors are novel regulators of the stress responsive gene OsDREB1B. J. Exp. Bot. 2012, 63, 3643–3656. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yang, Y.; Zhang, L. Zinc Finger-Homeodomain Transcriptional Factors (ZF-HDs) in Wheat (Triticum aestivum L.): Identification, Evolution, Expression Analysis and Response to Abiotic Stresses. Plants 2021, 10, 593. [Google Scholar] [CrossRef] [PubMed]
- Christenhusz, M.J.M.; Byng, J.W. The number of known plants species in the world and its annual increase. Phytotaxa 2016, 261, 201–217. [Google Scholar] [CrossRef]
- Zheng, S.G.; Hu, Y.D.; Zhao, R.X.; Yan, S.; Zhang, X.Q.; Zhao, T.M.; Chun, Z. Genome-wide researches and applications on Dendrobium. Planta 2018, 248, 769–784. [Google Scholar] [CrossRef]
- Teixeira da Silva, J.A.; Zeng, S.; Galdiano, R.F., Jr.; Dobránszki, J.; Cardoso, J.C.; Vendrame, W.A. In vitro conservation of Dendrobium germplasm. Plant Cell Rep. 2014, 33, 1413–1423. [Google Scholar] [CrossRef]
- Han, B.; Jing, Y.; Dai, J.; Zheng, T.; Gu, F.; Zhao, Q.; Zhu, F.; Song, X.; Deng, H.; Wei, P.; et al. A Chromosome-Level Genome Assembly of Dendrobium Huoshanense Using Long Reads and Hi-C Data. Genome Biol. Evol. 2020, 12, 2486–2490. [Google Scholar] [CrossRef]
- Niu, Z.; Zhu, F.; Fan, Y.; Li, C.; Zhang, B.; Zhu, S.; Hou, Z.; Wang, M.; Yang, J.; Xue, Q.; et al. The chromosome-level reference genome assembly for Dendrobium officinale and its utility of functional genomics research and molecular breeding study. Acta Pharm. Sin. B 2021, 11, 2080–2092. [Google Scholar] [CrossRef]
- Chao, Y.T.; Chen, W.C.; Chen, C.Y.; Ho, H.Y.; Yeh, C.H.; Kuo, Y.T.; Su, C.L.; Yen, S.H.; Hsueh, H.Y.; Yeh, J.H.; et al. Chromosome-level assembly, genetic and physical mapping of Phalaenopsis aphrodite genome provides new insights into species adaptation and resources for orchid breeding. Plant Biotechnol. J. 2018, 16, 2027–2041. [Google Scholar] [CrossRef]
- Zhang, G.Q.; Xu, Q.; Bian, C.; Tsai, W.C.; Yeh, C.M.; Liu, K.W.; Yoshida, K.; Zhang, L.S.; Chang, S.B.; Chen, F.; et al. The Dendrobium catenatum Lindl. genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci. Rep. 2016, 6, 19029. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Liu, D.-K.; Wang, Q.-Q.; Ke, S.; Li, Y.; Zhang, D.; Zheng, Q.; Zhang, C.; Liu, Z.-J.; Lan, S. Genome-wide identification and expression analysis of the GRAS gene family in Dendrobium chrysotoxum. Front. Plant Sci. 2022, 13, 1058287. [Google Scholar] [CrossRef]
- Huang, Y.; Zhao, X.; Zheng, Q.; He, X.; Zhang, M.-M.; Ke, S.; Li, Y.; Zhang, C.; Ahmad, S.; Lan, S.; et al. Genome-Wide Identification of TCP Gene Family in Dendrobium and Their Expression Patterns in Dendrobium chrysotoxum. Int. J. Mol. Sci. 2023, 24, 14320. [Google Scholar] [CrossRef] [PubMed]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zhang, W.; Zhang, Y.; Zhang, X.; Lang, D.; Zhang, X. The roles of methyl jasmonate to stress in plants. Funct. Plant Biol. FPB 2019, 46, 197–212. [Google Scholar] [CrossRef]
- Liang, T.; Jiang, C.; Yuan, J.; Othman, Y.; Xie, X.Q.; Feng, Z. Differential performance of RoseTTAFold in antibody modeling. Brief. Bioinform. 2022, 23, bbac152. [Google Scholar] [CrossRef]
- Liu, M.D.; Liu, H.; Liu, W.Y.; Ni, S.F.; Wang, Z.Y.; Geng, Z.H.; Zhu, K.Y.; Wang, Y.F.; Zhao, Y.H. Systematic Analysis of Zinc Finger-Homeodomain Transcription Factors (ZF-HDs) in Barley (Hordeum vulgare L.). Genes 2024, 15, 578. [Google Scholar] [CrossRef]
- Zhang, Y.T.; Zhang, Y.L.; Chen, S.X.; Yin, G.H.; Yang, Z.Z.; Lee, S.; Liu, C.G.; Zhao, D.D.; Ma, Y.K.; Song, F.Q.; et al. Proteomics of methyl jasmonate induced defense response in maize leaves against Asian corn borer. BMC Genom. 2015, 16, 224. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.B.; Wu, Y.; Wang, H.; Song, S.W.; Bai, T.H.; Jiao, J.; Song, C.H.; Pang, H.G.; Wang, M.M. Genome-Wide Investigation of the Zinc Finger-Homeodomain Family Genes Reveals Potential Roles in Apple Fruit Ripening. Front. Genet. 2021, 12, 783482. [Google Scholar] [CrossRef] [PubMed]
- Shi, B.; Haq, I.U.; Fiaz, S.; Alharthi, B.; Xu, M.L.; Wang, J.L.; Hou, W.H.; Feng, X.B. Genome-wide identification and expression analysis of the ZF-HD gene family in pea (Pisum sativum L.). Front. Genet. 2022, 13, 1089375. [Google Scholar] [CrossRef] [PubMed]
- Xing, L.; Peng, K.; Xue, S.; Yuan, W.; Zhu, B.; Zhao, P.; Wu, H.; Cheng, Y.; Fang, M.; Liu, Z. Genome-wide analysis of zinc finger-homeodomain (ZF-HD) transcription factors in diploid and tetraploid cotton. Funct. Integr. Genom. 2022, 22, 1269–1281. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, K.; Yamaguchi-Shinozaki, K. Regulons involved in osmotic stress-responsive and cold stress-responsive gene expression in plants. Physiol. Plant. 2006, 126, 62–71. [Google Scholar] [CrossRef]
- Zhu, D.; Mao, F.; Tian, Y.; Lin, X.; Gu, L.; Gu, H.; Qu, L.J.; Wu, Y.; Wu, Z. The Features and Regulation of Co-transcriptional Splicing in Arabidopsis. Mol. Plant 2020, 13, 278–294. [Google Scholar] [CrossRef] [PubMed]
- Sang, Y.; Liu, Q.; Lee, J.; Ma, W.; McVey, D.S.; Blecha, F. Expansion of amphibian intronless interferons revises the paradigm for interferon evolution and functional diversity. Sci. Rep. 2016, 6, 29072. [Google Scholar] [CrossRef]
- Jain, M.; Khurana, P.; Tyagi, A.K.; Khurana, J.P. Genome-wide analysis of intronless genes in rice and Arabidopsis. Funct. Integr. Genom. 2008, 8, 69–78. [Google Scholar] [CrossRef]
- Kong, H.; Landherr, L.L.; Frohlich, M.W.; Leebens-Mack, J.; Ma, H.; dePamphilis, C.W. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: Evidence for multiple mechanisms of rapid gene birth. Plant J. 2007, 50, 873–885. [Google Scholar] [CrossRef]
- Yu, J.; Ke, T.; Tehrim, S.; Sun, F.; Liao, B.; Hua, W. PTGBase: An integrated database to study tandem duplicated genes in plants. Database 2015, 2015, bav017. [Google Scholar] [CrossRef]
- Hernandez-Garcia, C.M.; Finer, J.J. Identification and validation of promoters and cis-acting regulatory elements. Plant Sci. 2014, 217, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-L.; Viswanath, K.K.; Tong, C.-G.; An, H.R.; Jang, S.; Chen, F.-C. Floral Induction and Flower Development of Orchids. Front. Plant Sci. 2019, 10, 1258. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhao, X.-W.; Li, Y.-Y.; Ke, S.-J.; Yin, W.-L.; Lan, S.; Liu, Z.-J. Advances and prospects of orchid research and industrialization. Hortic. Res. 2022, 9, uhac220. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Hou, H.; Li, X.; Xiang, J.; Yin, X.; Gao, H.; Zheng, Y.; Bassett, C.L.; Wang, X. Genome-wide identification and analysis of the SBP-box family genes in apple (Malus × domestica Borkh.). Plant Physiol. Biochem. 2013, 70, 100–114. [Google Scholar] [CrossRef] [PubMed]
- Shalmani, A.; Fan, S.; Jia, P.; Li, G.; Muhammad, I.; Li, Y.; Sharif, R.; Dong, F.; Zuo, X.; Li, K.; et al. Genome Identification of B-BOX Gene Family Members in Seven Rosaceae Species and Their Expression Analysis in Response to Flower Induction in Malus domestica. Molecules 2018, 23, 1763. [Google Scholar] [CrossRef] [PubMed]
- Kumar, G.; Arya, P.; Gupta, K.; Randhawa, V.; Acharya, V.; Singh, A.K. Comparative phylogenetic analysis and transcriptional profiling of MADS-box gene family identified DAM and FLC-like genes in apple (Malusx domestica). Sci. Rep. 2016, 6, 20695. [Google Scholar] [CrossRef] [PubMed]
- Shalmani, A.; Muhammad, I.; Sharif, R.; Zhao, C.; Ullah, U.; Zhang, D.; Jing, X.-Q.; Amin, B.; Jia, P.; Tahir, M.M.; et al. Zinc Finger-Homeodomain Genes: Evolution, Functional Differentiation, and Expression Profiling Under Flowering-Related Treatments and Abiotic Stresses in Plants. Evol. Bioinform. 2019, 15, 1176934319867930. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, G.Q.; Zhang, D.; Liu, X.D.; Xu, X.Y.; Sun, W.H.; Yu, X.; Zhu, X.; Wang, Z.W.; Zhao, X.; et al. Chromosome-scale assembly of the Dendrobium chrysotoxum genome enhances the understanding of orchid evolution. Hortic. Res. 2021, 8, 183. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.-Q.; Liu, K.-W.; Li, Z.; Lohaus, R.; Hsiao, Y.-Y.; Niu, S.-C.; Wang, J.-Y.; Lin, Y.-C.; Xu, Q.; Chen, L.-J.; et al. The Apostasia genome and the evolution of orchids. Nature 2017, 549, 379–383, Erratum in Nature 2020, 583, E30. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef] [PubMed]
- Enany, S. Structural and functional analysis of hypothetical and conserved proteins of Clostridium tetani. J. Infect. Public Health 2014, 7, 296–307. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.-H. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef] [PubMed]
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M.C. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef]
- Geourjon, C.; Deléage, G. SOPMA: Significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Comput. Appl. Biosci. 1995, 11, 681–684. [Google Scholar] [CrossRef]




| Name | Gene ID | AA 1 | GRAVY 2 | pI 3 | Mw 4 (kDa) | AI 5 | II 6 | Subcellular Localization 7 |
|---|---|---|---|---|---|---|---|---|
| DchZHD1 | Maker81525 | 216 | −0.593 | 8.73 | 23.70 | 57.55 | 60.85 | Nucleus. |
| DchZHD2 | Maker28129 | 216 | −0.593 | 8.73 | 23.70 | 57.55 | 60.85 | Nucleus. |
| DchZHD3 | Maker69363 | 217 | −0.767 | 8.96 | 24.50 | 57.60 | 83.88 | Nucleus. |
| DchZHD4 | Maker78792 | 314 | −0.787 | 8.25 | 34.50 | 51.56 | 66.90 | Nucleus. |
| DchZHD5 | Maker102972 | 221 | −0.871 | 8.68 | 25.19 | 48.55 | 69.86 | Nucleus. |
| DchZHD6 | Maker82648 | 297 | −0.666 | 6.37 | 32.51 | 59.19 | 53.96 | Nucleus. |
| DchZHD7 | Maker74870 | 286 | −0.598 | 9.07 | 31.28 | 59.34 | 57.57 | Nucleus. |
| DchMIF8 | Maker100805 | 126 | −0.917 | 9.04 | 13.68 | 39.60 | 67.20 | Mitochondrial. |
| DchMIF9 | Maker57367 | 129 | −0.788 | 7.61 | 14.24 | 46.20 | 64.53 | Mitochondrial. |
| DchZHD10 | Maker92643 | 249 | −0.691 | 8.99 | 27.41 | 60.00 | 61.99 | Nucleus. |
| DchZHD11 | Maker54517 | 450 | −0.471 | 8.87 | 49.31 | 67.62 | 48.02 | Cytoplasmic. |
| DchZHD12 | Maker53183 | 249 | −0.623 | 8.99 | 27.72 | 59.16 | 61.03 | Nucleus. |
| DchZHD13 | Maker57691 | 232 | −0.570 | 9.57 | 25.57 | 70.13 | 57.09 | Nucleus. |
| DchZHD14 | Maker77088 | 198 | −0.622 | 9.24 | 22.43 | 60.10 | 70.26 | Cytoplasmic. |
| DchZHD15 | Maker61228 | 85 | −0.326 | 8.70 | 9.38 | 55.18 | 60.76 | Chloroplast. |
| DchZHD16 | Maker61267 | 293 | −0.670 | 7.63 | 31.52 | 60.00 | 57.07 | Nucleus. |
| DchZHD17 | Maker58363 | 273 | −0.666 | 7.05 | 28.88 | 49.52 | 49.81 | Nucleus. |
| DhuZHD1 | Dhu000007045 | 198 | −0.648 | 9.12 | 22.64 | 56.16 | 72.97 | Nucleus. |
| DhuZHD2 | Dhu000019223 | 198 | −0.648 | 9.12 | 22.64 | 56.16 | 72.97 | Nucleus. |
| DhuZHD3 | Dhu000028169 | 146 | −0.770 | 9.65 | 17.02 | 46.16 | 81.27 | Nucleus. |
| DhuZHD4 | Dhu000024291 | 858 | 13.510 | 9.64 | 92.47 | 80.65 | 47.66 | Cytoplasmic. |
| DhuZHD5 | Dhu000009472 | 280 | −0.386 | 5.48 | 30.32 | 73.86 | 46.94 | Nucleus. |
| DhuZHD6 | Dhu000013154 | 390 | −0.381 | 5.36 | 41.72 | 66.54 | 48.07 | Cytoplasmic. |
| DhuMIF7 | Dhu000014526 | 260 | −0.955 | 5.39 | 28.60 | 45.12 | 68.88 | Nucleus. |
| DhuZHD8 | Dhu000009060 | 250 | −0.638 | 8.79 | 27.01 | 57.80 | 54.54 | Nucleus. |
| DhuZHD9 | Dhu000025178 | 626 | −0.977 | 10.17 | 70.34 | 57.68 | 65.98 | Nucleus. |
| DhuZHD10 | Dhu000014745 | 216 | −0.584 | 8.73 | 23.75 | 58.01 | 62.13 | Nucleus. |
| DhuZHD11 | Dhu000004498 | 216 | −0.581 | 8.73 | 23.74 | 58.01 | 62.27 | Nucleus. |
| DhuZHD12 | Dhu000014369 | 125 | −0.942 | 9.04 | 13.54 | 37.60 | 70.65 | Mitochondrial. |
| DhuZHD13 | Dhu000016048 | 219 | −0.762 | 8.54 | 24.84 | 52.10 | 59.08 | Nucleus. |
| DcaZHD1 | rna-XM_020822518.2 | 218 | −0.855 | 8.77 | 24.54 | 56.01 | 84.44 | Nucleus. |
| DcaZHD2 | rna-XM_020823161.2 | 263 | −0.719 | 8.92 | 29.55 | 58.56 | 66.36 | Nucleus. |
| DcaZHD3 | rna-XM_020825047.1 | 222 | −0.725 | 8.55 | 25.09 | 50.99 | 59.33 | Nucleus. |
| DcaMIF4 | rna-XM_020824991.2 | 129 | −0.736 | 7.60 | 14.16 | 49.22 | 65.82 | Mitochondrial. |
| DcaZHD5 | rna-XM_020824023.2 | 216 | −0.595 | 8.73 | 23.72 | 57.13 | 62.13 | Nucleus. |
| DcaZHD6 | rna-XM_020838304.2 | 249 | −0.646 | 8.92 | 26.89 | 57.67 | 50.52 | Nucleus. |
| DcaMIF7 | rna-XM_020825579.2 | 125 | −0.962 | 9.04 | 13.51 | 36.08 | 70.65 | Mitochondrial. |
| DcaZHD8 | rna-XM_020842089.1 | 85 | −0.420 | 8.10 | 9.68 | 52.82 | 51.73 | Chloroplast. |
| DcaZHD9 | rna-XM_020842002.2 | 293 | −0.683 | 7.64 | 31.54 | 57.99 | 56.76 | Nucleus. |
| DcaZHD10 | rna-XM_020831316.1 | 200 | −0.684 | 8.17 | 21.60 | 45.00 | 67.27 | Nucleus. |
| DcaZHD11 | rna-XM_020837938.2 | 249 | −0.650 | 8.99 | 27.69 | 57.99 | 59.00 | Nucleus. |
| DcaZHD12 | rna-XM_020837921.2 | 249 | −0.650 | 8.99 | 27.69 | 57.99 | 59.00 | Nucleus. |
| DcaZHD13 | rna-XM_020837895.2 | 249 | −0.650 | 8.99 | 27.69 | 57.99 | 59.00 | Nucleus. |
| DcaZHD14 | rna-XM_020837890.2 | 249 | −0.650 | 8.99 | 27.69 | 57.99 | 59.00 | Nucleus. |
| DcaZHD15 | rna-XM_028698070.1 | 249 | −0.650 | 8.99 | 27.69 | 57.99 | 59.00 | Nucleus. |
| DcaZHD16 | rna-XM_020837900.2 | 249 | −0.650 | 8.99 | 27.69 | 57.99 | 59.00 | Nucleus. |
| DcaZHD17 | rna-XM_020825507.2 | 258 | −0.748 | 9.15 | 28.94 | 58.29 | 63.93 | Nucleus. |
| DcaZHD18 | rna-XM_020846562.2 | 199 | −0.611 | 9.24 | 22.74 | 54.42 | 74.46 | Nucleus. |
| DcaZHD19 | rna-XM_020841238.2 | 285 | −0.604 | 9.07 | 31.21 | 60.56 | 58.07 | Nucleus. |
| DcaZHD20 | rna-XM_020831512.2 | 264 | −0.762 | 6.59 | 28.25 | 47.01 | 59.03 | Nucleus. |
| DcaZHD21 | rna-XM_020848621.2 | 273 | −0.675 | 7.05 | 28.91 | 49.16 | 49.50 | Nucleus. |
| DcaZHD22 | rna-XM_020845498.2 | 312 | −0.747 | 8.53 | 34.04 | 52.56 | 65.46 | Nucleus. |
| DcaZHD23 | rna-XM_020823387.2 | 147 | −0.784 | 9.79 | 17.17 | 46.53 | 81.65 | Nucleus. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, X.; Zhao, X.; Zheng, Q.; Zhang, M.-M.; Huang, Y.; Liu, Z.-J.; Lan, S. Whole-Genome Analysis of ZF-HD Genes among Three Dendrobium Species and Expression Patterns in Dendrobium chrysotoxum. Horticulturae 2024, 10, 610. https://doi.org/10.3390/horticulturae10060610
He X, Zhao X, Zheng Q, Zhang M-M, Huang Y, Liu Z-J, Lan S. Whole-Genome Analysis of ZF-HD Genes among Three Dendrobium Species and Expression Patterns in Dendrobium chrysotoxum. Horticulturae. 2024; 10(6):610. https://doi.org/10.3390/horticulturae10060610
Chicago/Turabian StyleHe, Xin, Xuewei Zhao, Qinyao Zheng, Meng-Meng Zhang, Ye Huang, Zhong-Jian Liu, and Siren Lan. 2024. "Whole-Genome Analysis of ZF-HD Genes among Three Dendrobium Species and Expression Patterns in Dendrobium chrysotoxum" Horticulturae 10, no. 6: 610. https://doi.org/10.3390/horticulturae10060610
APA StyleHe, X., Zhao, X., Zheng, Q., Zhang, M.-M., Huang, Y., Liu, Z.-J., & Lan, S. (2024). Whole-Genome Analysis of ZF-HD Genes among Three Dendrobium Species and Expression Patterns in Dendrobium chrysotoxum. Horticulturae, 10(6), 610. https://doi.org/10.3390/horticulturae10060610







