Identification of Resistance QTLs to Black Leaf Streak Disease (Due to Pseudocercospora fijiensis) in Diploid Bananas (Musa acuminata)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Artificial Inoculation and Disease Evaluation
2.3. Analysis of Phenotypic Data
2.4. Genotyping and Linkage Mapping
2.5. QTL Analysis
3. Results and Discussion
3.1. Analysis of Parental Meiosis
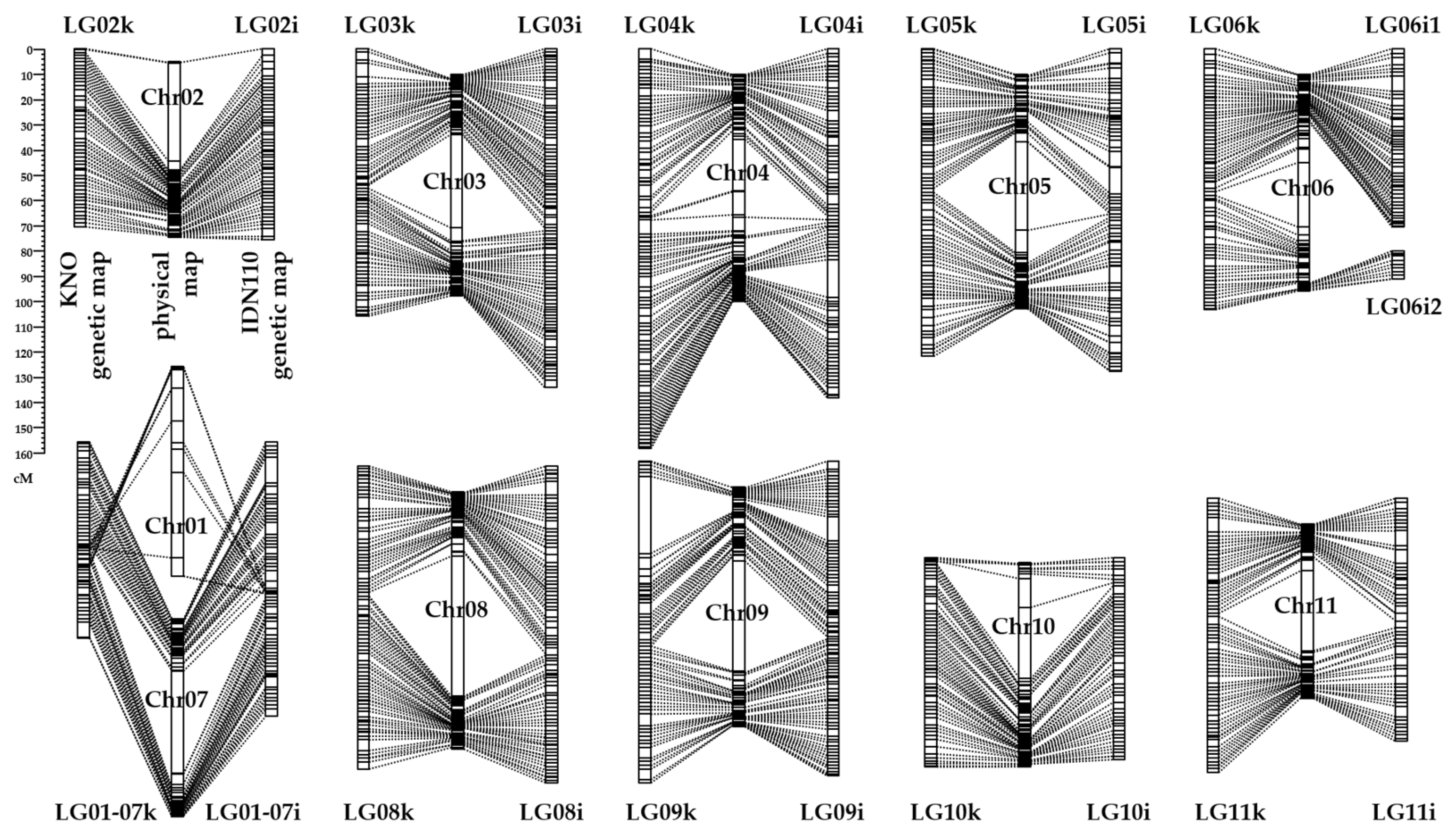
3.2. Linkage Maps
3.3. Reaction of the Progeny to P. fijiensis Inoculation
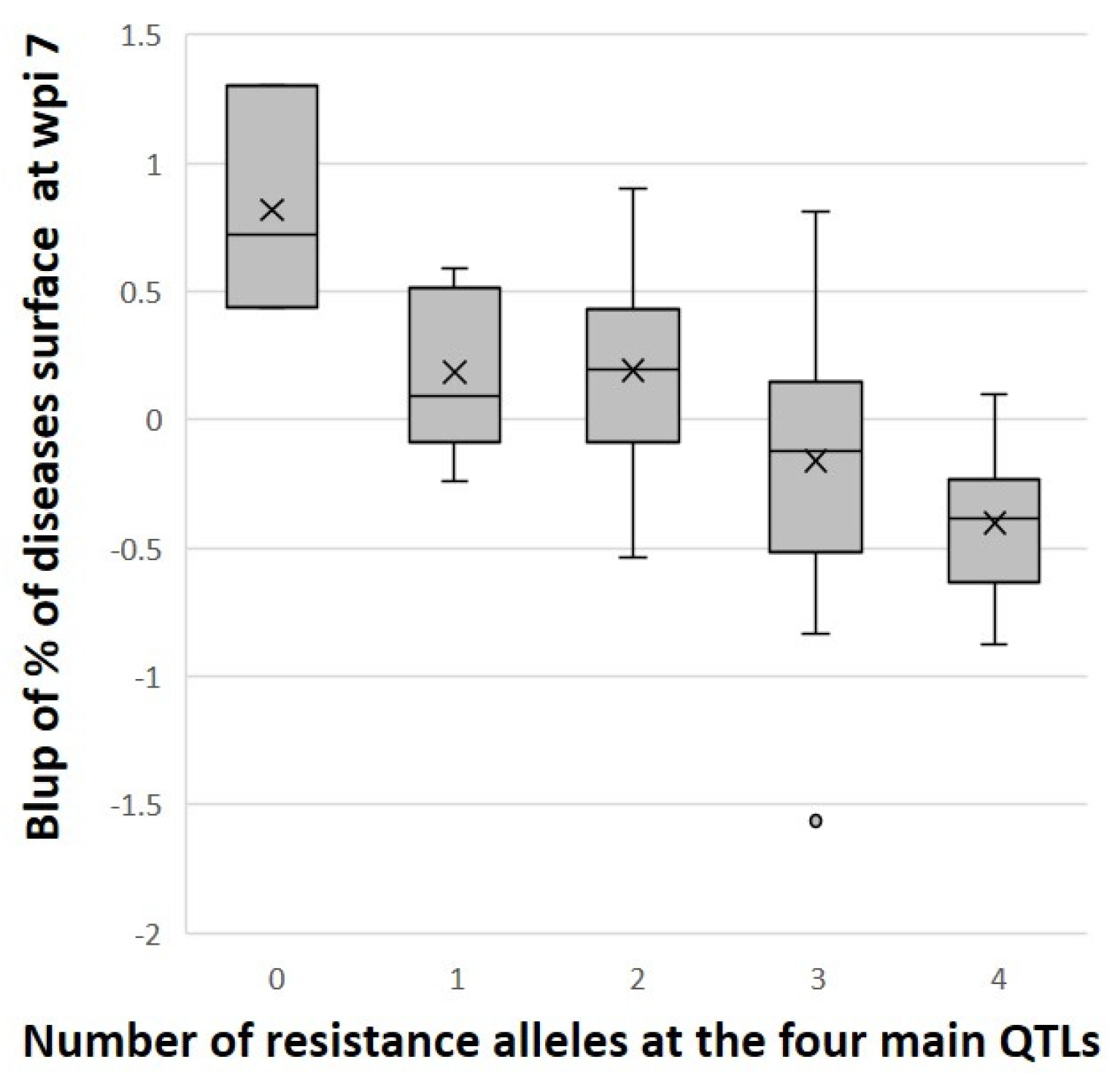
3.4. Identified QTLs
3.5. Putative Candidate Genes in the Four Main QTLs Detected on IDN110
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Simmonds, N.W.; Shepherd, K. The Taxonomy and Origins of the Cultivated Bananas. J. Linn. Soc. Lond. Bot. 1955, 55, 302–312. [Google Scholar] [CrossRef]
- Perrier, X.; de Langhe, E.; Donohue, M.; Lentfer, C.J.; Vrydaghs, L.; Bakry, F.; Carreel, F.; Hippolyte, I.; Horry, J.-P.; Jenny, C.; et al. Multidisciplinary Perspectives on Banana (Musa Spp.) Domestication. Proc. Natl. Acad. Sci. USA 2011, 108, 11311–11318. [Google Scholar] [CrossRef] [PubMed]
- Lescot, T. Banana Genetic Diversity. FruiTrop 2020, 269, 98–102. [Google Scholar]
- Churchill, A.C.L. Mycosphaerella fijiensis, the Black Leaf Streak Pathogen of Banana: Progress towards Understanding Pathogen Biology and Detection, Disease Development, and the Challenges of Control. Mol. Plant Pathol. 2011, 12, 307–328. [Google Scholar] [CrossRef] [PubMed]
- Crous, P.W.; Braun, U.; Hunter, G.C.; Wingfield, M.J.; Verkley, G.J.M.; Shin, H.-D.; Nakashima, C.; Groenewald, J.Z. Phylogenetic Lineages in Pseudocercospora. Stud. Mycol. 2013, 75, 37–114. [Google Scholar] [CrossRef] [PubMed]
- Guzman, M.; Pérez Vicente, L.; Carlier, J.; Abadie, C.; Lapeyre de Bellaire, L.; Carreel, F.; Marin, D.H.; Romero, R.A.; Gauhl, F.; Pasberg-Gauhl, C.; et al. Black Leaf Streak. In Handbook of Diseases of Banana, Abacà and Enset; CABI: Boston, MA, USA, 2019; pp. 41–115. ISBN 978-1-78064-719-7. [Google Scholar]
- Noar, R.D.; Thomas, E.; Daub, M.E. Genetic Characteristics and Metabolic Interactions between Pseudocercospora fijiensis and Banana: Progress toward Controlling Black Sigatoka. Plants 2022, 11, 948. [Google Scholar] [CrossRef]
- Pennisi, E. Armed and Dangerous. Science 2010, 327, 804–805. [Google Scholar] [CrossRef]
- Ramsey, M.D.; Daniells, J.W.; Anderson, V.J. Effects of Sigatoka Leaf Spot (Mycospherella musicola Leach) on Fruit Yield, Field Ripening and Greenlife of Bananas in North Queensland. Sci. Hortic. 1990, 41, 305–313. [Google Scholar] [CrossRef]
- Chillet, M.; Abadie, C.; Hubert, O.; Chilin-Charles, Y.; de Lapeyre de Bellaire, L. Sigatoka Disease Reduces the Greenlife of Bananas. Crop Prot. 2009, 28, 41–45. [Google Scholar] [CrossRef]
- Castelan, F.; Saraiva, L.; Lange, F.; de Lapeyre de Bellaire, L.; Cordenunsi, B.; Chillet, M. Effects of Black Leaf Streak Disease and Sigatoka Disease on Fruit Quality and Maturation Process of Bananas Produced in the Subtropical Conditions of Southern Brazil. Crop Prot. 2011, 35, 127–131. [Google Scholar] [CrossRef]
- Lapeyre de Bellaire, L.; Fouré, E.; Abadie, C.; Carlier, J. Black Leaf Streak Disease Is Challenging the Banana Industry. Fruits 2010, 65, 327–342. [Google Scholar] [CrossRef][Green Version]
- Fouré, E.; De Lapeyre De Bellaire, L. Maladies et Ravageurs de La Banane. FruiTrop 2020, 269, 104–112. [Google Scholar]
- Chong, P.; Essoh, J.N.; Arango Isaza, R.E.; Keizer, P.; Stergiopoulos, I.; Seidl, M.F.; Guzman, M.; Sandoval, J.; Verweij, P.E.; Scalliet, G.; et al. A World-wide Analysis of Reduced Sensitivity to DMI Fungicides in the Banana Pathogen Pseudocercospora fijiensis. Pest Manag. Sci. 2021, 77, 3273–3288. [Google Scholar] [CrossRef] [PubMed]
- Rieux, A.; Soubeyrand, S.; Bonnot, F.; Klein, E.K.; Ngando, J.E.; Mehl, A.; Ravigne, V.; Carlier, J.; Bellaire, L.d.L.d. Long-Distance Wind-Dispersal of Spores in a Fungal Plant Pathogen: Estimation of Anisotropic Dispersal Kernels from an Extensive Field Experiment. PLoS ONE 2014, 9, e103225. [Google Scholar] [CrossRef] [PubMed]
- Soares, J.M.S.; Rocha, A.J.; Nascimento, F.S.; Santos, A.S.; Miller, R.N.G.; Ferreira, C.F.; Haddad, F.; Amorim, V.B.O.; Amorim, E.P. Genetic Improvement for Resistance to Black Sigatoka in Bananas: A Systematic Review. Front. Plant Sci. 2021, 12, 657916. [Google Scholar] [CrossRef]
- Salmon, F.; Bakry, F.; Efile, J.C.; Ricci, S.; Toniutti, L.; Horry, J.P. Banana Breeding at CIRAD: Creating Resistant New Cultivars to Avoid the Use of Pesticides. Acta Hortic. 2023, 1367, 201–208. [Google Scholar] [CrossRef]
- Fouré, E.; Mouliom Pefoura, A.; Mourichon, X. Etude de La Sensibilité Variétale Des Bananiers et Des Plantains à Mycosphaerella fijiensis Morelet Au Cameroun. Caractérisation de La Résistance Au Champ de Bananiers Appartenant à Divers Groupes Génétiques. Fruits 1990, 45, 339–345. [Google Scholar]
- Fullerton, R.A.; Olsen, T.L. Pathogenic Variability in Mycosphaerella fijiensis Morelet, Cause of Black Sigatoka in Banana and Plantain. N. Z. J. Crop Hortic. Sci. 1995, 23, 39–48. [Google Scholar] [CrossRef][Green Version]
- Mouliom-Pefoura, A. First Observation of the Breakdown of High Resistance in Yangambi Km 5 (Musa Sp.) to the Black Leaf Streak Disease in Cameroon. Plant Dis. 1999, 83, 78. [Google Scholar] [CrossRef]
- Niks, R.E.; Qi, X.; Marcel, T.C. Quantitative Resistance to Biotrophic Filamentous Plant Pathogens: Concepts, Misconceptions, and Mechanisms. Annu. Rev. Phytopathol. 2015, 53, 445–470. [Google Scholar] [CrossRef]
- Nelson, R.; Wiesner-Hanks, T.; Wisser, R.; Balint-Kurti, P. Navigating Complexity to Breed Disease-Resistant Crops. Nat. Rev. Genet. 2018, 19, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Rowe, P.; Rosales, F. Musa breeding at FHIA. In The Improvement and Testing of Musa: A Global Partnership, Proceedings of the First Global Conference of the IMTP, San Pedro Sula, Honduras, 27–30 April 1994; Bioversity International: Rome, Italy, 1994; pp. 117–129. [Google Scholar]
- Bakry, F.; Carreel, F.; Jenny, C.; Horry, J.-P. Genetic Improvement of Banana. In Breeding Plantation Tree Crops: Tropical Species; Mohan Jain, S., Priyadarshan, P.M., Eds.; Springer: New York, NY, USA, 2009; pp. 3–50. ISBN 978-0-387-71199-7. [Google Scholar]
- Dumartinet, T.; Abadie, C.; Bonnot, F.; Carreel, F.; Roussel, V.; Habas, R.; Martinez, R.T.; Perez-Vicente, L.; Carlier, J. Pattern of Local Adaptation to Quantitative Host Resistance in a Major Pathogen of a Perennial Crop. Evol. Appl. 2020, 13, 824–836. [Google Scholar] [CrossRef]
- Cheesman, E.E. Principles of Banana Breeding. Trop. Agric. 1934, 11, 132–137. [Google Scholar]
- Shepherd, K. Banana Breeding in the West Indies. Pest Artic. News Summ. 1968, 14, 370–379. [Google Scholar] [CrossRef]
- Rowe, P.R.; Richardson, D.L. Breeding Bananas for Disease Resistance Fruit Quality and Yield; Tropical Agriculture Research Services (SIATSA): La Lima, Honduras, 1975; pp. 1–23. [Google Scholar]
- Tézenas du Montcel, H. Genetic Improvement of Bananas for Resistance to Diseases and Pests: Improvement Strategies. Fruits 1993, 48, 11–14. [Google Scholar]
- Brown, A.; Carpentier, S.C.; Swennen, R. Breeding climate-resilient bananas. In Genomic Designing of Climate-Smart Fruit Crops; Kole, C., Ed.; Springer International Publishing: Cham, Switzerland, 2020; pp. 91–115. ISBN 978-3-319-97946-5. [Google Scholar]
- Bakry, F.; Horry, J.-P.; Jenny, C. Making Banana Breeding More Effective. Available online: https://agritrop.cirad.fr/597567/ (accessed on 1 December 2023).
- Ortiz, R.; Vuylsteke, D. Inheritance of Black Sigatoka Disease Resistance in Plantain-Banana (Musa Spp) Hybrids. Theor. Appl. Genet. 1994, 89, 146–152. [Google Scholar] [CrossRef] [PubMed]
- Craenen, K.; Ortiz, R. Effect of the Bs1 Gene in Plantain-Banana Hybrids on Response to Black Sigatoka. Theor. Appl. Genet. 1997, 95, 497–505. [Google Scholar] [CrossRef]
- D’Hont, A.; Denoeud, F.; Aury, J.-M.; Baurens, F.-C.; Carreel, F.; Garsmeur, O.; Noel, B.; Bocs, S.; Droc, G.; Rouard, M.; et al. The Banana Musa acuminata Genome and the Evolution of Monocotyledonous Plants. Nature 2012, 488, 213–219. [Google Scholar] [CrossRef]
- Dupouy, M.; Baurens, F.-C.; Derouault, P.; Hervouet, C.; Cardi, C.; Cruaud, C.; Istace, B.; Labadie, K.; Guiougou, C.; Toubi, L.; et al. Two Large Reciprocal Translocations Characterized in the Disease Resistance-Rich Burmannica Genetic Group of Musa acuminata. Ann. Bot. 2019, 124, 319–329. [Google Scholar] [CrossRef]
- Martin, G.; Baurens, F.; Hervouet, C.; Salmon, F.; Delos, J.; Labadie, K.; Perdereau, A.; Mournet, P.; Blois, L.; Dupouy, M.; et al. Chromosome Reciprocal Translocations Have Accompanied Subspecies Evolution in Bananas. Plant J. 2020, 104, 1698–1711. [Google Scholar] [CrossRef]
- Belser, C.; Baurens, F.-C.; Noel, B.; Martin, G.; Cruaud, C.; Istace, B.; Yahiaoui, N.; Labadie, K.; Hribova, E.; Dolezel, J.; et al. Telomere-to-Telomere Gapless Chromosomes of Banana Using Nanopore Sequencing. Commun. Biol. 2021, 4, 1047. [Google Scholar]
- Raboin, L.M.; Carreel, F.; Noyer, J.-L.; Baurens, F.-C.; Horry, J.P.; Bakry, F.; Tézenas du Montcel, H.; Ganry, J.; Lanaud, C.; Lagoda, P.J.L. Diploid Ancestors of Triploid Export Banana Cultivars: Molecular Identification of 2n Restitution Gamete Donors and n Gamete Donors. Mol. Breed. 2005, 16, 333–341. [Google Scholar] [CrossRef]
- Martin, G.; Baurens, F.-C.; Labadie, K.; Hervouet, C.; Salmon, F.; Marius, F.; Paulo-de-la-Reberdiere, N.; Van Den Houwe, I.; Aury, J.-M.; D’Hont, A.; et al. Shared Pedigree Relationships and Transmission of Unreduced Gametes in Cultivated Banana. Ann. Bot. 2023, 131, 1149–1161. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Vicente, L.; Carreel, F.; Roussel, V.; Carlier, J.; Abadie, C. Guidelines for the Evaluation of Resistance to Pseudocercospora Leaf Spots of Banana. In Practical Guidelines for Early Screening and Field Evaluation of Banana against Fusarium Wilt, Pseudocercospora Leaf Spots and Drought; Dita, M., Ed.; Bioversity International: Rome, Italy, 2021; pp. 21–63. ISBN 978-92-9255-192-6. [Google Scholar]
- Kimunye, J.; Were, E.; Swennen, R.; Viljoen, A.; Mahuku, G. Sources of Resistance to Pseudocercospora fijiensis, the Cause of Black Sigatoka in Banana. Plant Pathol. 2021, 70, 1651–1664. [Google Scholar] [CrossRef] [PubMed]
- Roux-Cuvelier, M.; Grisoni, M.; Bellec, A.; Bloquel, E.; Charron, C.; Delalande, M.; Delmas, M.; Didier, A.; Durel, C.-E.; Duval, C.-H.; et al. Conservation of Horticultural Genetic Resources in France. Chron. Hortic. 2021, 61, 21–36. [Google Scholar]
- Bakry, F. Zygotic Embryo Rescue in Bananas. Fruits 2008, 63, 111–115. [Google Scholar] [CrossRef][Green Version]
- IPSDK Explorer. Reactiv’IP (Grenoble, France). 2022. Available online: https://www.reactivip.com/ (accessed on 23 January 2023).
- Alves, K.d.S.; Del Ponte, E.M. Epifitter, Analysis and Simulation of Plant Disease Progress Curves. R package Version 0.3.0. 2021. Available online: https://cran.r-project.org/web/packages/epifitter/index.html (accessed on 23 January 2023).
- Akaike, H. A New Look at the Statistical Model Identification. IEEE Trans. Autom. Control. 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Ratkowsky, D.A. Nonlinear Regression Modeling, Self-Starting Nls Weibull Growth Curve Model; Marcel Dekker Inc.: New York, NY, USA, 1983. [Google Scholar]
- Pinheiro, J.; Bates, D.; DebRoy, S.; Sarkar, D.; Eispack; Heisterkamp, S.; Van Willigen, B.; Ranke, J.; R Core Team. Nlme, Linear and Nonlinear Mixed Effects Models; R Package Version 3.1-160. 2022. Available online: https://cran.r-project.org/web/packages/nlme/index.html (accessed on 11 November 2022).
- Wickham, H.; Chang, W.; Henry, L.; Pedersen, T.L.; Takahashi, K.; Wilke, C.; Woo, K.; Yutani, H.; Dunnington, D.; van den Brand, T.; et al. Ggplot2, Create Elegant Data Visualisations Using the Grammar of Graphics; R Package Version 3.5.0. 2024. Available online: https://cran.r-project.org/web/packages/ggplot2/index.html (accessed on 24 February 2024).
- Brooks, M.E.; Kristensen, K.; van Benthem, K.J.; Magnusson, A.; Berg, C.W.; Nielsen, A.; Skaug, H.J.; Mächler, M.; Bolker, B.M. glmmTMB Balances Speed and Flexibility among Packages for Zero-Inflated Generalized Linear Mixed Modeling. R J. 2017, 9, 378–400, R Package Version 1.1.4. 2022. Available online: https://cran.r-project.org/web/packages/glmmTMB/index.html (accessed on 11 November 2022). [CrossRef]
- Hartig, F.; Lohse, L. DHARMa: Residual Diagnostics for Hierarchical (Multi-Level/Mixed) Regression Models; R Package Version 0.4.6. 2022. Available online: https://cran.r-project.org/web/packages/DHARMa/index.html (accessed on 23 January 2023).
- Cormier, F.; Lawac, F.; Maledon, E.; Gravillon, M.-C.; Nudol, E.; Mournet, P.; Vignes, H.; Chaïr, H.; Arnau, G. A Reference High-Density Genetic Map of Greater Yam (Dioscorea alata L.). Theor. Appl. Genet. 2019, 132, 1733–1744. [Google Scholar] [CrossRef]
- Herten, K.; Hestand, M.S.; Vermeesch, J.R.; Van Houdt, J.K. GBSX: A Toolkit for Experimental Design and Demultiplexing Genotyping by Sequencing Experiments. BMC Bioinform. 2015, 16, 73. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt Removes Adapter Sequences from High-Throughput Sequencing Reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Garsmeur, O.; Droc, G.; Antonise, R.; Grimwood, J.; Potier, B.; Aitken, K.; Jenkins, J.; Martin, G.; Charron, C.; Hervouet, C.; et al. A Mosaic Monoploid Reference Sequence for the Highly Complex Genome of Sugarcane. Nat. Commun. 2018, 9, 2638. [Google Scholar] [CrossRef] [PubMed]
- Van Ooijen, J.W. JoinMap® 5, Software for the Calculation of Genetic Linkage Maps in Experimental Populations of Diploid Species; Kyazma BV: Wageningen, The Netherlands, 2018. [Google Scholar]
- Martin, G.; Baurens, F.-C.; Droc, G.; Rouard, M.; Cenci, A.; Kilian, A.; Hastie, A.; Doležel, J.; Aury, J.-M.; Alberti, A.; et al. Improvement of the Banana “Musa acuminata” Reference Sequence Using NGS Data and Semi-Automated Bioinformatics Methods. BMC Genom. 2016, 17, 12. [Google Scholar] [CrossRef] [PubMed]
- Rami, J.-F. (CIRAD, Paris, France). Personal communication, 2017. [Google Scholar]
- Van Ooijen, J.W. MapQTL® 6, Software for Mapping of Quantitative Trait Loci in Experimental Populations of Diploid Species; Kyazma BV: Wageningen, The Netherlands, 2009. [Google Scholar]
- Calle García, J.; Guadagno, A.; Paytuvi-Gallart, A.; Saera-Vila, A.; Amoroso, C.G.; D’Esposito, D.; Andolfo, G.; Aiese Cigliano, R.; Sanseverino, W.; Ercolano, M.R. PRGdb 4.0: An Updated Database Dedicated to Genes Involved in Plant Disease Resistance Process. Nucleic Acids Res. 2022, 50, D1483–D1490. [Google Scholar] [CrossRef] [PubMed]
- Dievart, A.; Gottin, C.; Périn, C.; Ranwez, V.; Chantret, N. Origin and Diversity of Plant Receptor-Like Kinases. Annu. Rev. Plant Biol. 2020, 71, 131–156. [Google Scholar] [CrossRef] [PubMed]
- Perrier, X.; Bakry, F.; Carreel, F.; Jenny, C.; Horry, J.-P.; Lebot, V.; Hippolyte, I. Combining Biological Approaches to Shed Light on the Evolution of Edible Bananas. Ethnobot. Res. Appl. 2009, 7, 199–216. [Google Scholar] [CrossRef]
- Martin, G.; Carreel, F.; Coriton, O.; Hervouet, C.; Cardi, C.; Derouault, P.; Roques, D.; Salmon, F.; Rouard, M.; Sardos, J.; et al. Evolution of the Banana Genome (Musa acuminata) Is Impacted by Large Chromosomal Translocations. Mol. Biol. Evol. 2017, 34, 2140–2152. [Google Scholar] [CrossRef] [PubMed]
- Biabiany, S.; Araou, E.; Cormier, F.; Martin, G.; Carreel, F.; Hervouet, C.; Salmon, F.; Efile, J.-C.; Lopez-Lauri, F.; D’Hont, A.; et al. Detection of Dynamic QTLs for Traits Related to Organoleptic Quality during Banana Ripening. Sci. Hortic. 2022, 293, 110690. [Google Scholar] [CrossRef]
- Brazier, T.; Glémin, S. Diversity and Determinants of Recombination Landscapes in Flowering Plants. PLoS Genet. 2022, 18, e1010141. [Google Scholar] [CrossRef]
- Martin, G.; Cottin, A.; Baurens, F.-C.; Labadie, K.; Hervouet, C.; Salmon, F.; Paulo-de-la-Reberdiere, N.; Van den Houwe, I.; Sardos, J.; Aury, J.-M.; et al. Interspecific Introgression Patterns Reveal the Origins of Worldwide Cultivated Bananas in New Guinea. Plant J. 2023, 113, 802–818. [Google Scholar] [CrossRef]
- Landry, C.; Bonnot, F.; Ravigné, V.; Carlier, J.; Rengifo, D.; Vaillant, J.; Abadie, C. A Foliar Disease Simulation Model to Assist the Design of New Control Methods against Black Leaf Streak Disease of Banana. Ecol. Model. 2017, 359, 383–397. [Google Scholar] [CrossRef]
- French, E.; Kim, B.-S.; Iyer-Pascuzzi, A.S. Mechanisms of Quantitative Disease Resistance in Plants. Semin. Cell Dev. Biol. 2016, 56, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Sekhwal, M.K.; Li, P.; Lam, I.; Wang, X.; Cloutier, S.; You, F.M. Disease Resistance Gene Analogs (RGAs) in Plants. Int. J. Mol. Sci. 2015, 16, 19248–19290. [Google Scholar] [CrossRef] [PubMed]
- Anuradha, C.; Chandrasekar, A.; Backiyarani, S.; Uma, S. MusaRgeneDB: An Online Comprehensive Database for Disease Resistance Genes in Musa Spp. 3 Biotech 2022, 12, 222. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Pei, T.; Jiang, J.; Yang, H.; Zhang, H.; Li, J.; Xu, X. Understanding the Mechanisms of Resistance to Tomato Leaf Mold: A Review. Hortic. Plant J. 2022, 8, 667–675. [Google Scholar] [CrossRef]
- Isaza, R.E.A.; Diaz-Trujillo, C.; Dhillon, B.; Aerts, A.; Carlier, J.; Crane, C.F.; de Jong, T.V.; de Vries, I.; Dietrich, R.; Farmer, A.D.; et al. Combating a Global Threat to a Clonal Crop: Banana Black Sigatoka Pathogen Pseudocercospora fijiensis (Synonym Mycosphaerella fijiensis) Genomes Reveal Clues for Disease Control. PLoS Genet. 2016, 12, e1005876. [Google Scholar] [CrossRef] [PubMed]
- Stergiopoulos, I.; van den Burg, H.A.; Okmen, B.; Beenen, H.G.; van Liere, S.; Kema, G.H.J.; de Wit, P.J.G.M. Tomato Cf Resistance Proteins Mediate Recognition of Cognate Homologous Effectors from Fungi Pathogenic on Dicots and Monocots. Proc. Natl. Acad. Sci. USA 2010, 107, 7610–7615. [Google Scholar] [CrossRef] [PubMed]
- Pinheiro, T.D.M.; Rego, E.C.S.; Alves, G.S.C.; Fonseca, F.C.D.A.; Cotta, M.G.; Antonino, J.D.; Gomes, T.G.; Amorim, E.P.; Ferreira, C.F.; Costa, M.M.D.C.; et al. Transcriptome Profiling of the Resistance Response of Musa acuminata Subsp. burmannicoides, Var. Calcutta 4 to Pseudocercospora Musae. Int. J. Mol. Sci. 2022, 23, 13589. [Google Scholar] [CrossRef] [PubMed]
- Peraza-Echeverria, S.; Dale, J.L.; Harding, R.M.; Smith, M.K.; Collet, C. Characterization of Disease Resistance Gene Candidates of the Nucleotide Binding Site (NBS) Type from Banana and Correlation of a Transcriptional Polymorphism with Resistance to Fusarium oxysporum f.Sp. Cubense Race 4. Mol. Breed. 2008, 22, 565–579. [Google Scholar] [CrossRef]
- Zhang, L.; Cenci, A.; Rouard, M.; Zhang, D.; Wang, Y.; Tang, W.; Zheng, S.-J. Transcriptomic Analysis of Resistant and Susceptible Banana Corms in Response to Infection by Fusarium oxysporum f. Sp. Cubense Tropical Race 4. Sci. Rep. 2019, 9, 8199. [Google Scholar] [CrossRef]
- Saintenac, C.; Lee, W.-S.; Cambon, F.; Rudd, J.J.; King, R.C.; Marande, W.; Powers, S.J.; Bergès, H.; Phillips, A.L.; Uauy, C.; et al. Wheat Receptor-Kinase-like Protein Stb6 Controls Gene-for-Gene Resistance to Fungal Pathogen Zymoseptoria tritici. Nat. Genet. 2018, 50, 368–374. [Google Scholar] [CrossRef]
- Saintenac, C.; Cambon, F.; Aouini, L.; Verstappen, E.; Ghaffary, S.M.T.; Poucet, T.; Marande, W.; Berges, H.; Xu, S.; Jaouannet, M.; et al. A Wheat Cysteine-Rich Receptor-like Kinase Confers Broad-Spectrum Resistance against Septoria Tritici Blotch. Nat. Commun. 2021, 12, 433. [Google Scholar] [CrossRef] [PubMed]
- Breit-McNally, C.; Laflamme, B.; Singh, R.A.; Desveaux, D.; Guttman, D.S. ZAR1: Guardian of Plant Kinases. Front. Plant Sci. 2022, 13, 981684. [Google Scholar] [CrossRef]
- Monaghan, J.; Zipfel, C. Plant Pattern Recognition Receptor Complexes at the Plasma Membrane. Curr. Opin. Plant Biol. 2012, 15, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, H.A.; Rodriguez-Arango, E.; Morales, J.G.; Kema, G.; Arango, R.E. Defense Gene Expression Associated with Biotrophic Phase of Mycosphaerella fijiensis M. Morelet Infection in Banana. Plant Dis. 2016, 100, 1170–1175. [Google Scholar] [CrossRef] [PubMed]
- Torres, J.M.; Calderón, H.; Rodríguez-Arango, E.; Morales, J.G.; Arango, R. Differential Induction of Pathogenesis-Related Proteins in Banana in Response to Mycosphaerella fijiensis Infection. Eur. J. Plant Pathol. 2012, 133, 887–898. [Google Scholar] [CrossRef]
- Portal, O.; Izquierdo, Y.; De Vleesschauwer, D.; Sánchez-Rodríguez, A.; Mendoza-Rodríguez, M.; Acosta-Suárez, M.; Ocaña, B.; Jiménez, E.; Höfte, M. Analysis of Expressed Sequence Tags Derived from a Compatible Mycosphaerella fijiensis–Banana Interaction. Plant Cell Rep. 2011, 30, 913–928. [Google Scholar] [CrossRef] [PubMed]
- Sreedharan, S.; Shekhawat, U.K.S.; Ganapathi, T.R. MusaSAP1, a A20/AN1 Zinc Finger Gene from Banana Functions as a Positive Regulator in Different Stress Responses. Plant Mol. Biol. 2012, 80, 503–517. [Google Scholar] [CrossRef] [PubMed]
- Passos, M.A.N.; de Cruz, V.O.; Emediato, F.L.; de Teixeira, C.C.; Azevedo, V.C.R.; Brasileiro, A.C.M.; Amorim, E.P.; Ferreira, C.F.; Martins, N.F.; Togawa, R.C.; et al. Analysis of the Leaf Transcriptome of Musa acuminata during Interaction with Mycosphaerella musicola: Gene Assembly, Annotation and Marker Development. BMC Genom. 2013, 14, 78. [Google Scholar] [CrossRef] [PubMed]
- Sánchez Timm, E.; Hidalgo Pardo, L.; Pacheco Coello, R.; Chávez Navarrete, T.; Navarrete Villegas, O.; Santos Ordóñez, E. Identification of Differentially-Expressed Genes in Response to Mycosphaerella fijiensis in the Resistant Musa Accession “Calcutta-4” Using Suppression Subtractive Hybridization. PLoS ONE 2016, 11, e0160083. [Google Scholar] [CrossRef][Green Version]
- Uma, S.; Backiyarani, S.; Saravanakumar, A.S.; Chandrasekar, A.; Thangavelu, R.; Saraswathi, M.S. Identification of Mycosphaerella Eumusae Responsive Unique Genes/Transcripts from a Resistant Banana Cultivar. Acta Hortic. 2016, 1114, 111–118. [Google Scholar] [CrossRef]
- Rodriguez, H.A.; Hidalgo, W.F.; Sanchez, J.D.; Menezes, R.C.; Schneider, B.; Arango, R.E.; Morales, J.G. Differential Regulation of Jasmonic Acid Pathways in Resistant (Calcutta 4) and Susceptible (Williams) Banana Genotypes during the Interaction with Pseudocercospora fijiensis. Plant Pathol. 2020, 69, 872–882. [Google Scholar] [CrossRef]
- Uwimana, B.; Mwanje, G.; Batte, M.; Akech, V.; Shah, T.; Vuylsteke, M.; Swennen, R. Continuous Mapping Identifies Loci Associated with Weevil Resistance [Cosmopolites Sordidus (Germar)] in a Triploid Banana Population. Front. Plant Sci. 2021, 12, 753241. [Google Scholar] [CrossRef]
- Álvarez-López, D.; Herrera-Valencia, V.A.; Góngora-Castillo, E.; García-Laynes, S.; Puch-Hau, C.; López-Ochoa, L.A.; Lizama-Uc, G.; Peraza-Echeverria, S. Genome-Wide Analysis of the LRR-RLP Gene Family in a Wild Banana (Musa acuminata Ssp. malaccensis) Uncovers Multiple Fusarium Wilt Resistance Gene Candidates. Genes 2022, 13, 638. [Google Scholar] [CrossRef] [PubMed]
- Tian, D.; Qin, L.; Verma, K.K.; Wei, L.; Li, J.; Li, B.; Zhou, W.; He, Z.; Wei, D.; Huang, S.; et al. Transcriptomic and Metabolomic Differences between Banana Varieties Which Are Resistant or Susceptible to Fusarium Wilt. PeerJ 2023, 11, e16549. [Google Scholar] [CrossRef] [PubMed]
- Uwimana, B.; Nakato, G.V.; Kanaabi, R.; Nasuuna, C.; Mwanje, G.; Mahuku, G.S.; Akech, V.; Vuylsteke, M.; Swennen, R.; Shah, T. Identification of the Loci Associated with Resistance to Banana Xanthomonas Wilt (Xanthomonas Vasicola Pv. Musacearum) Using DArTSeq Markers and Continuous Mapping. Horticulturae 2024, 10, 87. [Google Scholar] [CrossRef]
- Lheureux, F.; Carreel, F.; Jenny, C.; Lockhart, B.E.L.; Iskra Caruana, M.L. Identification of Genetic Markers Linked to Banana Streak Disease Expression in Inter-Specific Musa Hybrids. Theor. Appl. Genet. 2003, 106, 594–598. [Google Scholar] [CrossRef] [PubMed]
- Iskra-Caruana, M.-L.; Baurens, F.-C.; Gayral, P.; Chabannes, M. A Four-Partner Plant–Virus Interaction: Enemies Can Also Come from Within. Mol. Plant Microbe Interact 2010, 23, 1394–1402. [Google Scholar] [CrossRef] [PubMed]
- Chabannes, M.; Baurens, F.-C.; Duroy, P.-O.; Bocs, S.; Vernerey, M.-S.; Rodier-Goud, M.; Gayral, P.; Iskra Caruana, M.L. Three Infectious Viral Species Lying in Wait in the Banana Genome. J. Virol. 2013, 87, 8624–8637. [Google Scholar] [CrossRef]
- Umber, M.; Pichaut, J.-P.; Farinas, B.; Laboureau, N.; Janzac, B.; Plaisir-Pineau, K.; Pressat, G.; Baurens, F.-C.; Chabannes, M.; Duroy, P.-O.; et al. Marker-Assisted Breeding of Musa balbisiana Genitors Devoid of Infectious Endogenous Banana Streak Virus Sequences. Mol. Breed. 2016, 36, 74. [Google Scholar] [CrossRef]
- Sardos, J.; Rouard, M.; Hueber, Y.; Cenci, A.; Hyma, K.E.; van den Houwe, I.; Hribova, E.; Courtois, B.; Roux, N. A Genome-Wide Association Study on the Seedless Phenotype in Banana (Musa Spp.) Reveals the Potential of a Selected Panel to Detect Candidate Genes in a Vegetatively Propagated Crop. PLoS ONE 2016, 11, 25. [Google Scholar] [CrossRef] [PubMed]
- Nyine, M.; Uwimana, B.; Akech, V.; Brown, A.; Ortiz, R.; Doležel, J.; Lorenzen, J.; Swennen, R. Association Genetics of Bunch Weight and Its Component Traits in East African Highland Banana (Musa Spp. AAA Group). Theor. Appl. Genet. 2019, 132, 3295–3308. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, F.; Martawi, N.M.; Poerba, Y.S.; de Jong, H.; Schouten, H.; Kema, G.H.J. Genetic Mapping of Fusarium Wilt Resistance in a Wild Banana Musa acuminata Ssp. malaccensis Accession. Theor. Appl. Genet. 2020, 133, 3409–3418. [Google Scholar] [CrossRef]
- Chen, A.; Sun, J.; Martin, G.; Gray, L.-A.; Hřibová, E.; Christelová, P.; Yahiaoui, N.; Rounsley, S.; Lyons, R.; Batley, J.; et al. Identification of a Major QTL-Controlling Resistance to the Subtropical Race 4 of Fusarium oxysporum f. Sp. Cubense in Musa acuminata Ssp. malaccensis. Pathogens 2023, 12, 289. [Google Scholar] [CrossRef]
- Chen, A.; Sun, J.; Viljoen, A.; Mostert, D.; Xie, Y.; Mangila, L.; Bothma, S.; Lyons, R.; Hřibová, E.; Christelová, P.; et al. Genetic Mapping, Candidate Gene Identification and Marker Validation for Host Plant Resistance to the Race 4 of Fusarium oxysporum f. Sp. Cubense Using Musa acuminata Ssp. malaccensis. Pathogens 2023, 12, 820. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Bastidas, F. Panama Disease in Banana, Spread, Screens and Genes. Ph.D. Thesis, Wageningen University, Wageningen, The Netherlands, 19 March 2019. [Google Scholar]
- Thangavelu, R.; Saraswathi, M.S.; Uma, S.; Loganathan, M.; Backiyarani, S.; Durai, P.; Raj, E.E.; Marimuthu, N.; Kannan, G.; Swennen, R. Identification of Sources Resistant to a Virulent Fusarium Wilt Strain (VCG 0124) Infecting Cavendish Bananas. Sci. Rep. 2021, 11, 3183. [Google Scholar] [CrossRef] [PubMed]
- Zaelani, A.; Herlina; Poerba, Y.S. Germplasm Screening of Diploid Musa Spp. Collection for Fusarium Wilt Disease Resistance Using Multiplex-SCAR Markers. In Proceedings of the 9th International Symposium on Innovative Bioproduction Indonesia on Biotechnology and Bioengineering 2022: Strengthening Bioeconomy through Applied Biotechnology, Bioengineering, and Biodiversity, Bogor, Indonesia, 23–24 November 2022; AIP Publishing: Long Island, NY, USA, 2023; p. 020006. [Google Scholar]
- Quénéhervé, P.; Valette, C.; Topart, P.; Tezenas du Montcel, H.; Salmon, F. Nematode Resistance in Bananas: Screening Results on Some Wild and Cultivated Accessions of Musa Spp. Euphytica 2008, 165, 123–136. [Google Scholar] [CrossRef]
- Cowger, C.; Brown, J.K.M. Durability of Quantitative Resistance in Crops: Greater Than We Know? Annu. Rev. Phytopathol. 2019, 57, 253–277. [Google Scholar] [CrossRef]


| RGA/QTL on | chr06 | chr07 | chr08 | chr09 | Total | Include Protein Classes of DRAGO3 Tool 1 |
|---|---|---|---|---|---|---|
| NBS encoding | 1 | 3 | 1 | 4 | 9 | CNL + NL + CN + N |
| RLK | 1 | 3 | 10 | 25 | 39 | RLK + LecRK |
| RLP | 1 | 1 | 1 | 10 | 13 | RLP + CL + Lec |
| Other-R | 5 | 10 | 3 | 19 | 37 | CK + K + L |
| Total | 8 | 17 | 15 | 58 | 98 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Carreel, F.; Martin, G.; Ravel, S.; Roussel, V.; Pages, C.; Habas, R.; Cantagrel, T.; Guiougou, C.; Delos, J.-M.; Hervouet, C.; et al. Identification of Resistance QTLs to Black Leaf Streak Disease (Due to Pseudocercospora fijiensis) in Diploid Bananas (Musa acuminata). Horticulturae 2024, 10, 608. https://doi.org/10.3390/horticulturae10060608
Carreel F, Martin G, Ravel S, Roussel V, Pages C, Habas R, Cantagrel T, Guiougou C, Delos J-M, Hervouet C, et al. Identification of Resistance QTLs to Black Leaf Streak Disease (Due to Pseudocercospora fijiensis) in Diploid Bananas (Musa acuminata). Horticulturae. 2024; 10(6):608. https://doi.org/10.3390/horticulturae10060608
Chicago/Turabian StyleCarreel, Françoise, Guillaume Martin, Sébastien Ravel, Véronique Roussel, Christine Pages, Rémy Habas, Théo Cantagrel, Chantal Guiougou, Jean-Marie Delos, Catherine Hervouet, and et al. 2024. "Identification of Resistance QTLs to Black Leaf Streak Disease (Due to Pseudocercospora fijiensis) in Diploid Bananas (Musa acuminata)" Horticulturae 10, no. 6: 608. https://doi.org/10.3390/horticulturae10060608
APA StyleCarreel, F., Martin, G., Ravel, S., Roussel, V., Pages, C., Habas, R., Cantagrel, T., Guiougou, C., Delos, J.-M., Hervouet, C., Mournet, P., D’Hont, A., Yahiaoui, N., & Salmon, F. (2024). Identification of Resistance QTLs to Black Leaf Streak Disease (Due to Pseudocercospora fijiensis) in Diploid Bananas (Musa acuminata). Horticulturae, 10(6), 608. https://doi.org/10.3390/horticulturae10060608







