Abstract
This study investigated the effects of cadmium (Cd) stress on the micropropagation and rooting dynamics of two myrtle (Myrtus communis L.) genotypes with different fruit colors under controlled in vitro conditions. We evaluated the response of these genotypes to varying concentrations of Cd (0, 100, 200, 300, 400, and 500 µM) to determine dose-dependent effects on plantlet multiplication and root formation. Our results demonstrate that the white-fruited (WF) genotype exhibits greater resilience than the black-fruited (BF) genotype across all concentrations, maintaining higher multiplication rates and shoot heights. For instance, the multiplication rate at 100 µM Cd was highest for WF at 6.73, whereas BF showed the lowest rate of 1.94 at 500 µM. Similarly, increasing Cd levels significantly impaired root length and the number of roots for both genotypes, illustrating the detrimental impact of Cd on root system development. Additionally, this study incorporated machine learning (ML) models to predict growth outcomes. The multilayer perceptron (MLP) model, including random forest (RF) and XGBoost, was used to analyze the data. The MLP model performed notably well, demonstrating the potential of advanced computational tools in accurately predicting plant responses to environmental stress. For example, the MLP model accurately predicted shoot height with an R2 value of 0.87 and root length with an R2 of 0.99, indicating high predictive accuracy. Overall, our findings provide significant insights into the genotypic differences in Cd tolerance and the utility of ML models in plant science. These results underscore the importance of developing targeted strategies to enhance plant resilience in contaminated environments.
1. Introduction
The myrtle plant (Myrtus communis L.) is an evergreen shrub endemic to the temperate Mediterranean and subtropical regions, classified within the Myrtaceae family, which comprises approximately 100 genera and 3000 species [1]. Myrtle essential oils, derived from its leaves and fruits, find extensive applications in the pharmaceutical industry. Additionally, the leaves are frequently consumed as herbal tea [2,3,4]. The plant thrives predominantly in the Mediterranean basin, forming part of spontaneous bush-cover landscapes. Myrtle is recognized to have two primary subspecies, communis and tarentina, which are further differentiated into varieties based on the color of the mature fruits. Varieties include “melanocarpa”, noted for its bluish-black skin, and “leucocarpa”, identified by its yellowish skin [5]. The species is monogenic with hermaphroditic flowers, which contributes to its reproductive versatility. The aromatic leaves of myrtle, together with its fruits—typically blackish-purple or white—ripen in the autumn months from October to December. These fruits exhibit a sugary yet astringent flavor and are primarily pollinated by insects [6]. In horticultural terms, myrtle represents a valuable species due to its diverse applications. Beyond its pharmaceutical significance, the plant is also utilized for afforestation in the fire-damaged Mediterranean coastal forests and is marketed as an ornamental shrub across Europe. In Türkiye, myrtle fruits are particularly sought after in local bazaars, underscoring their cultural and economic importance [7].
Recent studies have highlighted the significant concern of cadmium (Cd) contamination in soils and its subsequent accumulation in plants, emphasizing this heavy metal’s critical impact on human health [8,9]. Cd’s presence in the environment primarily stems from anthropogenic activities, including atmospheric deposition, the application of sewage sludge to agricultural fields, and fertilizers [10,11,12]. Despite not being an essential nutrient, Cd is readily absorbed by plant roots, where it can accumulate to toxic levels in plant tissues, adversely affecting plant growth and potentially entering the food chain. The detrimental effects of Cd on plant growth are well-documented, with research indicating that high Cd concentrations can lead to health issues in plants, animals, and humans, including diseases of the lungs, liver, and kidneys, as well as impairments in vision, anemia, and elevated blood pressure [10,13,14]. Several studies have assessed the impact of Cd on various plant species, highlighting its capacity to inhibit growth and development [8]. In vitro techniques have been utilized to simulate controlled exposure conditions to further explore Cd’s impacts on plants. Zhang et al. [15] investigated the effects of Cd on the growth and physiological traits of in vitro potato (Solanum tuberosum L.) plantlets, noting reductions in growth, photosynthesis, and chlorophyll content due to Cd exposure.
Similarly, Kaur and Bhandari [16] examined the impact of Cd stress on the in vitro growth and antioxidant defenses of Phyllanthus amarus Schum. and Thonn., finding significant reductions in growth, biomass, and key antioxidant enzymes like superoxide dismutase (SOD), catalase (CAT), and peroxidase (POD). Rahmati and Ghasemnezhad [17] also reported diminished growth and increased Cd accumulation in two Rosa sp. cultivars subjected to in vitro Cd stress. The application of traditional statistical approaches for decrypting the encrypted information within the extensive datasets of biological interactions presents significant challenges due to the intricate, noisy, and deceptive nature of the datasets, which encompass multifaceted processes [18]. Nonetheless, various machine learning (ML) models have recently been utilized to effectively achieve precision, forecasting, and enhancement tasks in plant tissue culture procedures. ML, a subset of artificial intelligence (AI), is extensively utilized in plant-related disciplines, such as plant breeding [19], drought stress [20,21,22], in vitro micropropagation [23,24,25,26], in vitro germination [27,28], and various other domains within plant science. By leveraging AI technologies, scholars can analyze and clarify extensive datasets acquired from multiple sources. The precise prediction of traits has been demonstrated through the utilization of decision tree-based ML algorithms like random forest (RF), extreme gradient boost (XGBoost), and artificial neural network (ANN)-based multilayer perceptron (MLP) models. The utilization of AI-driven ML models can surmount the previously mentioned challenges by scrutinizing and forecasting intricate and multivariate datasets [26,29] and comprehending complex interconnections [30,31]. Integrating ML techniques allows computers to autonomously learn and transform data into meaningful insights without human programming [32].
This study aimed to comprehensively examine the effects of Cd stress on the micropropagation dynamics of two distinct myrtle genotypes, each characterized by different fruit colors, under controlled in vitro conditions. Furthermore, this research aimed to integrate advanced machine learning (ML) methodologies to effectively predict the impact of Cd stress on myrtle’s growth parameters. By employing ML techniques such as ANN-based MLP and decision tree-based models like RF and XGBoost, the study seeks to elucidate the subtle differences in Cd tolerance between the genotypes and accurately model their growth responses. This dual approach not only aims to deepen our understanding of the physiological responses of myrtle to metal stress but also to enhance the precision of predictive modeling in plant science, thereby contributing to the development of targeted mitigation and adaptation strategies in contaminated or risk-prone environments.
2. Materials and Methods
2.1. Plant Material
Two distinct myrtle types were carefully chosen for this study based on extensive selection processes in the 2017 areas of Karaisalı, Adana, and Erdemli, Mersin, Türkiye. The authors complied with the IUCN Policy Statement on Research Involving Species at Risk of Extinction and the Convention on the Trade in Endangered Species of Wild Fauna and Flora for collecting the plant material. The plant material was collected with official permission, numbered 43368836-335.01-E.1189927, from the Ministry of Agriculture and Forestry, the Republic of Türkiye. Strict standards prioritizing important visual qualities, such as increased productivity, notable fruit size, and robust and solid plant structure, served as the basis for the selection process. The study used two genotypes: one variety had black fruits, and another had white fruits.
2.2. Sterilization and Culture Protocol
The research plant material’s shoot tips were carefully decontaminated before being used in the study. They were washed with tap water for ten minutes, submerged in 70% ethanol for three minutes, and left in a 20% sodium hypochlorite (NaClO) solution for ten minutes. The shot tips were triple-rinsed with sterile distilled water after asepsis. They were then put on a Murashige and Skoog (MS) (Duchefa Biochemie, Haarlem, The Netherlands) [33] nutrient medium supplemented with 30 g/L sucrose (Duchefa Biochemie, Haarlem, The Netherlands), 8 g/L agar (Duchefa Biochemie, Haarlem, The Netherlands), and 1 mg/L 6-benzylaminopurine (BAP) (Duchefa Biochemie, Haarlem, The Netherlands). Before autoclaving at 121 °C for 21 min, the pH was adjusted to 5.7 with 0.1 NaOH. The cultures were kept in controlled environments with a 16-h light/eight-hour dark photoperiod and a temperature of 25 °C.
2.3. Micropropagation under In Vitro Cd Stress
A controlled experiment was carried out to explore the influence of Cd stress on micropropagation myrtle plants. The MS nutrient media were prepared with varying concentrations of Cd (Merck KGaA, Darmstadt, Germany) (0, 100, 200, 300, 400, and 500 µM), supplemented with 1 mg/L of BAP. The plants were then cultured and maintained under controlled environmental conditions, consisting of a 16-h light/8-h dark photoperiod and a temperature of 25 °C. The growth parameters of the plants were carefully monitored throughout the experimental period, which spanned multiple subcultures conducted at four-week intervals for three cycles. Specifically, the micropropagation rate (MR), representing the number of plantlets per plant, and the plant height (PH) (in centimeters) were recorded at the end of each subculture period. This comprehensive evaluation aimed to determine the effect of varying levels of Cd stress on the myrtle plants, providing insights into potential impacts on micropropagation.
2.4. Rooting under In Vitro Cd Stress
To investigate the in vitro rooting behavior of myrtle plants under various Cd stress conditions, a range of concentrations (0, 100, 200, 300, 400, and 500 µM) of Cd were incorporated into MS media supplemented with 1 mg/L indole-3-butyric acid (IBA) (Duchefa Biochemie, Haarlem, The Netherlands). Simultaneously, plants previously grown in Cd-free control media were transferred to rooting media. The rooting response was carefully evaluated over an eight-week experimental period, during which key parameters such as rooting rate (RR) (expressed as a percentage), root length (RL) (measured in centimeters), and the number of roots (NoR) per plant were assessed. This study aimed to determine the effect of Cd stress on myrtle plants’ rooting dynamics and gain valuable insights into their physiological responses to heavy metal toxicity.
2.5. Data Analysis
The present study investigated the micropropagation and rooting of black and white fruited myrtle cultivars under in vitro conditions of Cd stress, using a factorial order random plot trial design. In this context, plants were subcultured every four weeks, with three subcultures. Three replications and 10 shoot tips per replication were used. Subsequently, the data collected underwent an analysis of variance (ANOVA). The least significant difference (LSD) test was employed to conduct post hoc pairwise comparisons of means for treatments deemed significant by ANOVA. Statistical analyses were executed using the R-programming language. In addition, the Pearson correlation coefficients for the parameters were computed using the corrplot package in R software version 4.3.1.
2.6. Modeling Procedure
The objective of the study was to assess the effects of various input variables—specifically different concentrations of Cd (0, 100, 200, 300, 400, and 500 µM) and two distinct genotypes—on crucial output variables, which include proliferation rate, shoot length, number of roots, and root length, for predictive modeling. Measurements were obtained from 10 explants for each Cd concentration, yielding 120 data vectors. For a particular combination of concentration and genotypes, each data vector represents the observed values of output variables, such as proliferation, shoot length, root number, and root length. One-hot encoding was used to represent the genotype to the input vector to show the existence of each genotype. Machine learning models, including artificial neural network-based multilayer perceptron (ANN-based MLP) and decision tree-based (RF and XGBoost), were employed to validate and predict the data collected in this study (Figure 1). ANN-based MLP is a feedforward neural network comprising a series of processing nodes arranged in a linear, feedforward configuration, with one or more fully integrated hidden level(s), input(s), and output(s). The biases and weights associated with the error are updated after the training is performed using backpropagation, as shown in Equation (1) [34,35].
where y = the observed value of data point n and n = number of samples.
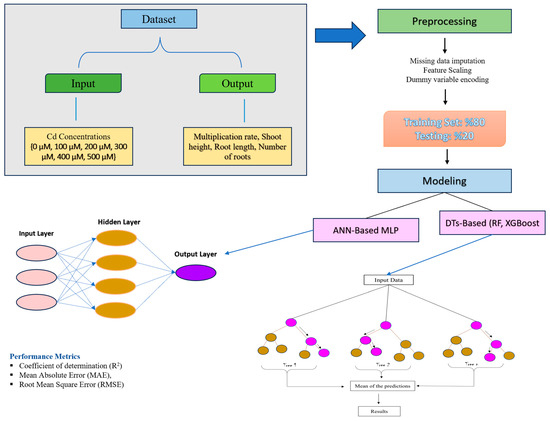
Figure 1.
Schematic diagram of modeling procedure.
As Pavlov [36] explained, Equation (2) summarizes the basic idea underpinning the entire operational system. Random forest, an ensemble learning technique composed of unpruned trees, was developed by Breiman [37] and has been effective in regression and classification tasks. Its efficiency and ease of implementation make it an effective tool for preventing overfitting, managing noise, and handling numerous features. During construction, the RF model incorporates randomness by introducing two sources into each tree, reducing correlation while preserving individual strengths.
where y = data point value and n = sampling size (number). ai and are coefficients associated with the predictor variables, k(x,xi) computes the similarity between the input vector x and the i-th data point xixi, and b is the bias term or intercept.
The XGBoost goal function that needs to be reduced is given in Equations (3) and (4), with an additional illustration of the XGBoost model at the jth iteration.
Equation (3), F is the function space of the tree model. fd represents an independent tree structure that classifies each individual i into one leaf. Equation (4), l is the convex loss function, and it measures the difference between observed y and predicted .
The dataset was divided into training and testing subsets using a 10-fold cross-validation method to thoroughly evaluate the predictive performance of the MLP and ML models. Cross-validation is typically employed when a machine learning task involves prediction, and the aim is to assess the accuracy of a predictive model during the training process [38]. The best model was determined using a grid search to identify the ideal hyperparameters. The coding implementation was facilitated by R-programming and using the Caret and Kernlab packages. A range of metrics was employed to assess and compare the accuracy and precision of the MLP and ML models. These metrics comprised the coefficient of determination (R2), which illustrates the extent of the relationship between the model and the dependent variable in Equation (5); the mean absolute error (MAE), which calculates the average discrepancy between the predicted and observed values in Equation (6) and the root mean square error (RMSE), which measures the degree of alignment between the regression line and the observed data points in Equation (7).
where = actual value, = Predicted value, = mean o the actual values, and n = sample count.
3. Results
3.1. Micropropagation Results
Table 1 depicts the influence of various Cd concentrations on the multiplication rates of two myrtle plant varieties, namely WF (white fruited) and BF (black fruited). The study examined Cd concentrations ranging from 0 µM to 500 µM, and the multiplication rates were documented for each Cd concentration level for both plant varieties (Figure 2 and Figure 3). The results exhibit a consistent pattern across both varieties, demonstrating a dose-dependent relationship between Cd exposure and myrtle plant multiplication. Notably, the WF variety generally displays higher multiplication rates than the BF variety across all Cd concentrations, indicating a potential difference in Cd tolerance between the two varieties. The multiplication rate of WF with 100 µM of Cd was the highest (6.73) after the control, while BF with 500 µM showed the lowest multiplication rate (1.94).

Table 1.
The effect of Cd concentrations on the multiplication rate of WF and BF myrtle varieties.

Figure 2.
Plantlets of BF myrtle genotype: (a) control and (b–f) 100, 200, 300, 400, and 500 µM Cd, respectively, from the third subculture.

Figure 3.
Plantlets of WF myrtle genotype: (a) control and (b–f) 100, 200, 300, 400, and 500 µM Cd, respectively, from the third subculture.
Statistical analysis reveals significant differences in multiplication rates between Cd concentrations and between myrtle varieties, as denoted by the low LSD (least significant difference) values (* indicating p < 0.05, ** indicating p < 0.01, and *** indicating p < 0.001). However, the interaction between genotype and Cd concentration was found to be non-significant, suggesting that the effect of Cd on multiplication rates is consistent across both WF and BF varieties. These findings emphasize the detrimental impact of Cd stress on myrtle plant growth and reproduction. Despite potential varietal differences in tolerance, increasing Cd concentrations adversely affect both WF and BF varieties.
A comprehensive overview of the influence of different Cd concentrations on the shoot height of two varieties of myrtle plants, namely “WF” and “BF”, is shown in Table 2. The table encompasses Cd concentrations ranging from 0 to 500 µM and measures the resulting shoot heights for each concentration level of Cd in both varieties. Observing the data in the table, it becomes evident that the shoot height diminishes as the Cd concentration escalates, indicating a negative impact of Cd on the vertical development of myrtle plants. This trend is consistent for WF and BF varieties, as taller shoot heights are observed at lower Cd concentrations, and progressively shorter heights are recorded as Cd concentration rises. Notably, the lowest shoot heights are registered at the highest Cd concentration of 500 µM for both varieties. Comparing the shoot heights of the varieties after the control, WF with 100 µM had the highest shoot height (6.14), while the BF with 500 µM had the lowest (1.96). While both WF and BF varieties show reduced shoot heights with increasing Cd concentration, their responses have subtle differences. Generally, WF plants tend to maintain slightly taller shoot heights than BF plants across all tested Cd concentrations. Statistical analysis reveals significant differences in shoot heights between Cd concentrations and myrtle varieties. Additionally, the interaction between genotype and Cd concentration is also significant, suggesting that the effect of Cd on shoot height varies depending on the myrtle variety. Overall, the findings underscore the detrimental impact of Cd stress on myrtle plant growth, emphasizing the necessity of implementing effective strategies to mitigate Cd-induced stress and safeguard plant health in contaminated environments.

Table 2.
The effect of Cd concentrations on shoot height (cm) of WF and BF myrtle varieties.
3.2. Rooting Results
The findings for both the WF and BF types show a consistent trend of decreased root length as Cd content rises. This implies that root system development in myrtle plants is inhibited by Cd exposure. Crucially, this decrease in root length is noticeable at all Cd doses examined, suggesting that Cd inhibits root growth in a dose-dependent manner Table 3. The LSD values (*** indicating p < 0.001) indicate a significant difference between the root lengths at different Cd doses. More specifically, roots get shorter at increased Cd concentrations; for both varieties, the lowest root lengths are found at the highest concentration of 500 µM. Furthermore, when root lengths are compared to the control (0 µM), BF exhibits the largest root length at 4.66, while WF records the lowest at 4.25. This comparison draws attention to the baseline root length before Cd exposure and demonstrates how significantly both varieties’ root lengths were reduced because of Cd stress. According to statistical analysis, there is no significant variation in root length between the kinds of myrtles (NS), suggesting that the reactions of the WF and BF varieties to Cd-induced stress are identical. This implies that myrtle plants are uniformly sensitive to Cd pollution, regardless of variety, highlighting the significance of implementing policies to reduce Cd contamination and safeguard plant health in impacted areas.

Table 3.
The effect of Cd concentrations on root length of WF and BF myrtle varieties.
Looking at the number of roots (Table 4), we can see that when the concentration of Cd rises, the WF and BF varieties both experience a decrease in the number of roots. This decrease indicates that Cd has a suppressive effect on the growth of roots in myrtle plants. This pattern is statistically significant, with fewer roots appearing as Cd concentration rises. Notably, the greatest Cd content significantly decreases the number of roots for both kinds. Additionally, no discernible variations exist between WF and BF cultivars regarding how they react to Cd, suggesting that both are susceptible to root inhibition brought on by Cd. A similar pattern can be seen when looking at the rooting rate, which measures the proportion of plants that can establish roots. The rooting rate decreases for WF and BF varieties as Cd content increases, suggesting that Cd interferes with myrtle plant root establishment. The adverse effects of increasing Cd levels on the rooting process are reaffirmed by significant disparities in rooting rates across Cd concentrations. Notably, for both varieties, the lowest rooting rates are found at the highest dose of Cd. Likewise, no appreciable variations exist in the rooting rate response of the WF and BF cultivars to Cd exposure (Table 5).

Table 4.
The effect of Cd concentrations on the number of roots WF and BF myrtle varieties.

Table 5.
The effect of Cd concentrations on the rooting rate of WF and BF myrtle varieties.
3.3. Correlation Analysis
The findings from the correlation analysis for the myrtle plants’ WF (Figure 4A) and BF (Figure 4B) varieties reveal strong positive relationships between several physiological parameters. The findings indicate that MR correlates strongly with SH, RR, RL, and NoR. Additionally, SH displays strong positive correlations with MR, RR, RL, and NoR. Furthermore, RR, RL, and NoR exhibit strong positive correlations with each other and with MR and SH. These correlations suggest coordinated growth patterns among these parameters within the WF variety, implying that changes in one parameter often coincide with changes in others.
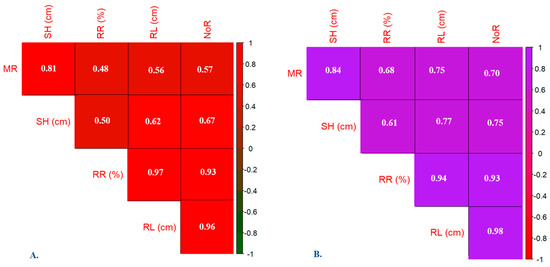
Figure 4.
Correlation triangle obtained for studied morphological traits of in vitro grown seedlings of myrtle genotypes: (A) white fruited (WF) variety and (B) black fruited (BF) variety. MR—multiplication rate; SH—shoot height; RR—root rate; RL—root length; NoR—number of roots.
3.4. Machine Learning Analysis
This study thoroughly examined ANN-based MLP and decision tree-based (RF, XGBoost) models’ performance in predicting various growth parameters of two myrtle varieties (Table 6). These parameters included the MR, SH, RL, and NoR. Regarding R2 values, which measure how well the models fit the data, MLP generally performed competitively across all parameters. For example, when predicting the MR, MLP achieved an R2 of 0.86, slightly lower than XGBoost’s 0.87 but higher than RF’s 0.83. Similarly, MLP scores 0.87 for SH, while RF and XGBoost perform slightly better at 0.89 and 0.91, respectively. For RL, MLP outperformed RF and XGBoost by a significant margin, with a remarkably high R2 of 0.99. However, the comparison is not solely based on the R2 values. Other metrics, such as mean absolute error (MAE) and root mean square error (RMSE), provide additional insights into the prediction accuracy. For instance, while the ANN might have slightly lower R2 values for certain parameters, it could have lower MAE and RMSE values, indicating more precise predictions regarding absolute errors.

Table 6.
Comparative performance evaluation of ANN and decision tree models for myrtle growth parameters.
A thorough analysis of the data presented in the table shows that the MLP model consistently demonstrates competitive or superior performance compared to the RF and XGBoost models across most of the parameters and metrics evaluated. For example, regarding R2 values, the MLP model frequently achieved comparable or higher scores than the RF and XGBoost models across various parameters. Additionally, when examining MAE and RMSE, the MLP model tended to have lower values, indicating better predictive accuracy regarding absolute and squared errors. While the RF and XGBoost models occasionally outperformed the MLP model in specific parameters or metrics, the overall trend suggests that the MLP model is the most reliable model for predicting myrtle growth parameters (MLP > XGBoost > RF). The model’s observed performance is attributed to the final hyperparameter values selected by grid search, as indicated by the performance indicators listed in Table 6. The MLP model’s final hyperparameter values were a regularization parameter of 0.01, three hidden layers of 100, 50, and 25 neurons each, and an ideal learning rate of 0.001. The final hyperparameter parameters for the random forest (RF) model were 100 trees with a maximum depth of 10 and a minimum of 5 samples per leaf. The final hyperparameter values for the extreme gradient boosting (XGBoost) model were 200 boosting rounds, a maximum tree depth of 5, and a learning rate of 0.1.
The graphical depictions of the actual and predicted scores generated by different models are presented in Figure 5. Each model’s performance is showcased by comparing the observed (actual) scores and the scores predicted by the respective model. These visual representations offer valuable insights into the effectiveness of each model in capturing the underlying patterns and trends in the data. Moreover, they enable a comprehensive evaluation of model performance, empowering stakeholders to make informed decisions regarding selecting and implementing the most appropriate predictive models for the specific task or application.
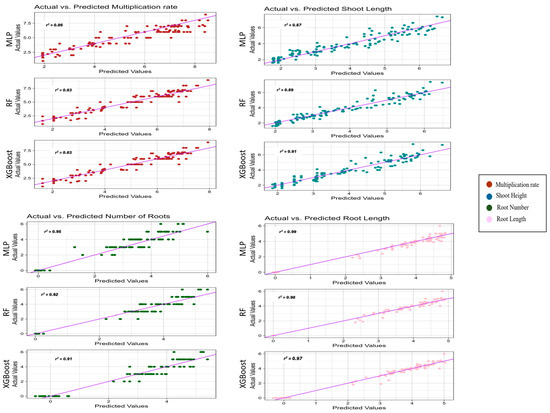
Figure 5.
Actual and predicted values of ANN-Based MLP and decision tree-based (RF, XGBoost) models.
4. Discussion
In our study, the influence of various Cd concentrations on the micropropagation of myrtle varieties WF and BF was examined. The results indicated a dose-dependent reduction in MR, SH, and rooting characteristics as Cd levels increased. This observation aligns with findings from several studies, highlighting the general phytotoxic effects of Cd across different plant species and experimental settings. The micropropagation results from our research revealed that even at lower concentrations of Cd (100 µM), there was a noticeable decrease in the multiplication rates and shoot heights, which further declined with higher concentrations. For instance, the BF variety exhibited the lowest multiplication rate at 500 µM of Cd. This outcome is consistent with Manquián-Cerda et al. [39], who observed a significant increase in oxidative stress markers like malondialdehyde (MDA) and hydrogen peroxide in blueberry plantlets under similar Cd exposures. These markers indicate cellular damage and oxidative stress, corroborating the decline in physiological functions observed in myrtle varieties under Cd stress.
Moreover, the significant differences in shoot heights and rooting characteristics observed between the WF and BF varieties under varying Cd concentrations suggest a genotype-specific response to Cd stress. This finding is particularly interesting as it adds to evidence suggesting varietal differences in Cd tolerance. For instance, in the study by Ullah et al. [40], chickpea cultivars exhibited differential growth responses and Cd accumulation patterns under hydroponic culture conditions, indicating varietal resilience to Cd toxicity. Similarly, our results underline the potential for identifying and utilizing genetically resilient plant varieties in areas prone to heavy metal contamination. The Baktemur [41] study on garlic also supports our observations, where increased Cd concentration led to a pronounced reduction in plant development metrics such as leaf and root growth. This indicates Cd’s broad-spectrum toxic effects on plant vitality, which we also noted in myrtle varieties. The complete inhibition of root formation at higher Cd concentrations reported in garlic underlines the severe impact of Cd, mirroring our findings where the highest Cd concentration resulted in the most substantial decline in rooting metrics for both myrtle varieties.
Furthermore, the comprehensive analysis by Torun et al. [42] on cherry rootstocks under in vitro conditions showed a similar trend of decreasing biomass with increasing Cd concentrations, reinforcing the general adverse effects of Cd we observed in our study. The specifics of Cd-induced physiological changes reported in their research, such as decreased dry matter production and symptoms of Cd toxicity, provide a comparative baseline that supports our conclusions about Cd’s detrimental effects on plant growth and viability. In conclusion, the corroborative findings across different studies enhance our understanding of Cd’s impact on plant physiology and stress response mechanisms. By drawing parallels between our results and those reported in the literature, it becomes evident that Cd contamination presents a consistent challenge to plant health across various species and conditions.
This highlights the necessity for continued research into species-specific responses and potential genetic resilience to Cd exposure, paving the way for developing effective phytoremediation strategies and improving crop tolerance to heavy metal stress. The findings of our study, focusing on the efficacy of ANN-based MLP and decision tree-based models like RF and XGBoost in predicting myrtle growth parameters, are in notable concordance with the outcomes reported by Şimşek [20], which emphasized the potency of RF in predicting water stress effects in strawberry cultivars using varying PEG concentrations. Similar to our observation where MLP exhibited a pronounced accuracy (R2 = 0.99 for root length), Şimşek et al. [23] also reported high accuracy for RF models in critical growth predictions, emphasizing the model’s utility in agricultural applications where precise prediction of plant responses to environmental stressors is crucial. The study by Şimşek et al. [23] on optimizing micropropagation protocols for lavender also corroborates our findings regarding integrating machine learning techniques to enhance plant propagation efficiencies. Their results highlighted the differential performance of various ML models, with MLP and XGBoost showing variable effectiveness across different plant characteristics. This parallels our observations of MLP’s robustness across multiple growth parameters in myrtle.
Moreover, the work by Tütüncü [43] on the impact of protein hydrolysates on primrose growth further aligns with our results, particularly in the use of MLP, which they found to be superior for modeling complex root traits, a finding echoed by our study where MLP consistently showed lower MAE and RMSE values, underscoring its precision in growth prediction. Lastly, the study by García-Pérez et al. [44], although focused on the phenolic compound production in Bryophyllum, offers an insightful comparison regarding the utility of machine learning to unravel complex biological responses under stress conditions. Their use of ANNs to identify key influencing factors on phenolic production mirrors our methodological approach, where machine learning models helped delineate the predictive reliability across different growth parameters in myrtle. It is evident that different models demonstrate unique strengths depending on the culture conditions and parameters being predicted. For instance, the study by Tarraf et al. [45] introduces a novel application of machine learning models like random forest (RF), support vector machines (SVM), Gaussian processes (GP), and multilayer perceptrons (MLP) in the in vitro propagation of saffron using a temporary immersion system (TIS). Their findings highlighted that RF models generally outperformed other algorithms across multiple growth parameters, such as the number of microcorms, shoots, and roots, indicating a robust predictive capability for this specific setup. In contrast, García-Pérez et al. [44] utilized neurofuzzy logic to elucidate the complex interplay between two phytohormones—auxin (IAA) and cytokinin (BAP)—in the organogenesis of unexploited medicinal plants from the Bryophyllum subgenus. This approach allowed for the discovery of nuanced interactions within the plant tissue culture, which could not have been easily deduced from traditional analytical methods. Here, the neurofuzzy model provided superior insights into how different concentrations of BAP and IAA influence various organogenetic outcomes, demonstrating the model’s applicability in more detailed hormonal studies. Kirtis et al. [46] emphasized the predictive power of machine learning models in analyzing the growth responses of desi chickpea to different concentrations of phytohormones in an MS medium. Their study successfully used models like RF, SVM, and XGBoost to predict shoot count and length, with the RF model showing exceptional accuracy. This example not only underscores the versatility of RF in another context but also points to the potential of integrating diverse machine learning models based on the specific traits and conditions studied. These studies collectively illustrate the growing relevance of machine learning in advancing plant tissue culture technologies. By integrating sophisticated computational models, researchers can better predict and enhance plant growth under in vitro conditions, potentially revolutionizing practices in agricultural biotechnology. However, the choice of model should be carefully considered based on the specific requirements of the study, such as the type of data available, the complexity of the biological interactions, and the precision needed in the predictions. While this study demonstrates the utility of ML models, including ANN-based MLP, decision tree-based RF, and XGBoost, for predicting the effects of Cd stress on myrtle genotypes, the relatively small sample size may impact the generalizability and reliability of the predictions. ML models typically require large datasets to capture the complexities and variations in biological data robustly. The limited number of samples available in this experimental setup raises concerns regarding overfitting, where models might adapt too closely to the specific dataset characteristics rather than learning generalizable patterns applicable in broader contexts. Acknowledging this limitation, further studies with expanded sample sizes are recommended to validate and refine the predictive capabilities of these ML models in plant stress physiology.
5. Conclusions
This study successfully delineated the impacts of varying Cd concentrations on the micropropagation and rooting dynamics of two distinct genotypes characterized by their fruit colors. The WF genotype consistently demonstrated higher resilience to Cd stress across all tested parameters than the BF genotype, suggesting inherent differences in Cd tolerance between the genotypes. These findings contribute to our understanding of Cd’s phytotoxic effects and underline the importance of selecting and breeding Cd-resistant plant varieties for cultivation in contaminated soils. Integrating machine learning models (ANN-based MLP, decision tree-based RF, and XGBoost) proved highly effective in predicting growth responses under Cd stress. This application highlights the potential of these computational tools to enhance predictive accuracy in plant physiological studies, providing a robust approach for future research focused on environmental stress mitigation. This research opens several avenues for advancing our knowledge of heavy metal stress management in plants. Future studies should explore the genetic bases of Cd tolerance, potentially leading to the development of genetically modified plants that can thrive in contaminated environments. Further refinement of machine learning models based on larger datasets could also improve the predictability and applicability of these techniques in real-world agricultural settings. By continuing to build on the methodologies and findings of this study, researchers can better equip themselves to address the challenges posed by heavy metal contamination in agriculture, ensuring food security and environmental sustainability.
Author Contributions
Conceptualization, M.T. and Ö.Ş.; data curation, M.A.I.; methodology, M.T., M.A.I., T.İ., D.D., Y.A.K. and Ö.Ş.; supervision, Ö.Ş.; visualization, M.A.I., T.İ. and D.D.; writing—original draft, M.T., M.A.I., D.D. and Ö.Ş.; writing—review and editing, T.İ., Y.A.K. and Ö.Ş. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
Data are contained within the article.
Acknowledgments
We would like to thank the Office of the Dean for Research at Erciyes University for providing the necessary infrastructure and laboratory facilities at the ArGePark research building.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Rezaee, A.; Kamali, K. A new commercial protocol for micropropagation of myrtus tree. Adv. Biores. 2014, 5, 73–79. [Google Scholar]
- Flamini, G.; Cioni, P.L.; Morelli, I.; Maccioni, S.; Baldini, R. Phytochemical typologies in some populations of Myrtus communis L. on caprione promontory (East Liguria, Italy). Food Chem. 2004, 85, 599–604. [Google Scholar] [CrossRef]
- Dönmez, D. Regeneration of plants from alginate-encapsulated shoot tips of myrtle (Myrtus communis L.). Erwerbs-Obstbau 2022, 64, 307–314. [Google Scholar] [CrossRef]
- Şimsek, Ö.; Açar, E.; Dönmez, D.; Şimsek, Ö.; Aka Kaçar, Y. Development of genic-SSR markers in myrtle by RNA-seq. Erwerbs-Obstbau 2022, 64, 475–483. [Google Scholar] [CrossRef]
- Medda, S.; Mulas, M. Fruit quality characters of myrtle (Myrtus communis L.) selections: Review of a domestication process. Sustainability 2021, 13, 8785. [Google Scholar] [CrossRef]
- Şimşek, O.; Donmez, D.; Saridas, M.A.; Paydas Kargi, S.; Aka Kacar, Y. Genetic relationship and polymorphism of Turkish myrtles (Myrtus communis L.) as revealed by inter simple sequence repeat (ISSR). Appl. Ecol. Environ. Res. 2020, 18, 1141–1149. [Google Scholar] [CrossRef]
- Şimşek, Ö.; Dönmez, D.; Sarıdaş, M.A.; Acar, E.; Kaçar, Y.A.; Kargı, S.P.; İzgü, T. In vitro and ex vitro propagation of Turkish myrtles through conventional and plantform bioreactor systems. PeerJ 2023, 11, e16061. [Google Scholar] [CrossRef]
- Dorris, M.; Motavalli, P.; Stevens, W. Cadmium effects on growth, photosynthesis, and nutrient use efficiency in tobacco. J. Plant Nutr. 2002, 25, 1–16. [Google Scholar]
- Tsadilas, C.D.; Karamanos, A.J.; Samaras, V. Effect of cadmium on seedling growth and nutrient uptake of common bean (Phaseolus vulgaris L.) in solution culture. Environ. Pollut. 2005, 133, 277–283. [Google Scholar]
- World Health Organization. Cadmium: Environmental Aspects; World Health Organization: Geneva, Switzerland, 1992. [Google Scholar]
- Manta, D.S.; Angelone, M.; Bellanca, A.; Neri, R.; Sprovieri, M. Heavy metals in urban soils: A case study from the city of Palermo (Sicily), Italy. Sci. Total Environ. 2002, 300, 229–243. [Google Scholar] [CrossRef] [PubMed]
- Komarnicki, G.J.K. Cadmium in soil and plants. Ecotoxicol. Environ. Saf. 2005, 61, 456–464. [Google Scholar]
- Marschner, H. Mineral Nutrition of Higher Plants; Academic Press: Cambridge, MA, USA, 1995. [Google Scholar]
- Jarup, L.; Berglund, M.; Elinder, C.G.; Nordberg, G.F. Health effects of cadmium exposure–a review of the literature and a risk estimate. Scand. J. Work Environ. Health 1998, 24, 1–51. [Google Scholar] [PubMed]
- Zhang, H.; Zong, N.; Zhang, Y.; Chen, M.; Feng, L. Effects of cadmium stress on the growth and physiological characteristics of potato (Solanum tuberosum L.) plantlets in vitro. Plant Growth Regul. 2019, 87, 427–437. [Google Scholar]
- Kaur, G.; Bhandari, K. In vitro cadmium stress-induced oxidative stress and changes in antioxidant defense system in Phyllanthus amarus Schum. & Thonn. Plant Growth Regul. 2018, 84, 245–256. [Google Scholar]
- Rahmati, M.; Ghasemnezhad, M. Effects of cadmium stress on growth, physiological and biochemical characteristics in two cultivars of Rosa sp. in vitro. J. Hortic. Postharvest Res. 2017, 1, 83–91. [Google Scholar]
- Pepe, M.; Hesami, M.; Jones, A.M.P. Machine learning-mediated development and optimization of disinfection protocol and scarification method for improved in vitro germination of cannabis seeds. Plants 2021, 10, 2397. [Google Scholar] [CrossRef] [PubMed]
- van Dijk, A.D.J.; Kootstra, G.; Kruijer, W.; de Ridder, D. Machine learning in plant science and plant breeding. iScience 2021, 24, 101822. [Google Scholar] [CrossRef] [PubMed]
- Şimşek, Ö. Machine learning offers insights into the impact of in vitro drought stress on strawberry cultivars. Agriculture 2024, 14, 294. [Google Scholar] [CrossRef]
- Jafari, M.; Shahsavar, A. The application of artificial neural networks in modeling and predicting the effects of melatonin on morphological responses of citrus to drought stress. PLoS ONE 2020, 15, e0240427. [Google Scholar] [CrossRef]
- Tahmasebi, A.; Niazi, A.; Akrami, S. Integration of meta-analysis, machine learning and systems biology approach for investigating the transcriptomic response to drought stress in Populus species. Sci. Rep. 2023, 13, 847. [Google Scholar] [CrossRef]
- Şimşek, Ö.; Dalda Şekerci, A.; Isak, M.A.; Bulut, F.; İzgü, T.; Tütüncü, M.; Dönmez, D. Optimizing Micropropagation and Rooting Protocols for Diverse Lavender Genotypes: A Synergistic Approach Integrating Machine Learning Techniques. Horticulturae 2024, 10, 52. [Google Scholar] [CrossRef]
- Jafari, M.; Daneshvar, M.H. Machine learning-mediated Passiflora caerulea callogenesis optimization. PLoS ONE 2024, 19, e0292359. [Google Scholar] [CrossRef]
- Demirel, F.; Uğur, R.; Popescu, G.C.; Demirel, S.; Popescu, M. Usage of Machine learning algorithms for establishing an effective protocol for the in vitro micropropagation ability of black chokeberry (Aronia melanocarpa (Michx.) Elliott). Horticulturae 2023, 9, 1112. [Google Scholar] [CrossRef]
- Özcan, E.; Atar, H.H.; Ali, S.A.; Aasim, M. Artificial neural network and decision tree–based models for prediction and validation of in vitro organogenesis of two hydrophytes—Hemianthus callitrichoides and Riccia fluitans. Vitr. Cell. Dev. Biol. Plant 2023, 59, 547–562. [Google Scholar] [CrossRef]
- Aasim, M.; Katırcı, R.; Akgur, O.; Yildirim, B.; Mustafa, Z.; Nadeem, M.A.; Yılmaz, G. Machine learning (ML) algorithms and artificial neural network for optimizing in vitro germination and growth indices of industrial hemp (Cannabis sativa L.). Ind. Crops Prod. 2022, 181, 114801. [Google Scholar] [CrossRef]
- Hesami, M.; Pepe, M.; Monthony, A.S.; Baiton, A.; Jones, A.M.P. Modeling and optimizing in vitro seed germination of industrial hemp (Cannabis sativa L.). Ind. Crops Prod. 2021, 170, 113753. [Google Scholar] [CrossRef]
- Kul, M.; Oskay, K.O.; Erden, F.; Akça, E.; Katırcı, R.; Köksaı, E.; Akıncı, E. Effect of process parameters on the electrodeposition of zinc on 1010 steel: Central composite design optimization. Int. J. Electrochem. Sci. 2020, 15, 9779–9795. [Google Scholar] [CrossRef]
- Jamshidi, S.; Yadollahi, A.; Arab, M.M.; Soltani, M.; Eftekhari, M.; Shiri, J. High throughput mathematical modeling and multi-objective evolutionary algorithms for plant tissue culture media formulation: Case study of pear rootstocks. PLoS ONE 2020, 15, e0243940. [Google Scholar] [CrossRef]
- Sadat-Hosseini, M.; Arab, M.M.; Soltani, M.; Eftekhari, M.; Soleimani, A.; Vahdati, K. Predictive modeling of Persian walnut (Juglans regia L.) in vitro proliferation media using machine learning approaches: A comparative study of ANN, KNN and GEP models. Plant Methods 2022, 18, 48. [Google Scholar] [CrossRef] [PubMed]
- Hesami, M.; Alizadeh, M.; Jones, A.M.P.; Torkamaneh, D. Machine learning: Its challenges and opportunities in plant system biology. Appl. Microbiol. Biotechnol. 2022, 106, 3507–3530. [Google Scholar] [CrossRef] [PubMed]
- Murashige, T.; Skoog, F. A revised medium for the rapid growth and bioassay with tobacco tissue cultures. Physiol. Plant 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Katırcı, R.; Yılmaz, E.K.; Kaynar, O.; Zontul, M. Automated evaluation of Cr-III coated parts using Mask RCNN and ML methods. Surf. Coat. Technol. 2021, 422, 127571. [Google Scholar] [CrossRef]
- Aasim, M.; Akin, F.; Ali, S.A. Synergizing LED Technology and Hydropriming for Intelligent Modeling and Mathematical Expressions to Optimize Chickpea Germination and Growth Indices. J. Plant Growth Regul. 2024, 1–20. [Google Scholar] [CrossRef]
- Pavlov, Y.L. Random Forests; CRC Press: Boca Raton, FL, USA, 2019; pp. 1–122. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Nti, I.K.; Nyarko-Boateng, O.; Aning, J. Performance of machine learning algorithms with different K values in K-fold cross-validation. Int. J. Inf. Technol. Comput. Sci. 2021, 13, 61–71. [Google Scholar] [CrossRef]
- Manquián-Cerda, K.; Escudey, M.; Zúñiga, G.; Arancibia-Miranda, N.; Molina, M.; Cruces, E. Effect of cadmium on phenolic compounds, antioxidant enzyme activity and oxidative stress in blueberry (Vaccinium corymbosum L.) plantlets grown in vitro. Ecotoxicol. Environ. Saf. 2016, 133, 316–326. [Google Scholar] [CrossRef] [PubMed]
- Ullah, S.; Khan, J.; Hayat, K.; Abdelfattah Elateeq, A.; Salam, U.; Yu, B.; Ma, Y.; Wang, H.; Tang, Z.-H. Comparative study of growth, cadmium accumulation and tolerance of three chickpea (Cicer arietinum L.) cultivars. Plants 2020, 9, 310. [Google Scholar] [CrossRef]
- Baktemur, G. The Effect of Some Heavy Metals on the Growth of Garlic under In Vitro Conditions. HortScience 2023, 58, 1–5. [Google Scholar] [CrossRef]
- Torun, A.A.; Kaçar, A.Y.; Çakmak, Ö.; Şimşek, Ö.; Erdem, H.; Yardım, P.; Tolay, İ. The effect of cadmium applied at increasing rates in cherry rootstock Maxma 14 on plant growth and cadmium uptake. J. Fac. Agric. Harran Univ. 2009, 13, 1–11. [Google Scholar]
- Tütüncü, M. Effects of Protein Hydrolysate Derived from Anchovy By-Product on Plant Growth of Primrose and Root System Architecture Analysis with Machine Learning. Horticulturae 2024, 10, 400. [Google Scholar] [CrossRef]
- García-Pérez, P.; Lozano-Milo, E.; Landin, M.; Gallego, P.P. Machine Learning unmasked nutritional imbalances on the medicinal plant Bryophyllum sp. cultured in vitro. Front. Plant Sci. 2020, 11, 576177. [Google Scholar] [CrossRef]
- Tarraf, W.; İzgü, T.; Şimşek, Ö.; Cicco, N.; Benelli, C. Saffron In Vitro Propagation: An Innovative Method by Temporary Immersion System (TIS), Integrated with Machine Learning Analysis. Horticulturae 2024, 10, 454. [Google Scholar] [CrossRef]
- Kirtis, A.; Aasim, M.; Katırcı, R. Application of artificial neural network and machine learning algorithms for modeling the in vitro regeneration of chickpea (Cicer arietinum L.). Plant Cell Tissue Organ Cult. 2022, 150, 141–152. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).