Comparative Study on Codon Usage Patterns across Chloroplast Genomes of Eighteen Taraxacum Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Sequence Retrieval and Filtering
- (1)
- The length of the CDS should be greater than 300 bp;
- (2)
- Each CDS must start with the start codon (ATG) and end with a stop codon (TAG, TGA, TAA);
- (3)
- The number of bases should be divisible by three;
- (4)
- CDSs must not include any internal stop codons or incorrect bases. The GC content at three positions (GC1, GC2, GC3) was then calculated using the CUSP program in EMBOSS explorer (http://emboss.toulouse.inra.fr/, (accessed on 20 March 2024)). Ultimately, a refined selection of CDSs, numbering between 48 to 65 for the 18 Taraxacum chloroplast genomes, was used for further analysis.
2.2. Analysis of Relative Synonymous Codon Usage (RSCU)
2.3. Identification of Putative Optimal Codons
2.4. Codon Usage Bias Analysis
2.5. ENc–GC3s Plot Analysis
2.6. PR2 Plot Analysis
2.7. Neutrality Plot Analysis
2.8. Correspondence Analysis (COA)
2.9. Statistical Analysis
3. Results
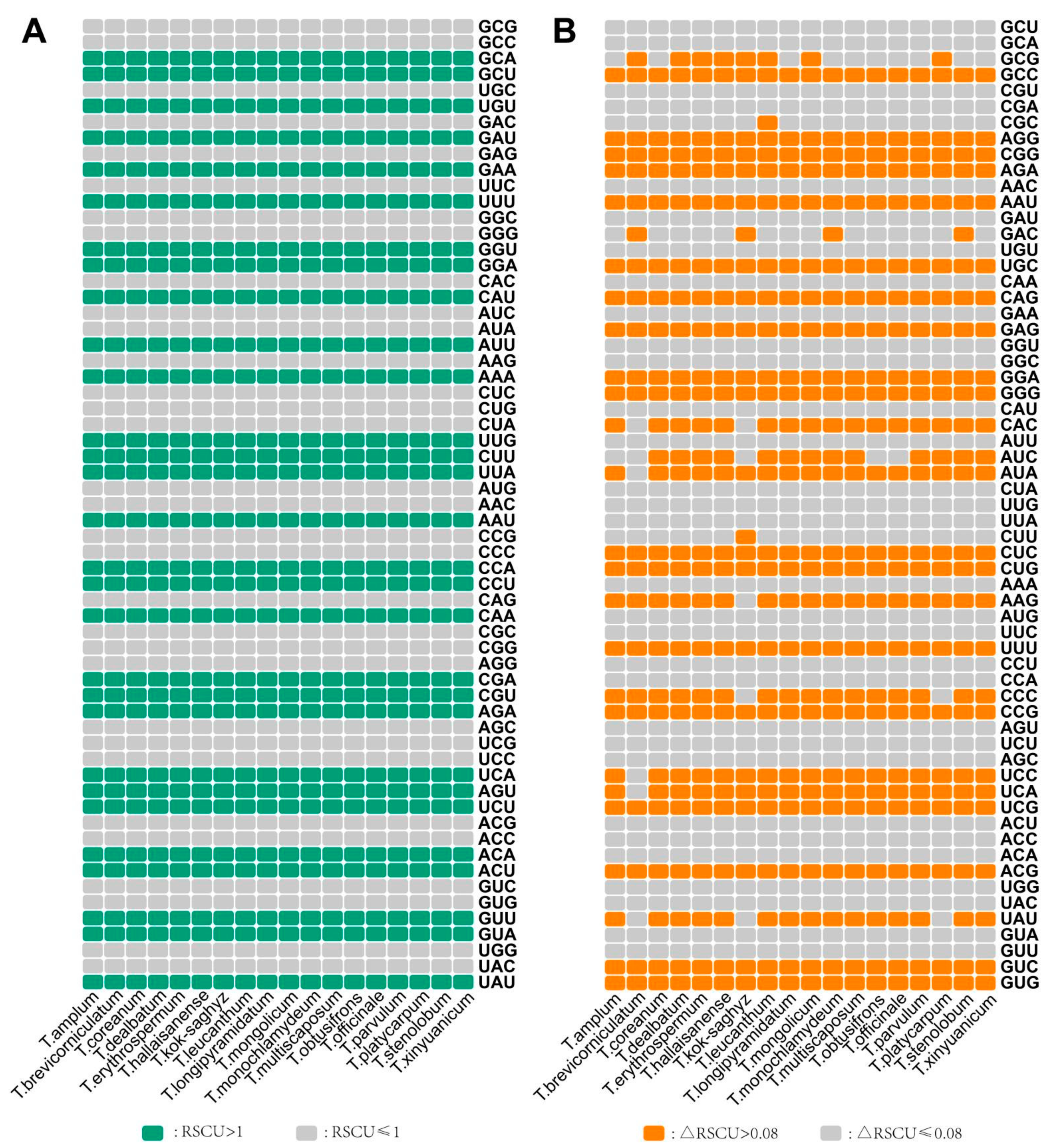
3.1. Characteristics of Codon Usage Bias Analysis
3.2. RSCU and Identification of Optimal Codons
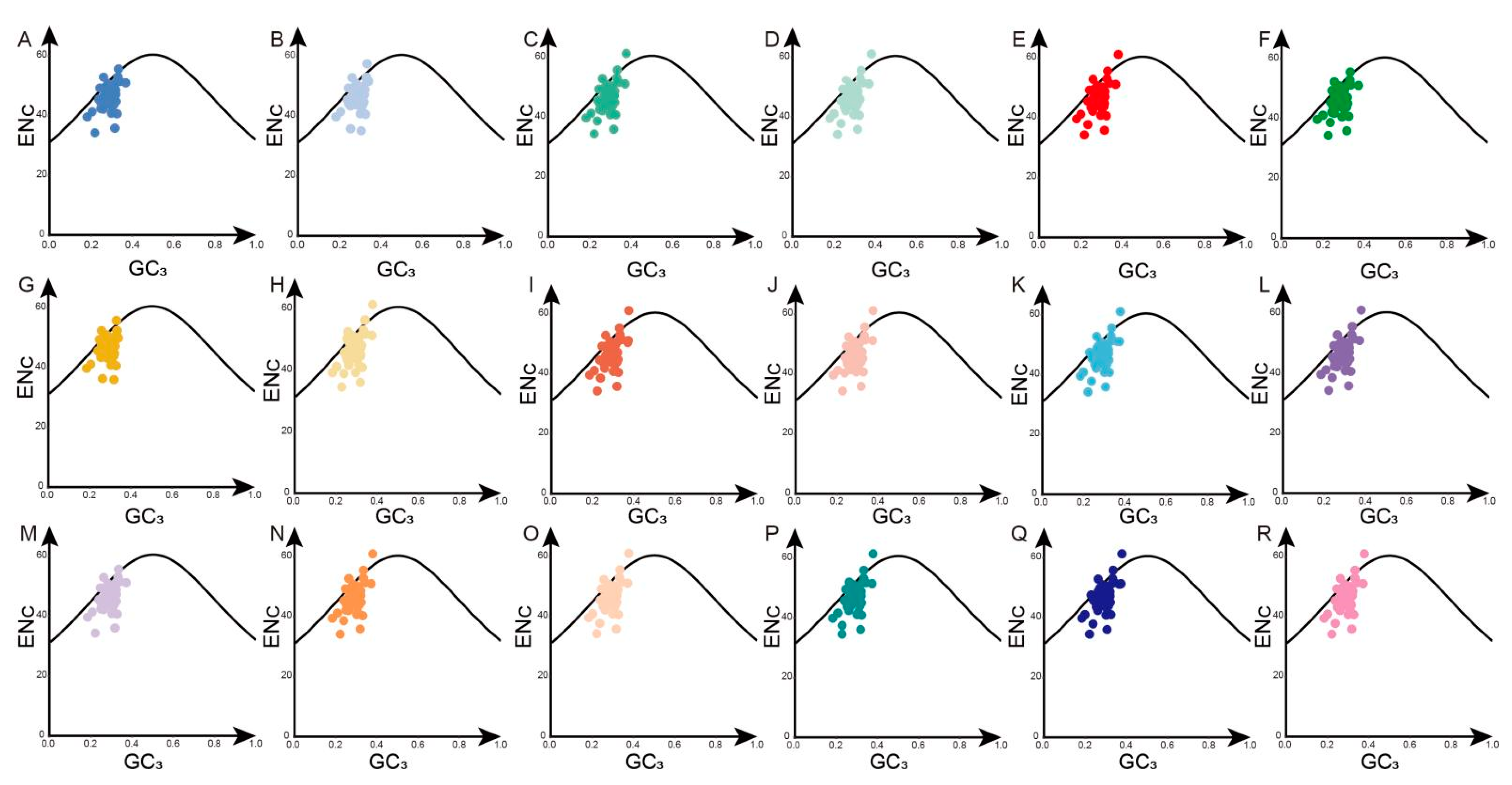
3.3. ENc Plot
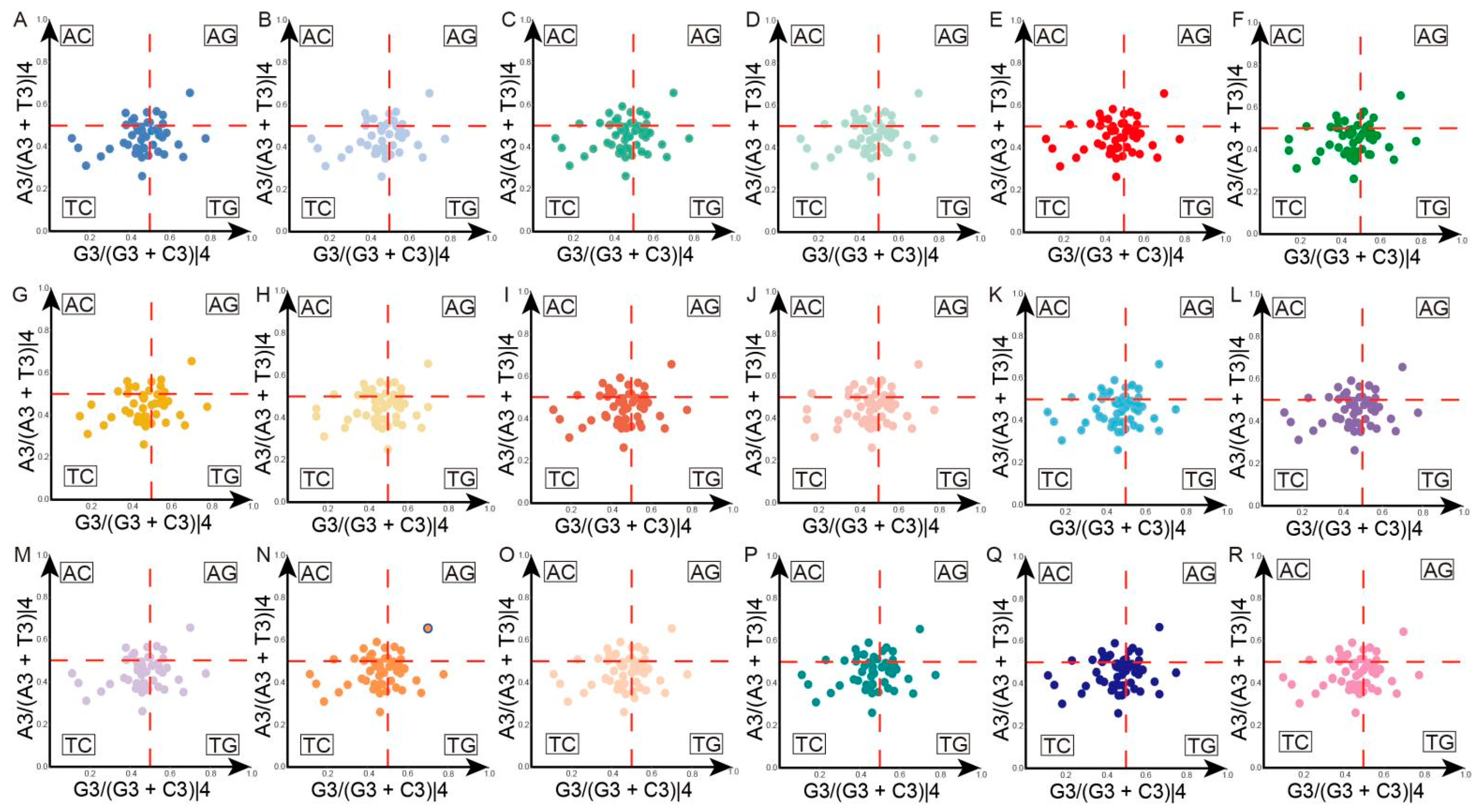
3.4. PR2 Plot
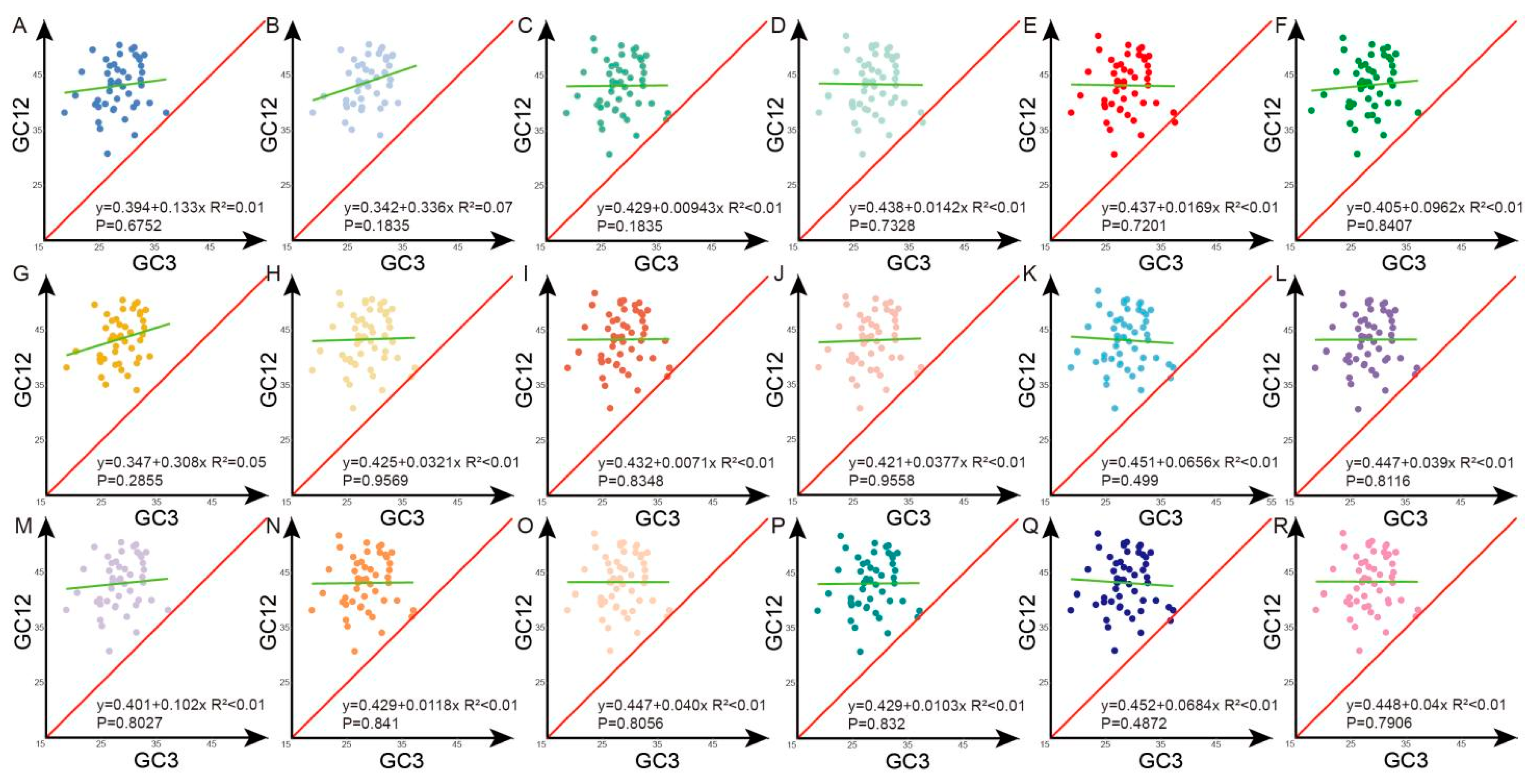
3.5. Neutrality Plot
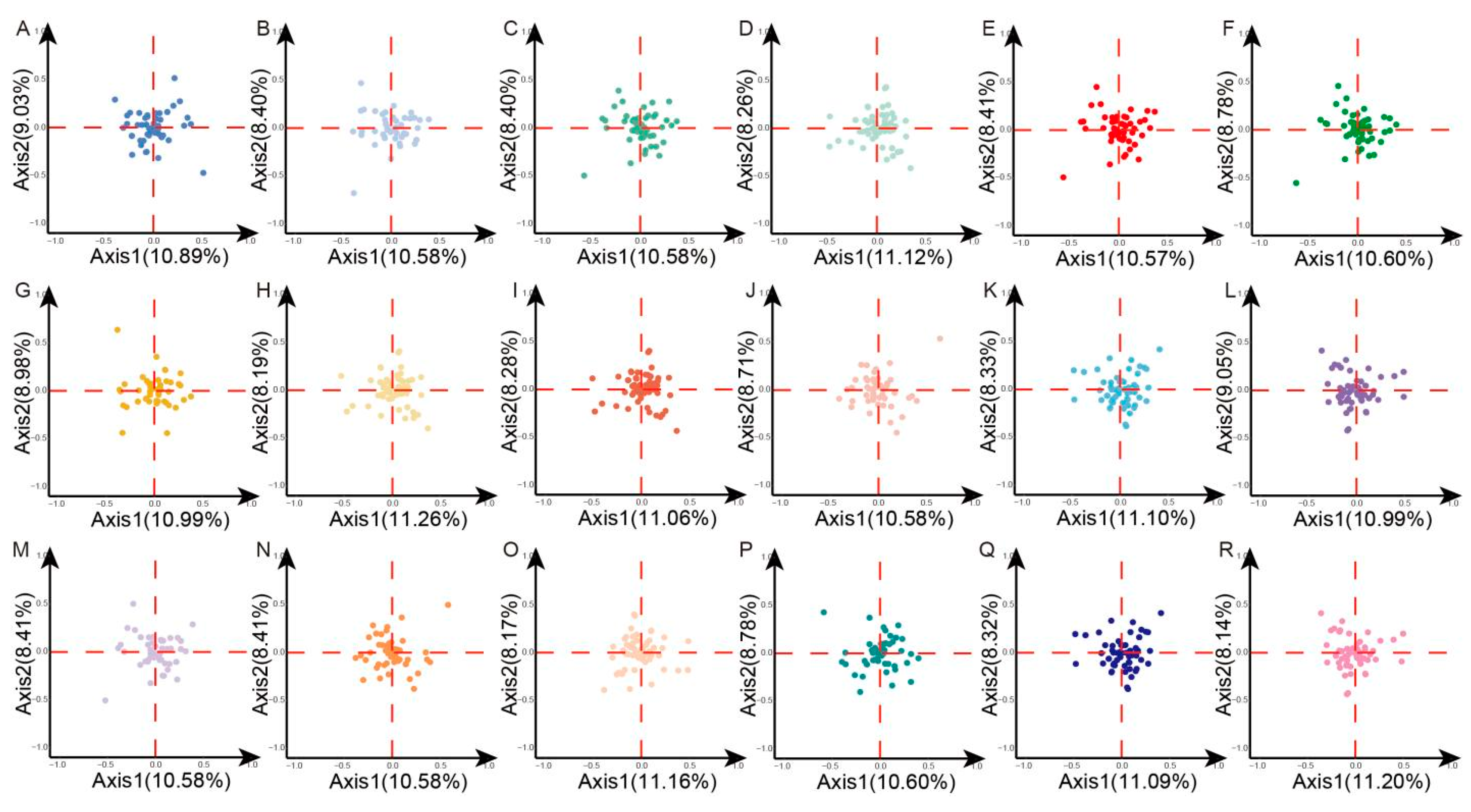
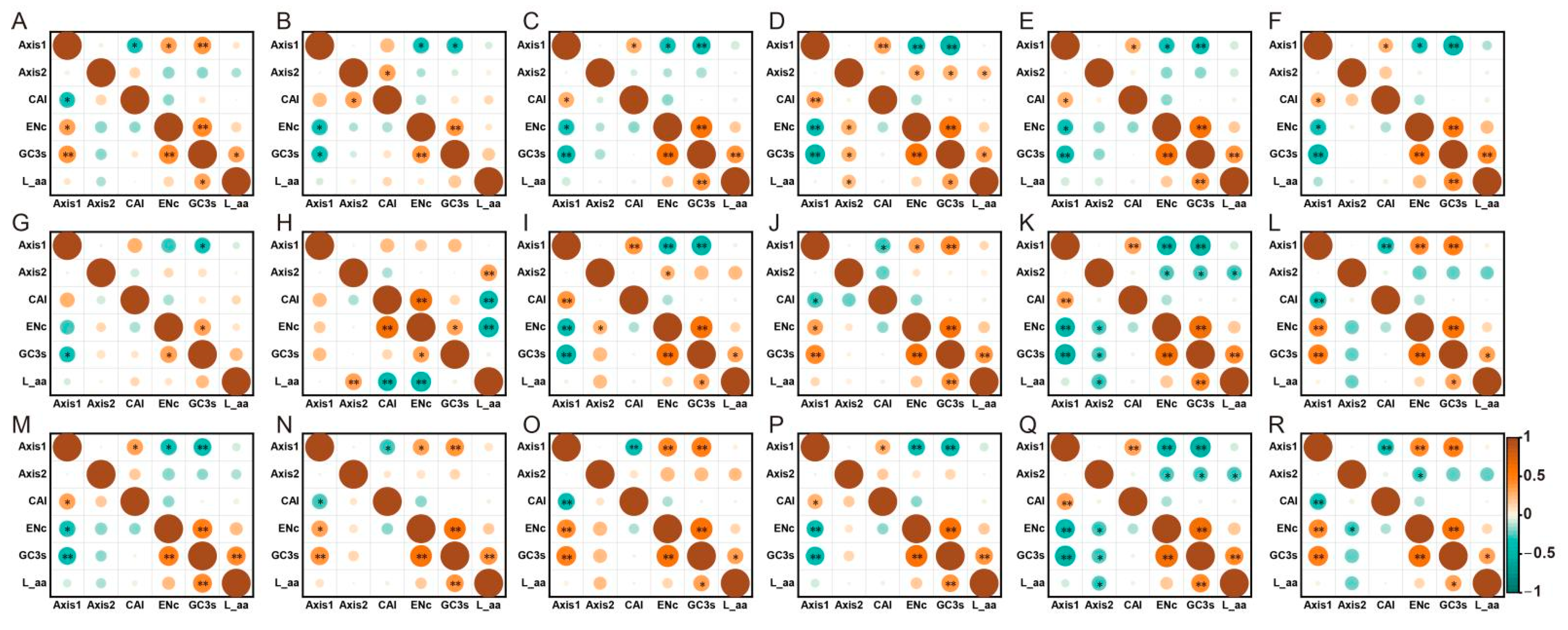
3.6. Correspondence Analysis (COA)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| A | Adenine |
| T | Thymine |
| C | Cytosine |
| G | Guanine |
| GC1, GC2, GC3 | The G + C content at the first, second, third codon positions |
| A3, T3, G3, C3 | The content of A, T, G, and C at the third codon position |
| GC12 | The average GC content at the first and second codon positions |
| RSCU | Relative synonymous codon usage |
| RFSC | Relative synonymous codon usage frequency |
| ENc | Effective number of codons |
| PR2 | Parity Rule 2 |
| COA | Correspondence analysis |
| GC3s | GC content at the third codon position of synonymous codons |
| L_aa | Total number of amino acids |
| CAI | Codon adaptation index |
| NCBI | National Center for Biotechnology Information |
References
- Manyanga, F.; Sithole, A. Nucleic Acids, Structure and Function for General Biochemistry, Biology and Biotechnology; Lulu.com: Morrisville, NC, USA, 2014. [Google Scholar]
- Zolyan, S. On the minimal elements of the genetic code and their semiotic functions (degeneracy, complementarity, wobbling). Biosystems 2023, 231, 104962. [Google Scholar] [CrossRef]
- Parvathy, S.T.; Udayasuriyan, V.; Bhadana, V. Codon usage bias. Mol. Biol. Rep. 2022, 49, 539–565. [Google Scholar] [CrossRef]
- Hu, H.; Dong, B.; Fan, X.; Wang, M.; Wang, T.; Liu, Q. Mutational bias and natural selection driving the synonymous codon usage of single-exon genes in rice (Oryza sativa L.). Rice 2023, 16, 11. [Google Scholar] [CrossRef]
- Lin, Y.-X.; Zhang, L.-B.; Zhang, X.-C.; He, Z.-R.; Wang, Z.-R.; Lu, S.-G.; Wu, S.-G.; Xing, F.-W.; Zhang, G.-M.; Liao, W.-B. Flora of China. Harv. Pap. Bot. 2013, 13, 301–302. [Google Scholar]
- Martinez, M.; Poirrier, P.; Chamy, R.; Prüfer, D.; Schulze-Gronover, C.; Jorquera, L.; Ruiz, G. Taraxacum officinale and related species—An ethnopharmacological review and its potential as a commercial medicinal plant. J. Ethnopharmacol. 2015, 169, 244–262. [Google Scholar] [CrossRef]
- Sharifi-Rad, M.; Roberts, T.H.; Matthews, K.R.; Bezerra, C.F.; Morais-Braga, M.F.B.; Coutinho, H.D.; Sharopov, F.; Salehi, B.; Yousaf, Z.; Sharifi-Rad, M. Ethnobotany of the genus Taraxacum—Phytochemicals and antimicrobial activity. Phytother. Res. 2018, 32, 2131–2145. [Google Scholar] [CrossRef]
- Zeisek, V. Taxonomic Principles, Reproductive Systems, Population Genetics and Relationships within Selected Groups of Genus Taraxacum (Asteraceae); Charles University: Prague, Czech Republic, 2018. [Google Scholar]
- Zhang, Y.; Iaffaldano, B.J.; Zhuang, X.; Cardina, J.; Cornish, K. Chloroplast genome resources and molecular markers differentiate rubber dandelion species from weedy relatives. BMC Plant Biol. 2017, 17, 34. [Google Scholar] [CrossRef]
- Hörandl, E. The classification of asexual organisms: Old myths, new facts, and a novel pluralistic approach. Taxon 2018, 67, 1066–1081. [Google Scholar] [CrossRef]
- Tan, Y.; Cao, J.; Tang, C.; Liu, K. Advances in Genome Sequencing and Natural Rubber Biosynthesis in Rubber-Producing Plants. Curr. Issues Mol. Biol. 2023, 45, 9342–9353. [Google Scholar] [CrossRef]
- Daniell, H.; Lin, C.-S.; Yu, M.; Chang, W.-J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 17, 1–29. [Google Scholar] [CrossRef]
- Zhao, J.; Chen, H.; Li, G.; Jumaturti, M.A.; Yao, X.; Hu, Y. Phylogenetics Study to Compare Chloroplast Genomes in Four Magnoliaceae Species. Curr. Issues Mol. Biol. 2023, 45, 9234–9251. [Google Scholar] [CrossRef]
- Ji, K.-k.; Song, X.; Chen, C.-G.; Li, G.; Xie, S.-Q. Codon usage profiling of chloroplast genome in Magnoliaceae. J. Agric. Sci. Technol. 2020, 22, 52–62. [Google Scholar]
- Fath, S.; Bauer, A.P.; Liss, M.; Spriestersbach, A.; Maertens, B.; Hahn, P.; Ludwig, C.; Schäfer, F.; Graf, M.; Wagner, R. Multiparameter RNA and codon optimization: A standardized tool to assess and enhance autologous mammalian gene expression. PLoS ONE 2011, 6, e17596. [Google Scholar] [CrossRef]
- Bahiri-Elitzur, S.; Tuller, T. Codon-based indices for modeling gene expression and transcript evolution. Comput. Struct. Biotechnol. J. 2021, 19, 2646–2663. [Google Scholar] [CrossRef]
- Wei, Z.; Zhu, S.-X.; Van den Berg, R.; Bakker, F.T.; Schranz, M.E. Phylogenetic relationships within Lactuca L. (Asteraceae), including African species, based on chloroplast DNA sequence comparisons. Genet. Resour. Crop Evol. 2017, 64, 55–71. [Google Scholar] [CrossRef]
- Das, J.K.; Roy, S. Comparative analysis of human coronaviruses focusing on nucleotide variability and synonymous codon usage patterns. Genomics 2021, 113, 2177–2188. [Google Scholar] [CrossRef]
- Wang, L.; Xing, H.; Yuan, Y.; Wang, X.; Saeed, M.; Tao, J.; Feng, W.; Zhang, G.; Song, X.; Sun, X. Genome-wide analysis of codon usage bias in four sequenced cotton species. PLoS ONE 2018, 13, e0194372. [Google Scholar] [CrossRef]
- OriginLab Corporation. Origin(Pro), Version 2021; OriginLab Corporation: Northampton, MA, USA, 2021. [Google Scholar]
- He, M.; Han, X.; Qin, X.; Bao, J.; Li, H.; Xie, Q.; Yang, Y.; Jin, X. Comparative chloroplast genome analyses provide new insights into phylogeny of Taraxacum and molecular markers for distinguishing rubber producing dandelions from their weedy relatives in China. Ind. Crop. Prod. 2024, 207, 117712. [Google Scholar] [CrossRef]
- Li, Q.; Luo, Y.; Sha, A.; Xiao, W.; Xiong, Z.; Chen, X.; He, J.; Peng, L.; Zou, L. Analysis of synonymous codon usage patterns in mitochondrial genomes of nine Amanita species. Front. Microbiol. 2023, 14, 1134228. [Google Scholar] [CrossRef]
- Xu, C.; Dong, J.; Tong, C.; Gong, X.; Wen, Q.; Zhuge, Q. Analysis of synonymous codon usage patterns in seven different citrus species. Evol. Bioinform. 2013, 9, EBO-S11930. [Google Scholar] [CrossRef]
- Kornberg, A.; Baker, T.A. DNA Replication; University Science Books: New York, NY, USA, 2005. [Google Scholar]
- Li, Z.; Huang, Z.; Wan, X.; Yu, J.; Dong, H.; Zhang, J.; Zhang, C.; Wang, S. Complete chloroplast genome sequence of Rhododendronmariesii and comparative genomics of related species in the family Ericaeae. Comp. Cytogenet. 2023, 17, 163. [Google Scholar] [CrossRef]
- Salih, M.; Hussein, R. Nuclear and Chloroplast Genome Diversity in Apomictic Microspecies of Taraxacum; University of Leicester: Leicester, UK, 2017. [Google Scholar]
- Liu, Q.; Li, X.; Li, M.; Xu, W.; Schwarzacher, T.; Heslop-Harrison, J.S. Comparative chloroplast genome analyses of Avena: Insights into evolutionary dynamics and phylogeny. BMC Plant Biol. 2020, 20, 406. [Google Scholar] [CrossRef]
- Freeland, J.R. Molecular Ecology; John Wiley & Sons: Hoboken, NJ, USA, 2020. [Google Scholar]
- Plotkin, J.B.; Dushoff, J.; Desai, M.M.; Fraser, H.B. Codon usage and selection on proteins. J. Mol. Evol. 2006, 63, 635–653. [Google Scholar] [CrossRef]
- Trotta, E. Selection on codon bias in yeast: A transcriptional hypothesis. Nucleic Acids Res. 2013, 41, 9382–9395. [Google Scholar] [CrossRef]
- Cutter, A.D.; Wasmuth, J.D.; Blaxter, M.L. The evolution of biased codon and amino acid usage in nematode genomes. Mol. Biol. Evol. 2006, 23, 2303–2315. [Google Scholar] [CrossRef]
- Hanson, G.; Coller, J. Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol. 2018, 19, 20–30. [Google Scholar] [CrossRef]
- Qian, W.; Yang, J.-R.; Pearson, N.M.; Maclean, C.; Zhang, J. Balanced codon usage optimizes eukaryotic translational efficiency. PLoS Genet. 2012, 8, e1002603. [Google Scholar] [CrossRef]
- Duan, H.; Zhang, Q.; Wang, C.; Li, F.; Tian, F.; Lu, Y.; Hu, Y.; Yang, H.; Cui, G. Analysis of codon usage patterns of the chloroplast genome in Delphinium grandiflorum L. reveals a preference for AT-ending codons as a result of major selection constraints. PeerJ 2021, 9, e10787. [Google Scholar] [CrossRef]
- Bhattacharyya, D.; Uddin, A.; Das, S.; Chakraborty, S. Mutation pressure and natural selection on codon usage in chloroplast genes of two species in Pisum L. (Fabaceae: Faboideae). Mitochondrial DNA Part A 2019, 30, 664–673. [Google Scholar] [CrossRef]
- Wang, Z.; Cai, Q.; Wang, Y.; Li, M.; Wang, C.; Wang, Z.; Jiao, C.; Xu, C.; Wang, H.; Zhang, Z. Comparative analysis of codon bias in the chloroplast genomes of theaceae species. Front. Genet. 2022, 13, 824610. [Google Scholar] [CrossRef]
- Zhou, Z.; Dang, Y.; Zhou, M.; Li, L.; Yu, C.-H.; Fu, J.; Chen, S.; Liu, Y. Codon usage is an important determinant of gene expression levels largely through its effects on transcription. Proc. Natl. Acad. Sci. USA 2016, 113, E6117–E6125. [Google Scholar] [CrossRef] [PubMed]
- Rocha, E.P. Codon usage bias from tRNA’s point of view: Redundancy, specialization, and efficient decoding for translation optimization. Genome Res. 2004, 14, 2279–2286. [Google Scholar] [CrossRef] [PubMed]
- Quax, T.E.; Claassens, N.J.; Söll, D.; van der Oost, J. Codon bias as a means to fine-tune gene expression. Mol. Cell 2015, 59, 149–161. [Google Scholar] [CrossRef] [PubMed]
- Novoa, E.M.; Pavon-Eternod, M.; Pan, T.; de Pouplana, L.R. A role for tRNA modifications in genome structure and codon usage. Cell 2012, 149, 202–213. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jiang, D.; Guo, K.; Zhao, L.; Meng, F.; Xiao, J.; Niu, Y.; Sun, Y. Comparative analysis of codon usage patterns in chloroplast genomes of ten Epimedium species. BMC Genom. Data 2023, 24, 3. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Xu, B.; Li, B.; Zhou, Q.; Wang, G.; Jiang, X.; Wang, C.; Xu, Z. Comparative analysis of codon usage patterns in chloroplast genomes of six Euphorbiaceae species. PeerJ 2020, 8, e8251. [Google Scholar] [CrossRef]
- Dhindsa, R.S.; Copeland, B.R.; Mustoe, A.M.; Goldstein, D.B. Natural selection shapes codon usage in the human genome. Am. J. Hum. Genet. 2020, 107, 83–95. [Google Scholar] [CrossRef]
| Species | Assembly | GC% | GC1% | GC2% | GC3% | CDSs Number (Before Filtering) | CDSs Number (After Filtering) | L_aa |
|---|---|---|---|---|---|---|---|---|
| T. amplum | KX499525.1 | 37.9% | 45.45% | 37.99% | 30.35% | 85 | 52 | 22,534 |
| T. brevicorniculatum | KX198559.1 | 39.0% | 48.00% | 40.02% | 28.85% | 81 | 48 | 16,259 |
| T. coreanum | MN689809.1 | 37.9% | 45.65% | 37.96% | 30.21% | 86 | 55 | 23,369 |
| T. dealbatum | CNA0052002 | 38.0% | 45.87% | 38.04% | 30.12% | 95 | 63 | 25,501 |
| T. erythrospermum | MN689810.1 | 38.0% | 45.62% | 37.96% | 30.28% | 86 | 55 | 23,363 |
| T. hallaisanense | MW067130.1 | 37.9% | 45.57% | 37.97% | 30.12% | 83 | 55 | 23,671 |
| T. kok-saghyz | KX198560.1 | 38.8% | 47.67% | 39.69% | 28.92% | 81 | 50 | 17,132 |
| T. leucanthum | CNA0052001 | 38.0% | 45.89% | 38.02% | 30.08% | 95 | 63 | 25,542 |
| T. longipyramidatum | CNA0052004 | 38.0% | 45.95% | 38.01% | 30.08% | 96 | 63 | 25,368 |
| T. mongolicum | KU736961.1 | 37.9% | 45.62% | 37.95% | 30.23% | 86 | 55 | 23,371 |
| T. monochlamydeum | CNA0052005 | 38.0% | 45.53% | 37.67% | 30.71% | 98 | 65 | 28,484 |
| T. multiscaposum | CNA0052003 | 38.0% | 45.86% | 38.02% | 30.05% | 95 | 63 | 25,671 |
| T. obtusifrons | KX499524.1 | 37.9% | 45.38% | 37.87% | 30.31% | 84 | 51 | 22,258 |
| T. officinale | KU361241.1 | 37.9% | 45.65% | 37.95% | 30.23% | 86 | 55 | 23,369 |
| T. parvulum | CNA0052008 | 38.0% | 45.86% | 38.01% | 30.04% | 96 | 63 | 25,524 |
| T. platycarpum | KU736960.1 | 37.9% | 45.64% | 37.94% | 30.27% | 86 | 55 | 23,369 |
| T. stenolobum | CNA0052006 | 38.0% | 45.53% | 37.67% | 30.71% | 98 | 65 | 28,484 |
| T. xinyuanicum | CNA0052007 | 38.0% | 45.87% | 38.00% | 30.09% | 96 | 63 | 25,694 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Wang, X.; Shi, Z. Comparative Study on Codon Usage Patterns across Chloroplast Genomes of Eighteen Taraxacum Species. Horticulturae 2024, 10, 492. https://doi.org/10.3390/horticulturae10050492
Yang Y, Wang X, Shi Z. Comparative Study on Codon Usage Patterns across Chloroplast Genomes of Eighteen Taraxacum Species. Horticulturae. 2024; 10(5):492. https://doi.org/10.3390/horticulturae10050492
Chicago/Turabian StyleYang, Yang, Xingliang Wang, and Zhenjie Shi. 2024. "Comparative Study on Codon Usage Patterns across Chloroplast Genomes of Eighteen Taraxacum Species" Horticulturae 10, no. 5: 492. https://doi.org/10.3390/horticulturae10050492
APA StyleYang, Y., Wang, X., & Shi, Z. (2024). Comparative Study on Codon Usage Patterns across Chloroplast Genomes of Eighteen Taraxacum Species. Horticulturae, 10(5), 492. https://doi.org/10.3390/horticulturae10050492






