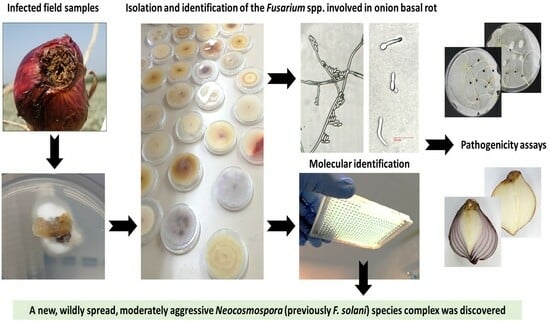
Impact of Fusarium Species Composition and Incidence on Onion Basal Rot in Northeastern Israel
Abstract
1. Introduction
2. Materials and Methods
2.1. Evaluation of Disease Severity
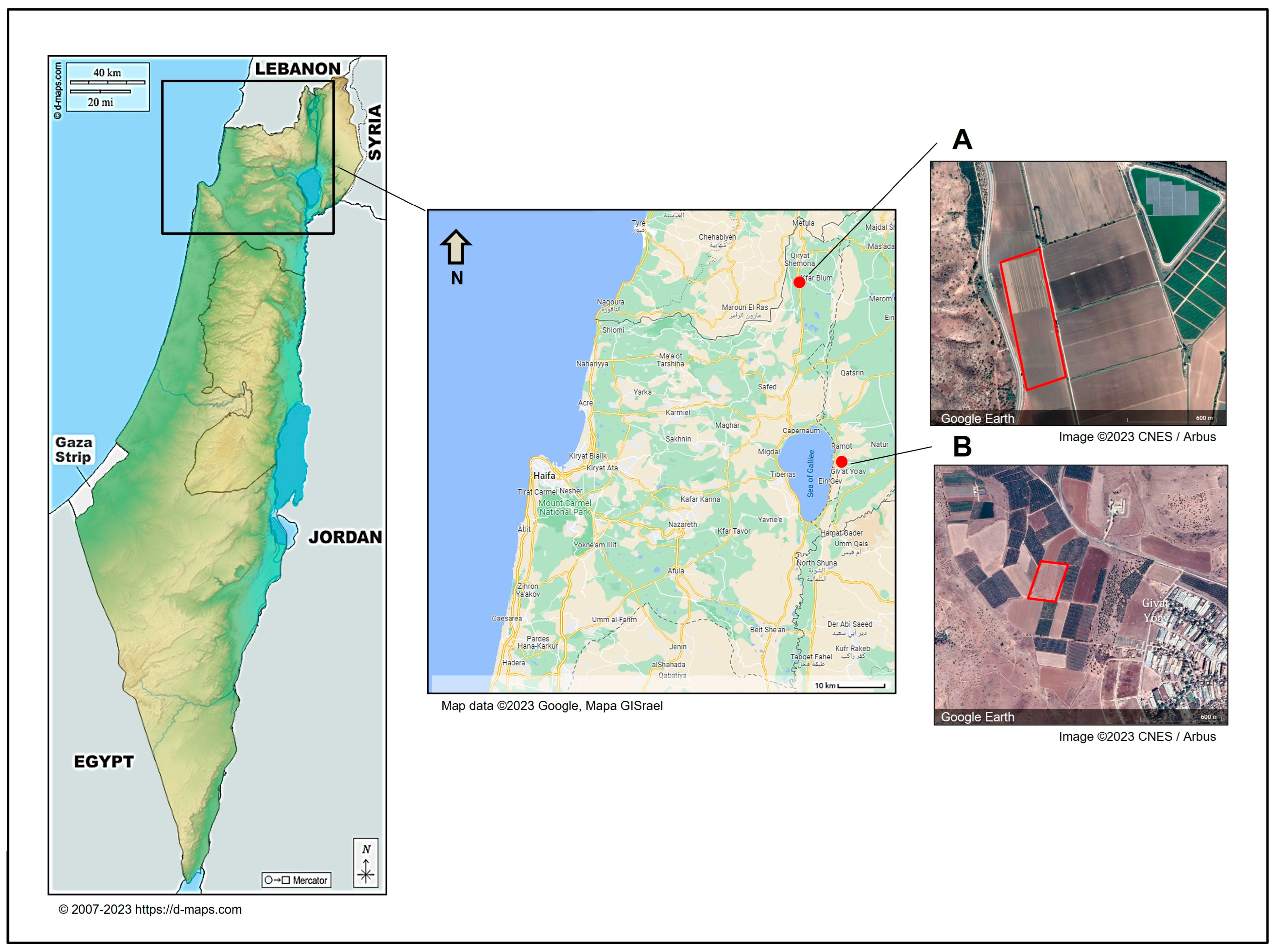
2.2. Isolation of Pathogens from Diseased Onion Plants
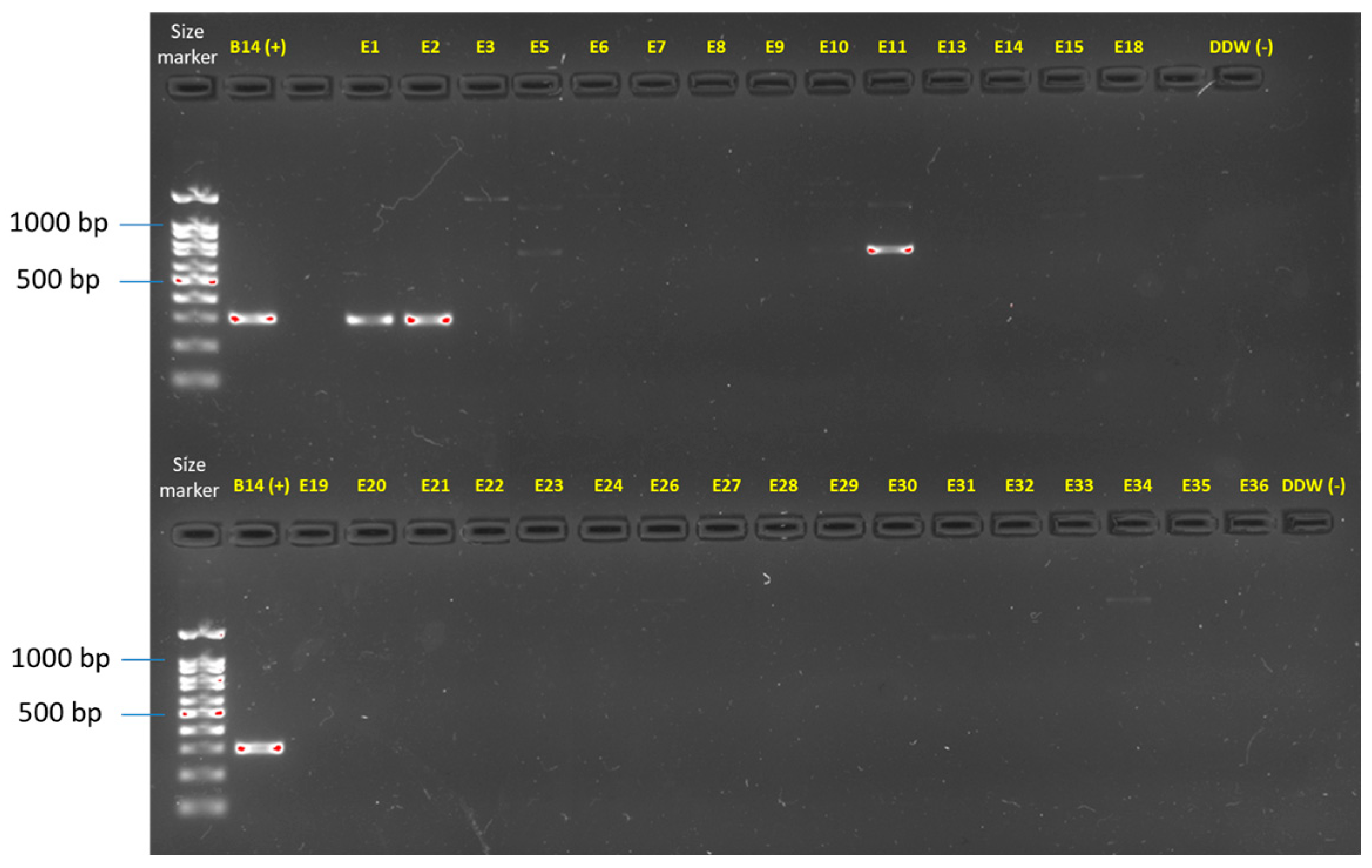
2.3. DNA Extraction and Molecular Identification of Fusarium spp.
| Primer | Gene | Sequence a | Fragment Length | Reference |
|---|---|---|---|---|
| E1/E2 | TEF1—Fusarium-specific | F-ATGGGTAAGGAGGACAAG | 680 bp | [24] |
| R-GGAAGTACCAGTGATCAT | ||||
| 7cF/11aR | RPB2—RNA polymerase second-largest subunit | F-ATGGGYAARCAAGCYATGGG | ~970 bp | [25] |
| R-GCRTGGATCTTRTCRTCSACC | ||||
| Fa/R8 | RPB1—RNA polymerase largest subunit | F-CAYAARGARTCYATGATGGGWC | 1607 bp | [26] |
| R-CAATGAGACCTTCTCGACCAGC | ||||
| SIX3 F/R | Fusarium oxysporum f. sp. cepae secreted in xylem genes 3 | F-ATGCGTTTCCTTCTGCTTATC | 306 bp | [21] |
| R-AGGTGCGACATCAATGACAG | ||||
| ISSR1 | Inter simple sequence repeat | F + R-AGAGAGAGAGAGAGA | Multiple lengths | [23] |
2.4. Identification of the Fusarium Species and Phylogenetic Relationships
2.5. Colony Morphology and Identification of the Fusarium Species
2.6. Germination Pathogenicity Assay
2.7. Onion Bulb Pathogenicity Assay
2.8. Statistical Analysis
3. Results
3.1. Evaluation of Disease Incidence
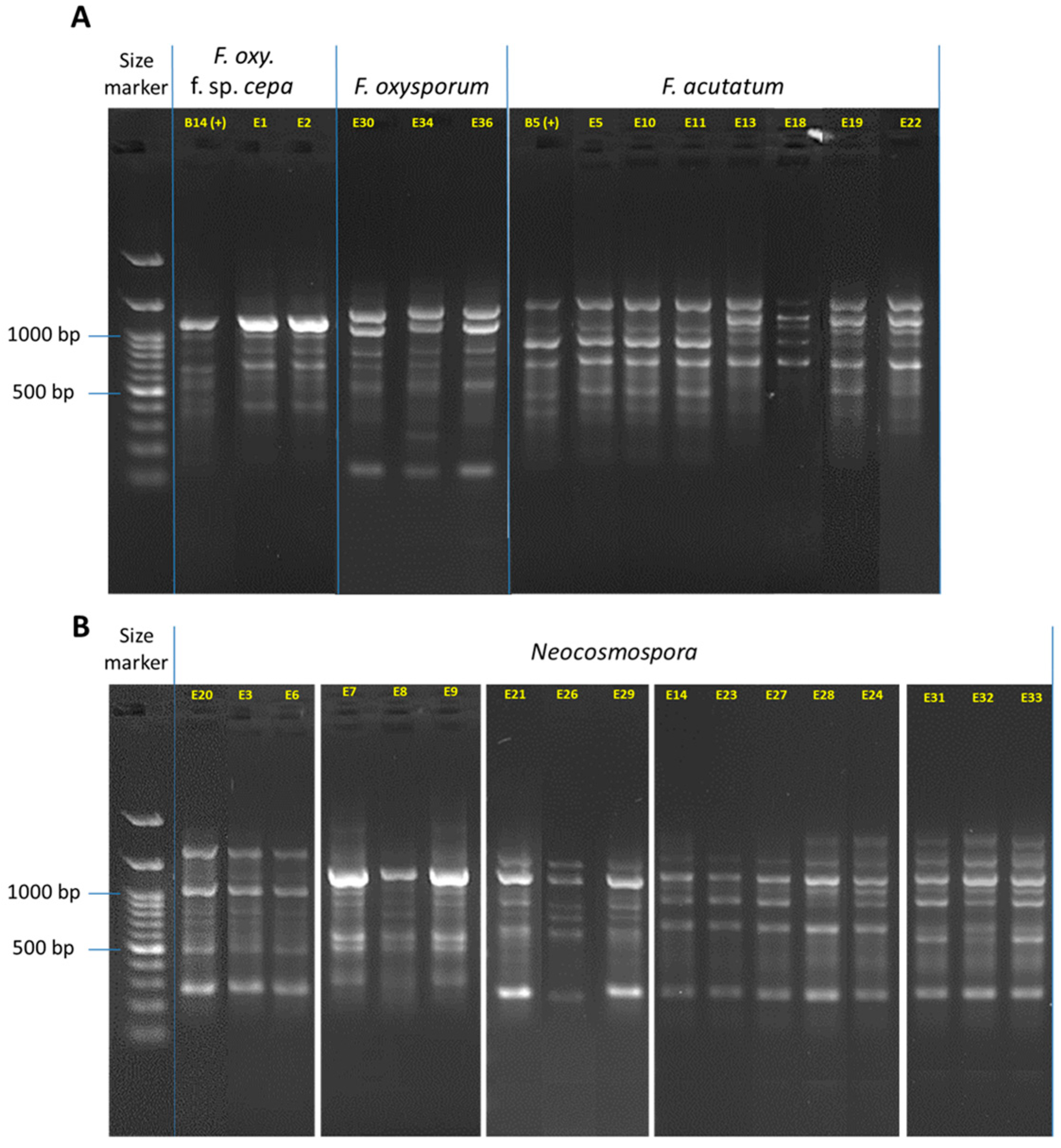
3.2. Isolation and Identification of the Fusarium Species from the Collected Onions
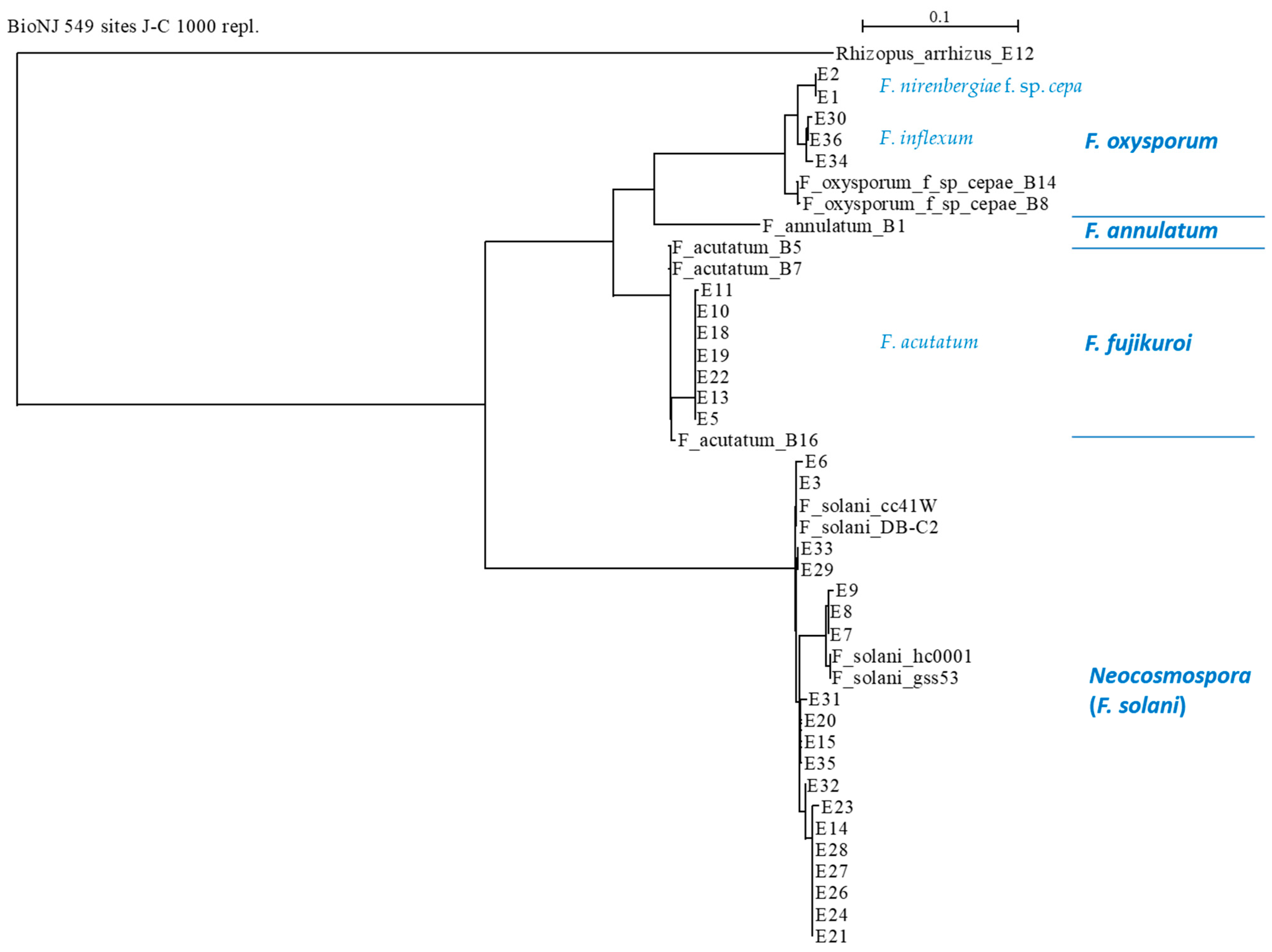
3.3. Phylogenetic Relationships between the Fusarium Species
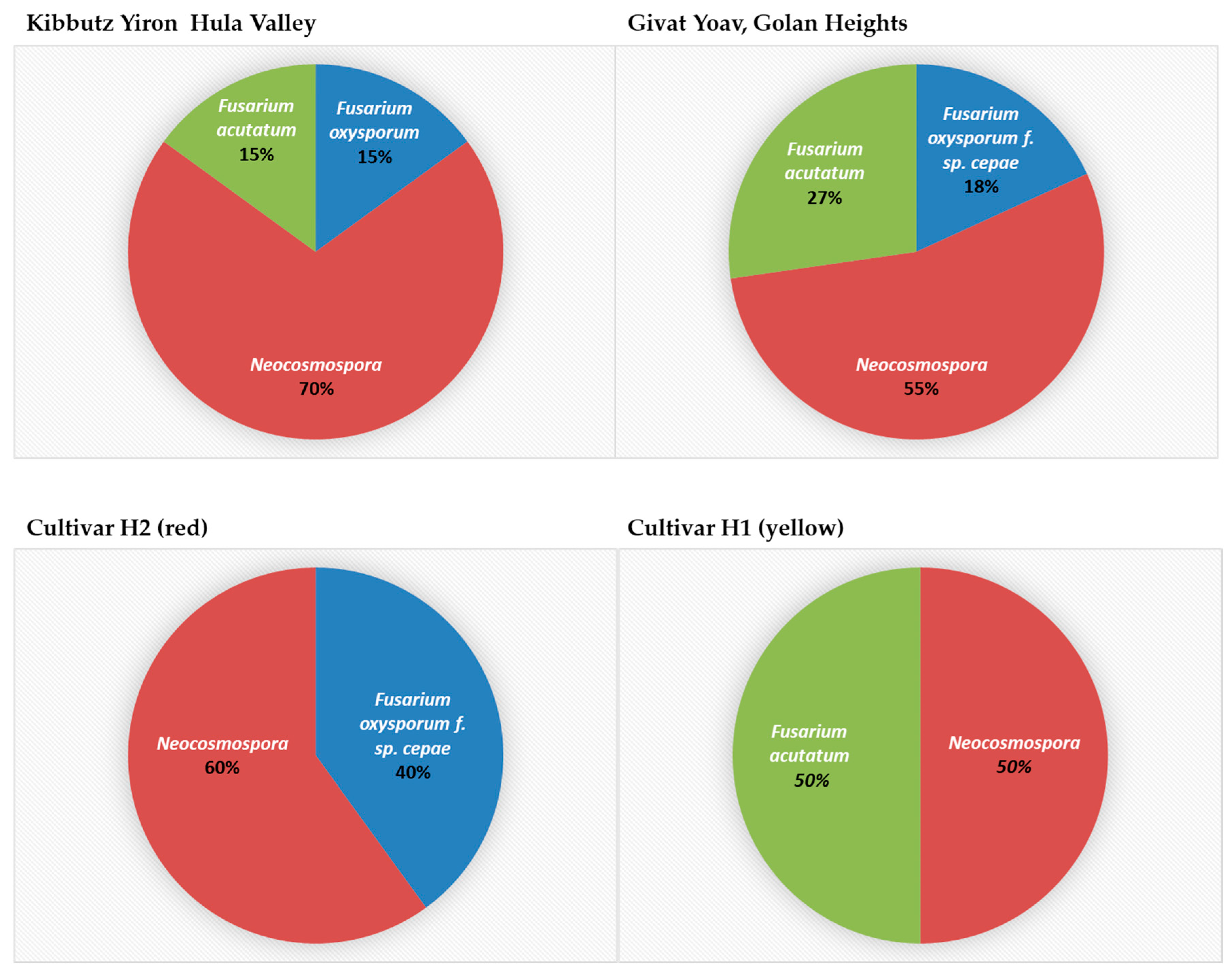
3.4. Geographic Distribution, Composition, and Incidence of the Fusarium Species Involved in Onion Basal Rot Disease in Northeastern Israel
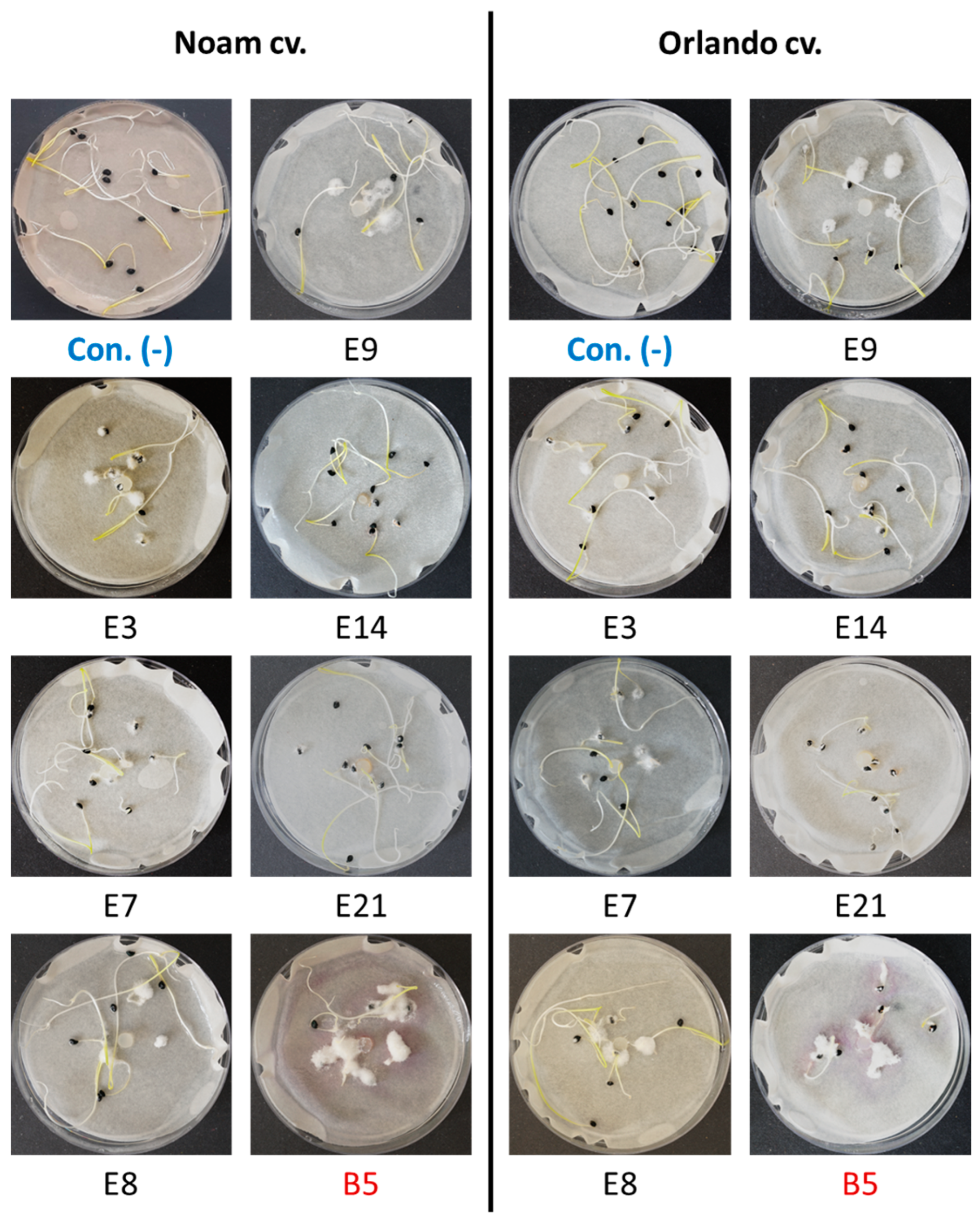
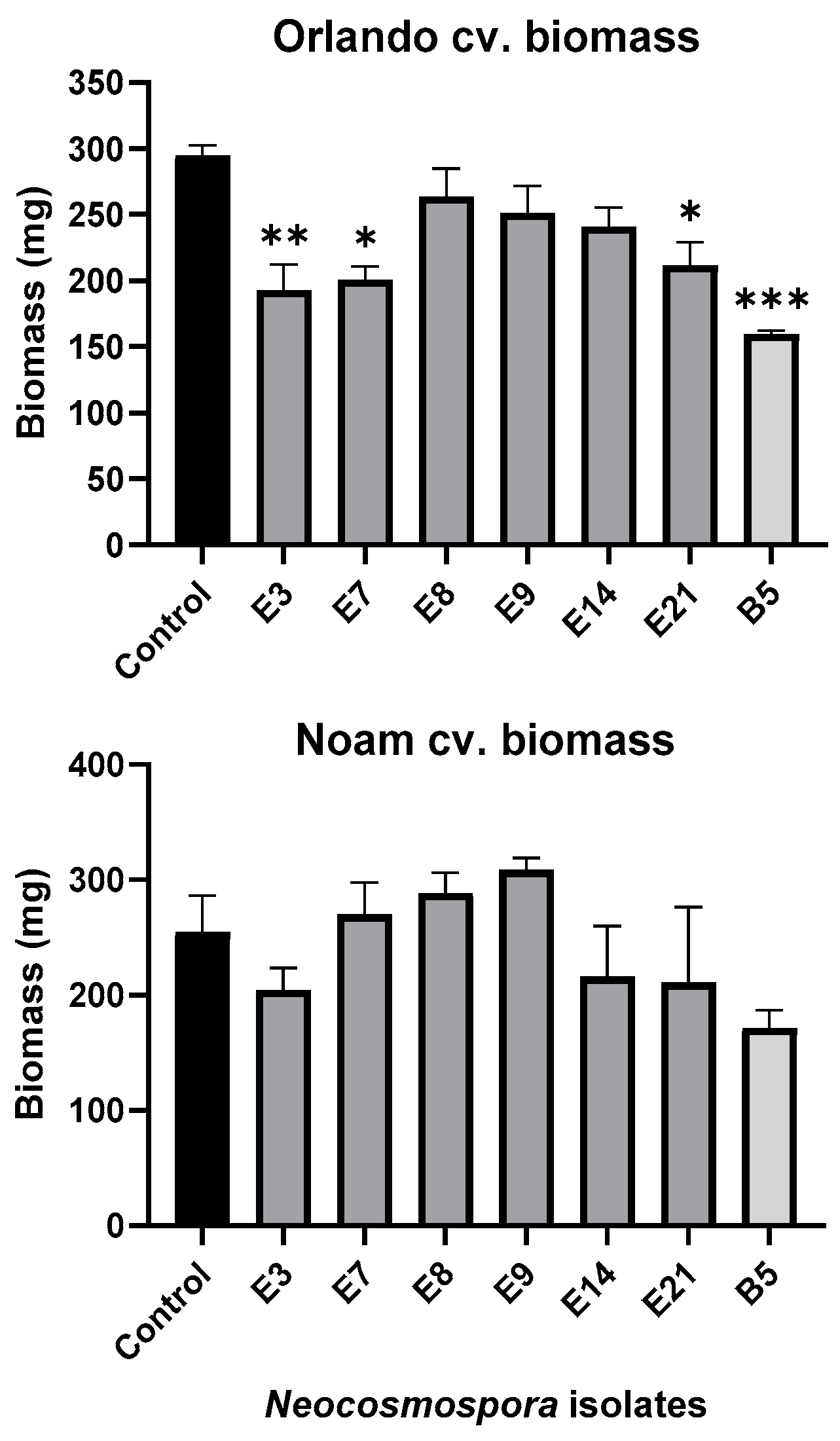
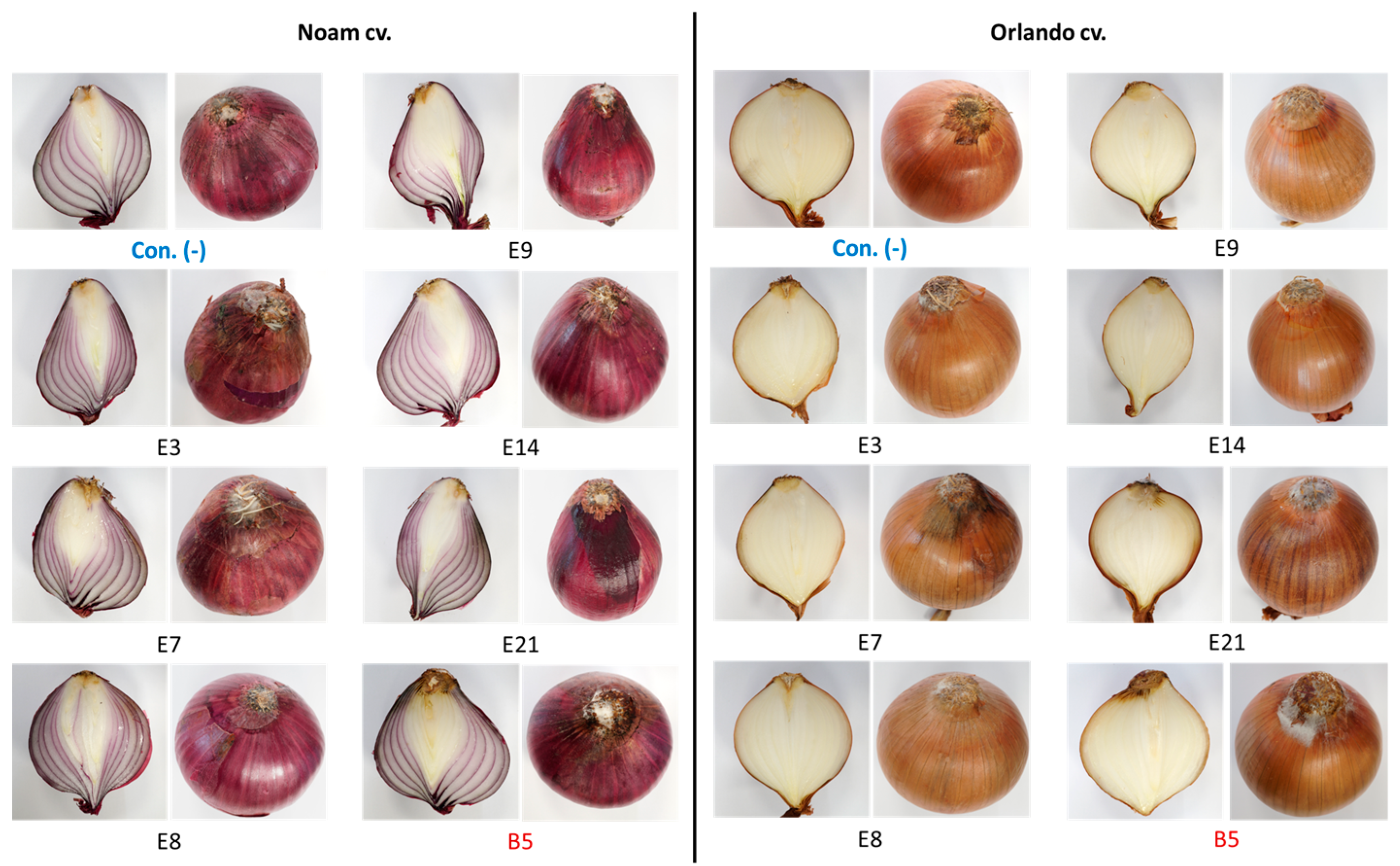
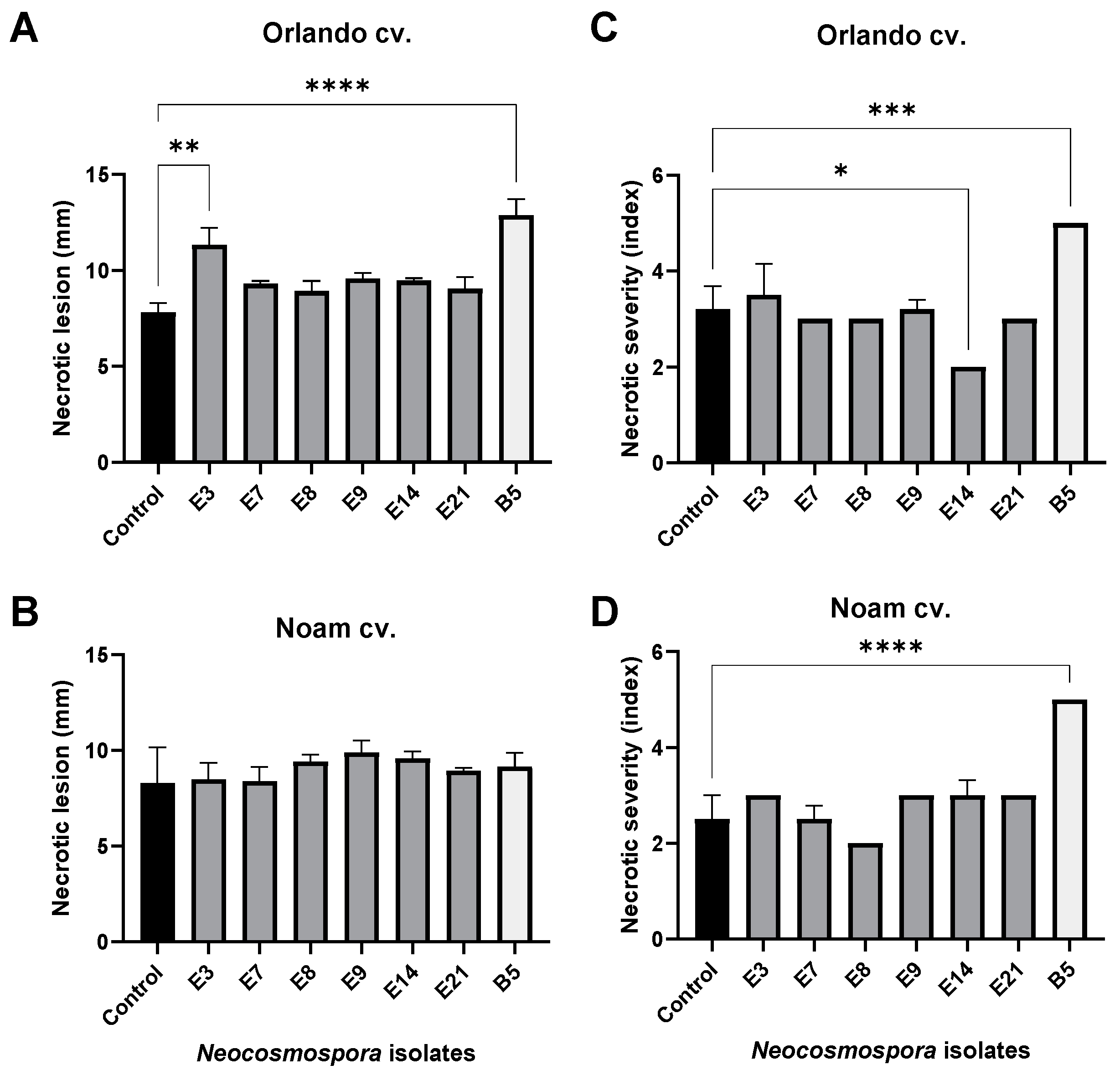
3.5. Pathogenicity Tests
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. GenBank Accession Numbers for the Nucleotide Sequences (https://www.ncbi.nlm.nih.gov/, Accessed on 1 April 2024)
References
- Le, D.; Ameye, M.; Landschoot, S.; Audenaert, K.; Haesaert, G. Phenology-regulated defence mechanisms as drivers for Fusarium basal rot in onion (Allium cepa). Plant Pathol. 2022, 71, 1440–1453. [Google Scholar] [CrossRef]
- Le, D.; Audenaert, K.; Haesaert, G. Fusarium basal rot: Profile of an increasingly important disease in Allium spp. Trop. Plant Pathol. 2021, 46, 241–253. [Google Scholar] [CrossRef]
- Klokočar-Šmit, Z.; Lević, J.; Maširević, S.; Gvozdanović-Varga, J.; Vasić, M.; Aleksić, S. Fusarium rot of onion and possible use of bioproduct. Zb. Matice Srp. Prir. Nauke. 2008, 144, 135–148. [Google Scholar] [CrossRef]
- Gunaratna, L.; Deshappriya, N.; Jayaratne, D.; Rajapaksha, R. Damping-off disease of big onion (Allium cepa L.) in Sri Lanka and evaluation of Trichoderma asperellum and Trichoderma virens for its control. Trop. Plant Res. 2019, 6, 2349–9265. [Google Scholar] [CrossRef]
- Le, D.; Ameye, M.; De Boevre, M.; De Saeger, S.; Audenaert, K.; Haesaert, G. Population, virulence, and mycotoxin profile of Fusarium spp. associated with basal rot of Allium spp. in Vietnam. Plant Dis. 2021, 105, 1942–1950. [Google Scholar] [CrossRef] [PubMed]
- Ghanbarzadeh, B.; Mohammadi Goltapeh, E.; Safaie, N. Identification of Fusarium species causing basal rot of onion in east Azerbaijan province, Iran and evaluation of their virulence on onion bulbs and seedlings. Arch. Phytopathol. Plant Prot. 2014, 47, 1050–1062. [Google Scholar] [CrossRef]
- Özer, N.; Köycü, N.; Chilosi, G.; Magro, P. Resistance to Fusarium basal rot of onion in greenhouse and field and associated expression of antifungal compounds. Phytoparasitica 2004, 32, 388–394. [Google Scholar] [CrossRef]
- Dauda, W.; Alao, S.; Zarafi, A.; Alabi, O. First report of die-back disease of onion (Allium cepa L.) induced by Fusarium equiseti (mart) sacc in Nigeria. Int. J. Plant Soil Sci. 2018, 21, 2320–7035. [Google Scholar] [CrossRef]
- Degani, O.; Dimant, E.; Gordani, A.; Graph, S.; Margalit, E. Prevention and control of Fusarium spp., the causal agents of onion (Allium cepa) basal rot. Horticulturae 2022, 8, 1071. [Google Scholar] [CrossRef]
- Galván, G.A.; Koning-Boucoiran, C.F.; Koopman, W.J.; Burger-Meijer, K.; González, P.H.; Waalwijk, C.; Kik, C.; Scholten, O.E. Genetic variation among Fusarium isolates from onion, and resistance to Fusarium basal rot in related Allium species. Eur. J. Plant Pathol. 2008, 121, 499–512. [Google Scholar] [CrossRef]
- Boehnke, B.; Karlovsky, P.; Pfohl, K.; Gamliel, A.; Isack, Y.; Dehne, H. Identification of different Fusarium spp. in Allium spp. in Germany. Commun. Agric. Appl. Biol. Sci. 2015, 80, 453–463. [Google Scholar] [PubMed]
- Gebremichael, D.E.; Haile, Z.M.; Negrini, F.; Sabbadini, S.; Capriotti, L.; Mezzetti, B.; Baraldi, E. RNA interference strategies for future management of plant pathogenic fungi: Prospects and challenges. Plants 2021, 10, 650. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, R.; Rajasekaran, K.; Cary, J.W. RNA interference (RNAi) as a potential tool for control of mycotoxin contamination in crop plants: Concepts and considerations. Front. Plant Sci. 2017, 8, 200. [Google Scholar] [CrossRef] [PubMed]
- Koch, A.; Biedenkopf, D.; Furch, A.; Weber, L.; Rossbach, O.; Abdellatef, E.; Linicus, L.; Johannsmeier, J.; Jelonek, L.; Goesmann, A. An RNAi-based control of Fusarium graminearum infections through spraying of long dsRNAs involves a plant passage and is controlled by the fungal silencing machinery. PLoS Pathog. 2016, 12, e1005901. [Google Scholar] [CrossRef] [PubMed]
- McDonald, T.; Brown, D.; Keller, N.P.; Hammond, T.M. RNA silencing of mycotoxin production in Aspergillus and Fusarium species. Mol. Plant. Microbe. Interact. 2005, 18, 539–545. [Google Scholar] [CrossRef]
- Abdel-Hadi, A.M.; Caley, D.P.; Carter, D.R.; Magan, N. Control of aflatoxin production of Aspergillus flavus and Aspergillus parasiticus using RNA silencing technology by targeting AFLD (nor-1) gene. Toxins 2011, 3, 647–659. [Google Scholar] [CrossRef]
- Ghag, S.B.; Shekhawat, U.K.; Ganapathi, T.R. Host-induced post-transcriptional hairpin RNA-mediated gene silencing of vital fungal genes confers efficient resistance against Fusarium wilt in banana. Plant Biotechnol. J. 2014, 12, 541–553. [Google Scholar] [CrossRef] [PubMed]
- Lebiush-Mordechai, S.; Erlich, O.; Maymon, M.; Freeman, S.; Ben-David, T.; Ofek, T.; Palevsky, E.; Tsror Lahkin, L. Bulb and root rot in lily (Lilium longiflorum) and onion (Allium cepa) in Israel. J Phytopathol. 2014, 162, 466–471. [Google Scholar] [CrossRef]
- Gamliel, A.; Gillett, D.; Minkovsky, N.; Benikhis, M.; Dobrynin, S.; Margalit, E. Fusarium proliferatum Disease Outburst in White Onions from Different Fields in the Southern Israel Arava Area; 30 October 2012. Available online: https://aravard.org.il/wp-content/uploads/2013/10/12VegOniFusTissueCult.pdf (accessed on 1 April 2024). (In Hebrew).
- Degani, O.; Kalman, B. Assessment of commercial fungicides against onion (Allium cepa) basal rot disease caused by Fusarium oxysporum f. sp. cepae and Fusarium acutatum. J. Fungi 2021, 7, 235. [Google Scholar] [CrossRef]
- Kalman, B.; Abraham, D.; Graph, S.; Perl-Treves, R.; Meller Harel, Y.; Degani, O. Isolation and identification of Fusarium spp., the causal agents of onion (Allium cepa) basal rot in northeastern Israel. Biology 2020, 9, 69. [Google Scholar] [CrossRef]
- Dimant, E.; Degani, O. Molecular real-time PCR monitoring of onion Fusarium basal rot chemical control. J. Fungi 2023, 9, 809. [Google Scholar] [CrossRef]
- Reddy, M.P.; Sarla, N.; Siddiq, E.A. Inter simple sequence repeat (ISSR) polymorphism and its application in plant breeding. Euphytica 2002, 128, 9–17. [Google Scholar] [CrossRef]
- O’Donnell, K.; Kistler, H.C.; Cigelnik, E.; Ploetz, R.C. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl. Acad. Sci. USA 1998, 95, 2044–2049. [Google Scholar] [CrossRef]
- Hong, S.-Y.; Kang, M.-R.; Cho, E.-J.; Kim, H.-K.; Yun, S.-H. Specific PCR detection of four quarantine Fusarium species in Korea. Plant Pathol. J. 2010, 26, 409–416. [Google Scholar] [CrossRef]
- O’Donnell, K.; Sutton, D.A.; Rinaldi, M.G.; Sarver, B.A.; Balajee, S.A.; Schroers, H.-J.; Summerbell, R.C.; Robert, V.A.; Crous, P.W.; Zhang, N. Internet-accessible DNA sequence database for identifying Fusaria from human and animal infections. J. Clin. Microbiol. 2010, 48, 3708–3718. [Google Scholar] [CrossRef]
- Gouy, M.; Guindon, S.; Gascuel, O. Seaview version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef]
- Cramer, C.S. Breeding and genetics of Fusarium basal rot resistance in onion. Euphytica 2000, 115, 159–166. [Google Scholar] [CrossRef]
- Bayraktar, H. Genetic diversity and population structure of Fusarium oxysporum f. sp. cepae, the causal agent of Fusarium basal plate rot on onion using RAPD markers. J. Agric. Sci. 2010, 16, 3. [Google Scholar]
- Caligiore Gei, P.F.; Valdez, J.G.; Piccolo, R.J.; Galmarini, C.R. Influence of Fusarium spp. Isolate and inoculum density on resistance screening tests in onion. Trop. Plant Pathol. 2014, 39, 19–27. [Google Scholar] [CrossRef]
- Nasr Esfahani, M. Genetic and virulence variation in Fusarium oxysporum f. sp. cepae causing root and basal rot of common onion in Iran. J. Phytopathol. 2018, 166, 572–580. [Google Scholar] [CrossRef]
- Brizuela, A.M.; Lalak-Kańczugowska, J.; Koczyk, G.; Stępień, Ł.; Kawaliło, M.; Palmero, D. Geographical origin does not modulate pathogenicity or response to climatic variables of Fusarium oxysporum associated with vascular wilt on asparagus. J. Fungi 2021, 7, 1056. [Google Scholar] [CrossRef] [PubMed]
- Khar, A.; Singh, H. Rapid methods for onion breeding. In Accelerated Plant Breeding, Volume 2: Vegetable Crops; Springer: Berlin/Heidelberg, Germany, 2020; pp. 77–99. [Google Scholar]
- Stankovic, S.; Levic, J.; Petrovic, T.; Logrieco, A.; Moretti, A. Pathogenicity and mycotoxin production by Fusarium proliferatum isolated from onion and garlic in Serbia. Eur. J. Plant Pathol. 2007, 118, 165–172. [Google Scholar] [CrossRef]
- Mishra, R.; Jaiswal, R.; Kumar, D.; Saabale, P.; Singh, A. Management of major diseases and insect pests of onion and garlic: A comprehensive review. Plant Breed. Crop Sci. 2014, 6, 160–170. [Google Scholar]
- Gupta, R.; Gupta, R. Effect of integrated disease management packages on diseases incidence and bulb yield of onion (Allium cepa L.). SAARC J. Agric. 2013, 11, 49–59. [Google Scholar] [CrossRef]
- Naik, D.; Burden, O. Chemical control of basal rot of onion in Zambia. Int. J. Pest Manag. 1981, 27, 455–460. [Google Scholar] [CrossRef]
- Haapalainen, M.; Latvala, S.; Kuivainen, E.; Qiu, Y.; Segerstedt, M.; Hannukkala, A. Fusarium oxysporum, F. proliferatum and F. redolens associated with basal rot of onion in Finland. Plant Pathol. 2016, 65, 1310–1320. [Google Scholar] [CrossRef]
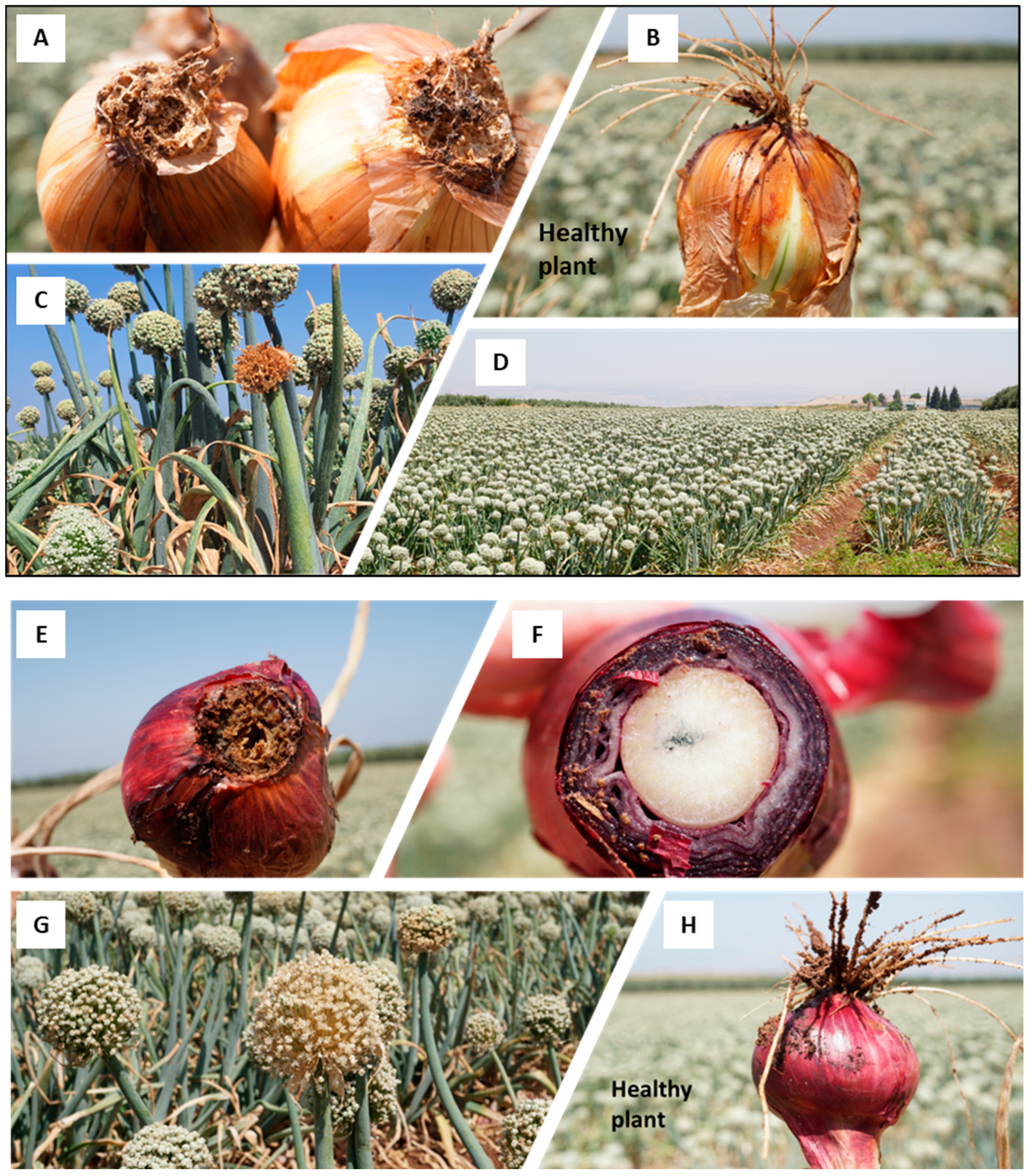



| Cultivar | Number of Inflorescences Sampled | Number of Diseased Plants | Percentage of Diseased Plants b |
|---|---|---|---|
| Red, female (Ha3 cv.) | 500 | 40 | 8% ± 0.22 c |
| Red, male (Ha2 cv.) | 500 | 12 | 2.4% ± 0.10 |
| Yellow, female (Ha1 cv.) | 1000 | 24 | 2.4% ± 0.13 |
| Average | - | 25.3 | 5% ± 0.77 |
| Isolate | Primer | Fusarium spp. | Gene | Accession | Similarity | Overlap | Onion Cultivar | Collection Site c | |
|---|---|---|---|---|---|---|---|---|---|
| E1 b | E1/E2 | Fusarium nirenbergiae, F. oxysporum SC d | TEF1/2 | JW 124027 | https://www.fusarium.org/details/23/1257 | 99.85% | 98.52% | Ha2 | Givat Yoav, Golan Heights |
| 7cf/11ar | Fusarium nirenbergiae, F. oxysporum SC | RPB2 | LC13757 | https://www.fusarium.org/details/23/1895 | 99.88% | 77.20% | |||
| E2 b | E1/E2 | Fusarium nirenbergiae, F. oxysporum SC | TEF1/2 | JW 124027 | https://www.fusarium.org/details/23/1257 | 99.85% | 98.37% | Ha2 | Givat Yoav, Golan Heights |
| 7cf/11ar | Fusarium nirenbergiae, F. oxysporum SC | RPB2 | LC13757 | https://www.fusarium.org/details/23/1895 | 99.64% | 85.73% | |||
| E3 | E1/E2 | Neocosmospora falciformis, Neocosmospora (previously the F. solani) SC | TEF1/2 | NRRL 32544 | https://www.fusarium.org/details/23/492 | 99.70% | 91.39% | Ha1 | Givat Yoav, Golan Heights |
| 7cf/11ar | Neocosmospora falciformis, Neocosmospora (previously the F. solani) SC | RPB2 | CBS 475.67 | https://www.fusarium.org/details/23/478 | 99.47% | 63.83% | |||
| E5 | E1/E2 | Fusarium acutatum, F. fujikuroi SC | TEF1/2 | CBS 138572 | https://www.fusarium.org/details/23/877 | 99.85% | 97.62% | Ha1 | Givat Yoav, Golan Heights |
| 7cf/11ar | Fusarium acutatum, F. fujikuroi SC | RPB2 | CBS 137545 | https://www.fusarium.org/details/23/875 | 99.77% | 89.20% | |||
| E6 | E1/E2 | Neocosmospora falciformis, Neocosmospora (previously the F. solani) SC | TEF1/2 | NRRL 32544 | https://www.fusarium.org/details/23/492 | 99.70% | 90.48% | Ha1 | Givat Yoav, Golan Heights |
| Fa/R8 | Neocosmospora falciformis, Neocosmospora (previously the F. solani) SC | RPB1 | CBS 121450 | https://www.fusarium.org/details/23/479 | 91.48% | 44.90% | |||
| E7 | E1/E2 | Neocosmospora gamtoosensis, Neocosmospora (previously the F. solani) SC | TEF1/2 | CBS 146502 | https://www.fusarium.org/details/23/525 | 100% | 87.29% | Ha1 | Givat Yoav, Golan Heights |
| 7cf/11ar | Neocosmospora solani, Neocosmospora (previously the F. solani) SC | RPB2 | CBS 117149 | https://www.fusarium.org/details/23/735 | 99.74% | 79.45% | |||
| E8 | E1/E2 | Neocosmospora gamtoosensis, Neocosmospora (previously the F. solani) SC | TEF1/2 | CBS 146502 | https://www.fusarium.org/details/23/525 | 100% | 87.66% | Ha2 | Givat Yoav, Golan Heights |
| Fa/R8 | Neocosmospora solani, Neocosmospora (previously the F. solani) SC | RPB1 | JW 191039 | https://www.fusarium.org/details/23/1337 | 97.98% | 71.44% | |||
| E9 | E1/E2 | Neocosmospora gamtoosensis, Neocosmospora (previously the F. solani) SC | TEF1/2 | CBS 146502 | https://www.fusarium.org/details/23/525 | 99.84% | 88.27% | Ha2 | Givat Yoav, Golan Heights |
| 7cf/11ar | Neocosmospora solani, Neocosmospora (previously the F. solani) SC | RPB2 | NRRL 43474 | https://www.fusarium.org/details/23/749 | 99.77% | 89.68% | |||
| E10 | E1/E2 | Fusarium acutatum, F. fujikuroi SC | TEF1/2 | CBS 137545 | https://www.fusarium.org/details/23/875 | 99.85% | 84.29% | Ha1 | Givat Yoav, Golan Heights |
| 7cf/11ar | Fusarium acutatum, F. fujikuroi SC | RPB2 | CBS 137545 | https://www.fusarium.org/details/23/875 | 99.19% | 89.41% | |||
| E11 | E1/E2 | Fusarium acutatum, F. fujikuroi SC | TEF1/2 | CBS 137545 | https://www.fusarium.org/details/23/875 | 99.70% | 96.22% | Ha1 | Givat Yoav, Golan Heights |
| 7cf/11ar | Fusarium acutatum, F. fujikuroi SC | RPB2 | CBS 137545 | https://www.fusarium.org/details/23/875 | 99.88% | 87.64% | |||
| E13 | E1/E2 | Fusarium acutatum, F. fujikuroi SC | TEF1/2 | CBS 402.97 | https://www.fusarium.org/details/23/31 | 100% | 77.12% | Orlando | Yiron, Galilee |
| 7cf/11ar | Fusarium acutatum, F. fujikuroi SC | RPB2 | CBS 137545 | https://www.fusarium.org/details/23/875 | 99.18% | 89.19% | |||
| E14 | E1/E2 | Neocosmospora falciformis, Neocosmospora (previously the F. solani) SC | TEF1/2 | NRRL 54966 | https://www.fusarium.org/details/23/509 | 99.69% | 89.62% | Orlando | Yiron, Galilee |
| 7cf/11ar | Neocosmospora falciformis, Neocosmospora (previously the F. solani) SC | RPB2 | NRRL 28563 | https://www.fusarium.org/details/23/486 | 98.09% | 73.39% | |||
| E15 | E1/E2 | Neocosmospora falciformis, Neocosmospora (previously the F. solani) SC | TEF1/2 | CBS 121450 | https://www.fusarium.org/details/23/479 | 99.70% | 93.93% | Orlando | Yiron, Galilee |
| 7cf/11ar | Neocosmospora falciformis, Neocosmospora (previously the F. solani) SC | RPB2 | NRRL 43441 | https://www.fusarium.org/details/23/501 | 98.03% | 66.10% | |||
| E18 | E1/E2 | Fusarium acutatum, F. fujikuroi SC | TEF1/2 | CBS 401.97 | https://www.fusarium.org/details/23/32 | 100% | 75.88% | Orlando | Yiron, Galilee |
| Fa/R8 | Fusarium acutatum, F. fujikuroi SC | RPB1 | CBS 137545 | https://www.fusarium.org/details/23/875 | 99.64% | 75.57% | |||
| E19 | E1/E2 | Fusarium acutatum, F. fujikuroi SC | TEF1/2 | CBS 401.97 | https://www.fusarium.org/details/23/32 | 100% | 74.57% | Orlando | Yiron, Galilee |
| 7cf/11ar | Fusarium acutatum, F. fujikuroi SC | RPB2 | CBS 137545 | https://www.fusarium.org/details/23/875 | 99.07% | 87.55% | |||
| E20 | E1/E2 | Neocosmospora falciformis, Neocosmospora (previously the F. solani) SC | TEF1/2 | BS 121450 | https://www.fusarium.org/details/23/479 | 99.70% | 94.05% | Orlando | Yiron, Galilee |
| 7cf/11ar | Neocosmospora falciformis, Neocosmospora (previously the F. solani) SC | RPB2 | CBS 475.67 | https://www.fusarium.org/details/23/478 | 99.47% | 78.73% | |||
| E21 | E1/E2 | Neocosmospora falciformis, Neocosmospora (previously the F. solani) SC | TEF1/2 | NRRL 54983 | https://www.fusarium.org/details/23/510 | 96% | 99.42% | Orlando | Yiron, Galilee |
| 7cf/11ar | Neocosmospora falciformis, Neocosmospora (previously the F. solani) SC | RPB2 | CBS 475.67 | https://www.fusarium.org/details/23/478 | 97.53% | 79.03% | |||
| E22 | E1/E2 | Fusarium acutatum, F. fujikuroi SC | TEF1/2 | CBS 402.97 | https://www.fusarium.org/details/23/31 | 100% | 75.88% | Orlando | Yiron, Galilee |
| 7cf/11ar | Fusarium acutatum, F. fujikuroi SC | RPB2 | CBS 137545 | https://www.fusarium.org/details/23/875 | 99.07% | 89.29% | |||
| E23 | E1/E2 | Neocosmospora falciformis, Neocosmospora (previously the F. solani) SC | TEF1/2 | NRRL 54966 | https://www.fusarium.org/details/23/509 | 99.22% | 90.24% | Orlando | Yiron, Galilee |
| 7cf/11ar | Neocosmospora falciformis, Neocosmospora (previously the F. solani) SC | RPB2 | CBS 475.67 | https://www.fusarium.org/details/23/478 | 97.86% | 70.14% | |||
| E24 | E1/E2 | Neocosmospora falciformis, Neocosmospora (previously the F. solani) SC | TEF1/2 | NRRL 54983 | https://www.fusarium.org/details/23/510 | 99.53% | 90.90% | Orlando | Yiron, Galilee |
| 7cf/11ar | Neocosmospora falciformis, Neocosmospora (previously the F. solani) SC | RPB2 | CBS 475.67 | https://www.fusarium.org/details/23/478 | 98.95% | 77.41% | |||
| E26 | E1/E2 | Neocosmospora falciformis, Neocosmospora (previously the F. solani) SC | TEF1/2 | NRRL 54983 | https://www.fusarium.org/details/23/510 | 99.69% | 90.51% | Orlando | Yiron, Galilee |
| 7cf/11ar | Neocosmospora falciformis, Neocosmospora (previously the F. solani) SC | RPB2 | NRRL 43441 | https://www.fusarium.org/details/23/501 | 99.66% | 91.81% | |||
| E27 | E1/E2 | Neocosmospora falciformis, Neocosmospora (previously the F. solani) SC | TEF1/2 | NRRL 54983 | https://www.fusarium.org/details/23/510 | 82% 84% | 99.56% 98.98% | Orlando | Yiron, Galilee |
| 7cf/11ar | Neocosmospora variasi, Neocosmospora (previously the F. solani) SC | RPB2 | CBS 146890 | https://www.fusarium.org/details/23/1313 | 93.20% | 51.97% | |||
| E28 | E1/E2 | Neocosmospora falciformis, Neocosmospora (previously the F. solani) SC | TEF1/2 | NRRL 54983 | https://www.fusarium.org/details/23/510 | 99.68% | 89.36% | Orlando | Yiron, Galilee |
| 7cf/11ar | Neocosmospora falciformis, Neocosmospora (previously the F. solani) SC | RPB2 | NRRL 54983 | https://www.fusarium.org/details/23/510 | 94.87% | 67.59% | |||
| E29 | E1/E2 | Neocosmospora falciformis, Neocosmospora (previously the F. solani) SC | TEF1/2 | CBS 121450 | https://www.fusarium.org/details/23/479 | 100% | 94.86% | Orlando | Yiron, Galilee |
| 7cf/11ar | Neocosmospora falciformis, Neocosmospora (previously the F. solani) SC | RPB2 | NRRL 28563 | https://www.fusarium.org/details/23/486 | 99.54% | 88.88% | |||
| E30 | E1/E2 | Fusarium fabacearum, F. oxysporum SC | TEF1/2 | CBS 144742 | https://www.fusarium.org/details/23/160 | 99.67% | 74.05% | Orlando | Yiron, Galilee |
| 7cf/11ar | Fusarium inflexum, F. oxysporum SC | RPB2 | CBS 716.74 | https://www.fusarium.org/details/23/207 | 99.77% | 89.27% | |||
| E31 | E1/E2 | Neocosmospora falciformis, Neocosmospora (previously the F. solani) SC | TEF1/2 | CBS 121450 | https://www.fusarium.org/details/23/479 | 99.55% | 80.94% | Orlando | Yiron, Galilee |
| 7cf/11ar | Neocosmospora ipomoeae, Neocosmospora (previously the F. solani) SC | RPB2 | CBS 833.97 | https://www.fusarium.org/details/23/534 | 97.92% | 52.91% | |||
| E32 | E1/E2 | Neocosmospora falciformis, Neocosmospora (previously the F. solani) SC | TEF1/2 | NRRL 43441 | https://www.fusarium.org/details/23/501 | 99.70% | 95.01% | Orlando | Yiron, Galilee |
| 7cf/11ar | Neocosmospora falciformis, Neocosmospora (previously the F. solani) SC | RPB2 | LC13827 | https://www.fusarium.org/details/23/2031 | 95.53% | 63.17% | |||
| E33 | E1/E2 | Neocosmospora falciformis, Neocosmospora (previously the F. solani) SC | TEF1/2 | CBS 121450 | https://www.fusarium.org/details/23/479 | 99.85% | 93.93% | Orlando | Yiron, Galilee |
| 7cf/11ar | Neocosmospora falciformis, Neocosmospora (previously the F. solani) SC | RPB2 | NRRL 43441 | https://www.fusarium.org/details/23/501 | 95.95% | 79.74% | |||
| E34 | E1/E2 | Fusarium fabacearum, F. oxysporum SC | TEF1/2 | CBS 144742 | https://www.fusarium.org/details/23/160 | 99.50% | 88.01% | Orlando | Yiron, Galilee |
| 7cf/11ar | Fusarium inflexum, F. oxysporum SC | RPB2 | CBS 716.74 | https://www.fusarium.org/details/23/207 | 99.77% | 87.36% | |||
| E35 | E1/E2 | Neocosmospora falciformis, Neocosmospora (previously the F. solani) SC | TEF1/2 | CBS 121450 | https://www.fusarium.org/details/23/479 | 99.70% | 94.18% | Orlando | Yiron, Galilee |
| 7cf/11ar | Neocosmospora falciformis, Neocosmospora (previously the F. solani) SC | RPB2 | CBS 475.67 | https://www.fusarium.org/details/23/478 | 96.22% | 75.36% | |||
| E36 | E1/E2 | Fusarium brevicatenulatum, F. fujikuroi SC | TEF1/2 | CBS 143874 | https://www.fusarium.org/details/23/375 | 89.44% | 93.70% | Orlando | Yiron, Galilee |
| 7cf/11ar | Fusarium inflexum, F. oxysporum SC | RPB2 | CBS 716.74 | https://www.fusarium.org/details/23/207 | 100% | 89.25% | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Degani, O.; Dimant, E.; Margalit, E. Impact of Fusarium Species Composition and Incidence on Onion Basal Rot in Northeastern Israel. Horticulturae 2024, 10, 373. https://doi.org/10.3390/horticulturae10040373
Degani O, Dimant E, Margalit E. Impact of Fusarium Species Composition and Incidence on Onion Basal Rot in Northeastern Israel. Horticulturae. 2024; 10(4):373. https://doi.org/10.3390/horticulturae10040373
Chicago/Turabian StyleDegani, Ofir, Elhanan Dimant, and Eliyahu Margalit. 2024. "Impact of Fusarium Species Composition and Incidence on Onion Basal Rot in Northeastern Israel" Horticulturae 10, no. 4: 373. https://doi.org/10.3390/horticulturae10040373
APA StyleDegani, O., Dimant, E., & Margalit, E. (2024). Impact of Fusarium Species Composition and Incidence on Onion Basal Rot in Northeastern Israel. Horticulturae, 10(4), 373. https://doi.org/10.3390/horticulturae10040373