Screening of Apple Cultivars for Scab Resistance in Kazakhstan
Abstract
1. Introduction
2. Materials and Methods
2.1. Disease Monitoring
2.2. Field Evaluation
2.3. Collection of Plant Materials, DNA Extraction, and Detection of Rvi Genes with Molecular Markers
2.4. Data analysis
3. Results
3.1. Field Evaluation of Apple Cultivars for Scab Resistance
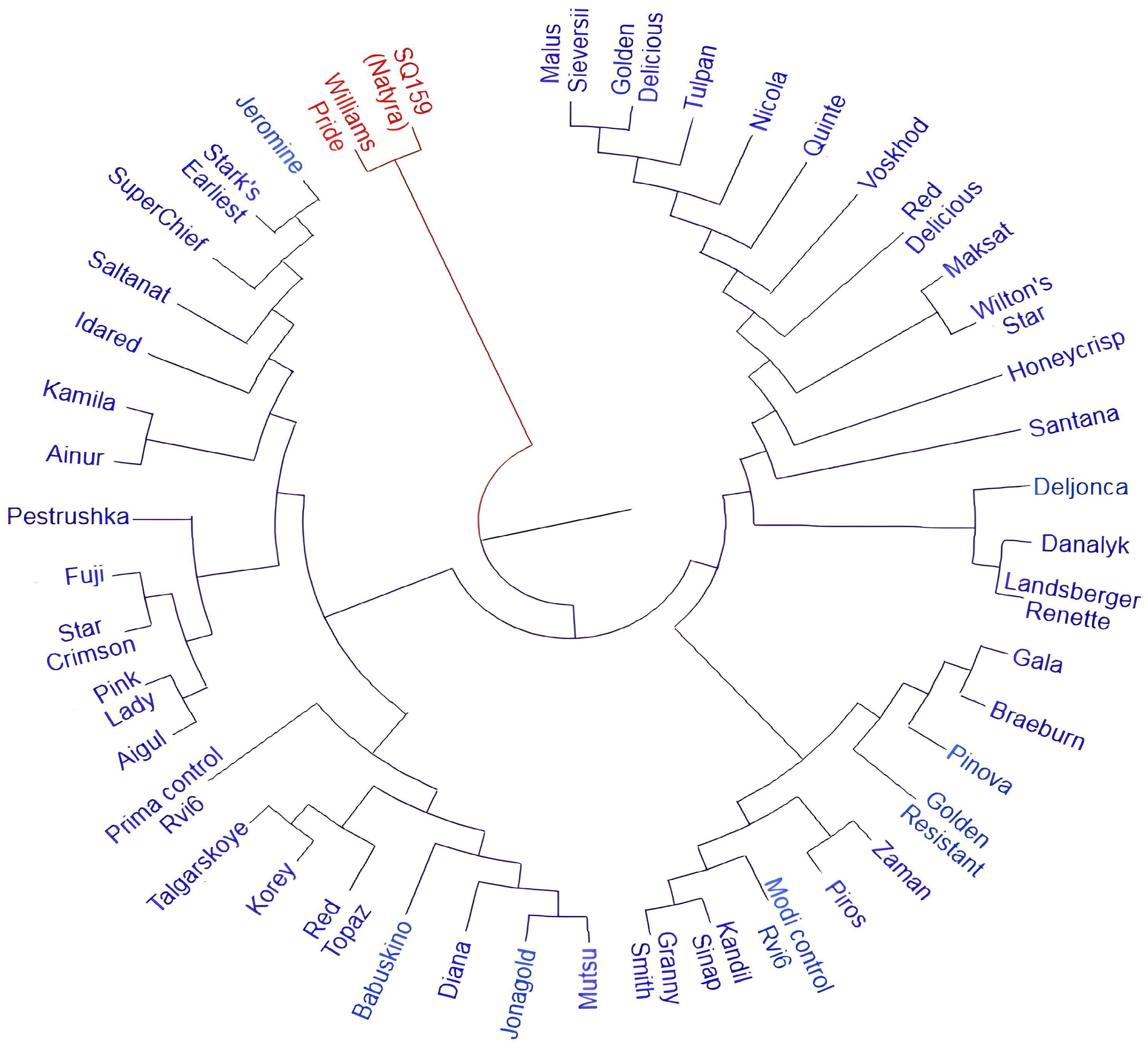
3.2. Molecular Screening of Apple Cultivars for Scab Resistance
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kairova, G.; Daulet, N.; Solomadin, M.; Sandybayev, N.; Orkara, S.; Beloussov, V.; Kerimbek, N.; Gritsenko, D.; Sapakhova, Z. Identification of Apple Cultivars Resistant to Fire Blight (Erwinia amylovora) Using Molecular Markers. Horticulturae 2023, 9, 1000. [Google Scholar] [CrossRef]
- Kairova, G.; Pozharskiy, A.; Daulet, N.; Solomadin, M.; Sandybayev, N.; Khusnitdinova, M.; Nizamdinova, G.; Sapakhova, Z.; Gritsenko, D. Evaluation of Fire Blight Resistance of Eleven Apple Rootstocks Grown in Kazakhstani Fields. Appl. Sci. 2023, 13, 11530. [Google Scholar] [CrossRef]
- Höfer, M.; Flachowsky, H.; Schröpfer, S.; Peil, A. Evaluation of Scab and Mildew Resistance in the Gene Bank Collection of Apples in Dresden-Pillnitz. Plants 2021, 10, 1227. [Google Scholar] [CrossRef]
- MacHardy, W. Inheritance of resistance to Venturia inaequalis. In Apple Scab, Biology, Epidemiology and Management; APS: St. Paul, MN, USA, 1996; pp. 61–103. [Google Scholar]
- Schubert, K.; Anja, R.; Uwe, B. A monograph of Fusicladium s. lat. (hyphomycetes). Schlechtendalia 2003, 9, 1–132. [Google Scholar]
- Beresford, R.; Wright, P.; Wood, P.; Park, N. Sensitivity of Venturia inaequalis to myclobutanil penconazole and dodine in relation to fungicide use in hawkes bay apple orchards. N. Z. Plant Prot. 2012, 65, 106–113. [Google Scholar] [CrossRef]
- Chapman, K.S.; Sundin, G.W.; Beckerman, J.L. Identification of resistance to multiple fungicides in field populations of Venturia inaequalis. Plant Dis. 2011, 95, 921–926. [Google Scholar] [CrossRef]
- Frederick, Z.A.; Villani, S.M.; Cooley, D.R.; Biggs, A.R.; Raes, J.J.; Cox, K.D. Prevalence and stability of qualitative QoI resistance in populations of Venturia inaequalis in the northeastern United States. Plant Dis. 2014, 98, 1122–1130. [Google Scholar] [CrossRef]
- Ayer, K.M.; Villani, S.M.; Choi, M.W.; Cox, K.D. Characterization of the VisdhC and VisdhD genes in Venturia inaequalis, and sensitivity to fluxapyroxad, pydiflumetofen, inpyrfluxam, and benzovindiflupyr. Plant Dis. 2019, 103, 1092–1100. [Google Scholar] [CrossRef]
- Beckerman, J.; Chatfield, J.; Draper, E. A 33-year Evaluation of Resistance and Pathogenicity in the Apple Scab–crabapples Pathosystem. HortSci. Horts 2009, 44, 599–608. [Google Scholar] [CrossRef]
- van den Bosch, F.; Blake, J.; Gosling, P.; Helps, J.C.; Paveley, N. Identifying when it is financially beneficial to increase or decrease fungicide dose as resistance develops: An evaluation from long-term field experiments. Plant Pathol. 2020, 69, 631–641. [Google Scholar] [CrossRef]
- Holb, I.J. Effect of six sanitation treatments on leaf litter density, ascospore production of Venturia inaequalis and scab incidence in integrated and organic apple orchards. Eur. J. Plant Pathol. 2006, 115, 293–307. [Google Scholar] [CrossRef]
- Holb, I.J. Effect of four non-chemical sanitation treatments on leaf infection by Venturia inaequalis in organic apple orchards. Eur. J. Hortic. Sci. 2007, 72, 60. [Google Scholar]
- tSaoir, S.M.A.; Cooke, L.R. The effects of leaf litter treatments, post-harvest urea and omission of early season fungicide sprays on the overwintering of apple scab on Bramley’s Seedling grown in a maritime environment. Ir. J. Agric. Food Res. 2010, 49, 55–66. [Google Scholar]
- Porsche, F.M.; Pfeiffer, B.; Kollar, A. A New Phytosanitary Method to Reduce the Ascospore Potential of Venturia inaequalis. Plant Dis. 2017, 101, 414–420. [Google Scholar] [CrossRef][Green Version]
- Acero, F.J.; Carbú, M.; El-Akhal, M.R.; Garrido, C.; González-Rodríguez, V.E.; Cantoral, J.M. Development of proteomics-based fungicides: New strategies for environmentally friendly control of fungal plant diseases. Int. J. Mol. Sci. 2011, 12, 12795–12816. [Google Scholar] [CrossRef]
- Bus, V.G.M.; Rikkerink, E.H.; Caffier, V.; Durel, C.-E.; Plummer, K.M. Revision of the nomenclature of the differential host-Pathogen interactions of Venturia inaequalis and Malus. Annu. Rev. Phytopathol. 2011, 49, 391–413. [Google Scholar] [CrossRef]
- Gross, B.L.; Kellogg, E.A.; Miller, A.J. Speaking of food: Connecting basic and applied plant science. Am. J. Bot. 2014, 101, 1597–1600. [Google Scholar] [CrossRef]
- Peace, C.P.; Luby, J.J.; van de Weg, W.E.; Bink, M.C.A.M.; Iezzoni, A.F. A strategy for developing representative germplasm sets for systematic QTL validation, demonstrated for apple, peach, and sweet cherry. Tree Gen. Genom. 2014, 10, 1679–1694. [Google Scholar] [CrossRef]
- Kabylbekova, B.; Kovalchuk, I.; Mukhitdinova, Z.; Turdiyev, T.; Kairova, G.; Madiyeva, G.; Reed, B.M. Reduced major minerals and increased minor nutrients improve micropropagation in three apple cultivars. In Vitr. Cell. Dev. Biol.-Plant 2020, 56, 335–349. [Google Scholar] [CrossRef]
- Gessler, C.; Patocchi, A.; Sansavini, S.; Tartarini, S.; Gianfranceschi, L. Venturia inaequalis resistance in apple. Crit. Rev. Plant Sci. 2006, 25, 473–503. [Google Scholar] [CrossRef]
- Liang, W.; Dondini, L.; De Franceschi, P.; Paris, R.; Silviero, S.; Tartarini, S. Genetic diversity, population structure and construction of a core collection of apple cultivars from Italian germplasm. Plant Mol. Biol. Rep. 2015, 33, 458–473. [Google Scholar] [CrossRef]
- Baumgartner, I.O.; Kellerhals, M.; Costa, F.; Dondini, L.; Pagliarani, G.; Gregori, R.; Tartarini, S.; Leumann, L.; Laurens, F.; Patocchi, A. Development of SNP-based assays for disease resistance and fruit quality traits in apple (Malus × domestica Borkh.) and validation in breeding pilot studies. Tree Genet. Genomes 2016, 12, 35. [Google Scholar] [CrossRef]
- Khajuria, Y.P.; Kaul, S.; Wani, A.A.; Dhar, M.K. Genetics of resistance in apple against Venturia inaequalis (Wint) Cke. Tree Genet. Genomes 2018, 14, 1–20. [Google Scholar] [CrossRef]
- Bus, V.; Rikkerink, E.; Aldwinckle, H.S.; Caffier, V.; Durel, C.E.; Gardiner, S.; Gessler, C.; Groenwold, R.; Laurens, F.; Cam, B.L.; et al. A proposal for the nomenclature of Venturia inaequalis races. Acta Hortic. 2009, 814, 739–746. [Google Scholar] [CrossRef]
- Soriano, J.M.; Madduri, M.; Schaart, J.G.; Burgh, A.; Kaauwen, M.V.; Tomić, L.; Groenwold, R.; Velasco, R.; Weg, E.V.; Schouten, H.J. Fine mapping of the gene Rvi18 (V25) for broad-spectrum resistance to apple scab, and development of a linked SSR marker suitable for marker-assisted breeding. Mol. Breed. 2014, 34, 2021–2032. [Google Scholar] [CrossRef]
- Patocchi, A.; Frei, A.; Frey, J.E.; Kellerhals, M. Towards improvement of marker assisted selection of apple scab resistant cultivars: Venturia inaequalis virulence surveys and standardization of molecular marker alleles associated with resistance genes. Mol. Breed. 2009, 24, 337–347. [Google Scholar] [CrossRef]
- Cheng, F.S.; Weeden, N.F.; Brown, S.K.; Aldwinckle, H.S.; Gardiner, S.E.; Bus, V.G. Development of a DNA marker for Vm, a gene conferring resistance to apple scab. Genome 1998, 41, 208–214. [Google Scholar] [CrossRef]
- Durel, C.E.; Freslon, V.; Denancé, C.; Laurens, F.; Lespinasse, Y.; Rat, E.; Parisi, L.; Bus, V.; Dapena de la Fuente, E.; Miñarro, M.; et al. Genetic localisation of new major and minor pest and disease factors in the apple genome. In Proceedings of the 3rd Rosaceae Genomics Conference, Napier, New Zealand, 19–22 March 2006. [Google Scholar]
- Soufflet-Freslon, V.; Gianfranceschi, L.; Patocchi, A.; Durel, C.-E. Inheritance studies of apple scab resistance and identification of Rvi14, a new major gene that acts together with other broad-spectrum QTL. Genome 2008, 51, 657–667. [Google Scholar] [CrossRef]
- Soriano, J.M.; Joshi, S.G.; van Kaauwen, M.; Noordijk, Y.; Groenwold, R.; Henken, B.; van de Weg, W.E.; Schouten, H.J. Identification and mapping of the novel apple scab resistance gene Vd3. Tree Genet. Genomes 2009, 5, 475–482. [Google Scholar] [CrossRef]
- Bus VGMRikkerink, E.H.A.; EW van de Weg Rusholme, R.L.; Gardiner, S.E.; Bassett, H.C.M.; Kodde, L.P.; Parisi, L.; Laurens, F.N.D.; Meulenbroek, E.; Plummer, K.M. The Vh2 and Vh4 scab resistance genes in two differential hosts derived from Russian apple R12740-7A map to the same linkage group of apple. Mol. Breed. 2005, 15, 103–116. [Google Scholar] [CrossRef]
- Bus, V.G.M.; Laurens, F.N.D.; van de Weg, W.E.; Rusholme, R.L.; Rikkerink, E.H.A.; Gardiner, S.E.; Bassett, H.C.M.; Kodde, L.P.; Plummer, K.M. The Vh8 locus of a new gene for gene interaction between Venturia inaequalis and the wild apple Malus sieversii is closely linked to the Vh2 locus in Malus pumila R12740-7A. New Phytol. 2005, 166, 1035–1049. [Google Scholar] [CrossRef]
- Jansch, M.; Broggini, G.A.; Weger, J.; Bus, V.G.; Gardiner, S.E.; Bassett, H.; Patocchi, A. Identification of SNPs linked to eight apple disease resistance loci. Mol. Breed. 2015, 35, 45. [Google Scholar] [CrossRef]
- Baldi, P.; Patocchi, A.; Zini, E.; Toller, C.; Velasco, R.; Komjanc, M. Cloning and linkage mapping of resistance gene homologues in apple. Theor. Appl. Genet. 2004, 109, 231–239. [Google Scholar] [CrossRef]
- Galli, P.; Broggini, G.A.L.; Kellerhals, M.; Gessler, C.; Potachi, A. High resolution genetic map of the Rvi15 (Vr2) apple scab resistance locus. Mol. Breed. 2010, 26, 561–572. [Google Scholar] [CrossRef][Green Version]
- Dayton, D.F.; Williams, E.B. Additional allelic genes in Malus for scab resistance of two reaction types. J. Am. Soc. Hortic. Sci. 1970, 95, 735–736. [Google Scholar] [CrossRef]
- Patocchi, A.; Walser, M.; Tartarini, S.; Broggini, G.A.L.; Gennari, F.; Sansavini, S.; Gessler, C. Identification by genome scanning approach (GSA) of a microsatellite tightly associated with the apple scab resistance gene Vm. Genome 2005, 48, 630–636. [Google Scholar] [CrossRef]
- Bandara, N.L.; Cova, V.; Tartarini, S.; Gessler, C.; Patocchi, A.; Cestaro, A.; Troggio, M.; Velasco, R.; Komjanc, M. Isolation of Rvi5 (vm) locus from Malus × domestica ‘Murray’. Acta Hortic. 2015, 1100, 21–24. [Google Scholar] [CrossRef]
- Vinatzer, B.A.; Zhang, H.B.; Sansavini, S. Construction and characterization of a bacterial artificial chromosome library of apple. Theor. Appl. Genet. 1998, 97, 1183–1190. [Google Scholar] [CrossRef]
- Patocchi, A.; Gianfranceschi, L.; Gessler, C. Towards the map-based cloning of Vf: Fine and physical mapping of the Vf region. Theor. Appl. Genet. 1999, 99, 1012–1017. [Google Scholar] [CrossRef]
- Xu, M.L.; Korban, S.S. Saturation mapping of the apple scab resistance gene Vf using AFLP markers. Theor. Appl. Genet. 2000, 101, 844–851. [Google Scholar] [CrossRef]
- Patrascu, B.; Pamfil, D.; Sestras, R.E.; Botez, C.; Gaboreanu, I.; Barbos, A.; Qin, C.; Rusu, R.A.; Bondrea, I.; Dirle, E. Marker assisted selection for response attack of Venturia inaequalis in different apple genotypes. Not. Bot. Hort. Agrobot. Cluj-Napoca 2006, 34, 121–132. [Google Scholar]
- Chizzali, C.; Gusberti, M.; Schouten, H.J.; Gessler, C.; Broggini, G.A. Cisgenic Rvi6 scab-resistant apple lines show no differences in Rvi6 transcription when compared with conventionally bred cultivars. Planta 2016, 243, 635–644. [Google Scholar] [CrossRef]
- Dayton, D.F.; Williams, E.B. Independent genes in Malus for resistance to Venturia inaequalis. Proc. Am. Soc. Hortic. Sci. 1968, 92, 89–94. [Google Scholar]
- Gygax, M.; Gianfranceschi, L.; Liebhard, R.; Kellerhals, M.; Gessler, C.; Patocchi, A. Molecular markers linked to the apple scab resistance gene Vbj derived from Malus baccata jackii. Theor. Appl. Genet. 2004, 109, 1702–1709. [Google Scholar] [CrossRef]
- Patocchi, A.; Bigler, B.; Liebhard, R.; Koller, B.; Gessler, C. Mapping of Vr 2, a third apple scab resistance gene of Russian seedling (R12740-7A). Plant Anim. Genomes XI Conf. Poster 2003, 540. [Google Scholar]
- Patocchi, A.; Bigler, B.; Koller, B.; Kellerhals, M.; Gessler, C. A new apple scab resistance gene. Theor. Appl. Genet. 2004, 109, 1087–1092. [Google Scholar] [CrossRef]
- Peil, A.; Howard, N.P.; Bühlmann-Schütz, S.; Hiller, I.; Schouten, H.; Flachowsky, H.; Patocchi, A. Rvi4 and Rvi15 are the same apple scab resistance genes. Mol. Breed. 2023, 43, 74. [Google Scholar] [CrossRef]
- Caffier, V.; Patocchi, A.; Expert, P.; Bellanger, M.N.; Durel, C.E.; Hilber-Bodmer, M.; Broggini, G.A.L.; Groenwold, R.; Bus, V.G. Virulence characterization of Venturia inaequlis reference isolates on the differential set of Malus hosts. Plant Dis. 2015, 99, 370–375. [Google Scholar] [CrossRef]
- Peil, A.; Patocchi, A.; Hanke, M.; Bus, V.G. Apple cultivar Regia possessing both Rvi2 and Rvi4 resistance genes is the source of a new race of Venturia inaequalis. Eur. J. Plant Pathol. 2018, 151, 533–539. [Google Scholar] [CrossRef]
- Papp, D.; Gangadharappa Harigondra, S.; Paredes, C.; Karacs-Végh, A.; Penksza, K.; T.-Járdi, I.; Papp, V. Strong genetic differentiation between generalist populations of Venturia inaequalis and populations from partially resistant apple cultivars carrying Rvi3 or Rvi5. Diversity 2022, 14, 1050. [Google Scholar] [CrossRef]
- Silfverberg-Dilworth, E.; Matasci, C.L.; Van de Weg, W.E.; Van Kaauwen, M.P.W.; Walser, M.; Kodde, L.P.; Soglio, V.; Gian-franceschi, L.; Durel, C.E.; Costa, F.; et al. Microsatellite markers spanning the apple (Malus domestica Borkh.) genome. Tree Genet. Genomes 2006, 2, 202–224. [Google Scholar] [CrossRef]
- Tartarini, S.; Gianfranceschi, L.; Sansavini, S.; Gessler, C. Development of reliable PCR markers for the selection of the Vf gene conferring scab resistance in apple. Plant Breed. 1999, 118, 183–186. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Yeh, F.C.; Yang, R.; Boyle, T.J.; Ye, Z.; Xiyan, J.M. PopGene32, Microsoft Windows-Based Freeware for Population Genetic Analysis, Version 1.32; Molecular Biology and Biotechnology Centre, University of Alberta: Edmonton, AB, Canada, 2000. [Google Scholar]
- Dice, I.R. Measures of the amount of ecologic association between species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Broggini, G.A.L.; Bus, V.G.; Parravicini, G.; Kumar, S.; Groenwold, R.; Gessler, C. Mapping of 14 avirulence genes in an EU-B04 × 1639 progeny of Venturia inaequalis. Fungal Genet. Biol. 2011, 48, 166–176. [Google Scholar] [CrossRef]
- Luby, J.J.; Forsline, P.; Aldwinckle, H.; Bus, V.; Geibel, M. Silk road apples collection, evaluation, and utilization of Malus sieversii from Central Asia. HortScience 2001, 36, 225–231. [Google Scholar] [CrossRef]
- Omasheva, M.Y.; Pozharskiy, A.S.; Maulenbay, A.D.; Ryabushkina, N.A.; Galiakparov, N.N. SSR genotyping of Kazakhstan apple cultivars: Identification of alleles associated with resistance to highly destructive pathogens. Biot. Theory Pract. 2016, 2, 46–58. [Google Scholar] [CrossRef]
- Sagi, S.; Svetlana, D.; Moldir, Z.; Aigul, M.; Zhanna, I.; Balnur, K. Physiological and phytopathological assessment scion-rootstock combinations for apple cv. Aport and M. sieversii. Res. Crop. 2022, 23, 795–800. [Google Scholar] [CrossRef]
- Win, J.; Greenwood, D.R.; Plummer, K.M. Characterisation of a protein from Venturia inaequalis that induces necrosis in Malus carrying the Vm resistance gene. Physiol. Mol. Plant Pathol. 2003, 62, 193–202. [Google Scholar] [CrossRef]
- Galli, P.; Broggini, G.A.L.; Gessler, C.; Patocchi, A. Phenotypic characterization of the Rvi15 (Vr2) apple scab resistance. Plant Pathol. 2010, 92, 219–226. [Google Scholar]
- Parisi, L.; Fouillet, V.; Schouten, H.J.; Groenwold, R.; Laurens, F.; Didelot, F.; Evans, K.; Fischer, C.; Gennari, F.; Kemp, H.; et al. Variability of the pathogenicity of Venturia inaequalis in Europe. Acta Hortic. 2004, 663, 107–113. [Google Scholar] [CrossRef]
- Patocchi, A.; Wehrli, A.; Dubuis, P.-H.; Auwerkerken, A.; Leida, C.; Cipriani, G.; Passey, T.; Staples, M.; Didelot, F.; Philion, V.; et al. Ten years of VINQUEST: First insight for breeding new apple cultivars with durable apple scab resistance. Plant Dis. 2020, 104, 2074–2081. [Google Scholar] [CrossRef] [PubMed]
- Peil, A.; Kellerhals, M.; Höfer, M.; Flachowsky, H. Apple breeding from the origin to genetic engineering. Agric. Food Sci. Biol. Environ. Sci. 2011, 5, 118–138. [Google Scholar]
| Breeding Program | Geographical Site * | Coordinates | Precipitation May–August (mm) | Number of Trees for Sampling ** |
|---|---|---|---|---|
| Kazygurt district, Kyzylkiyan rural district, “Akniyet Agro Orchard” LLP | Turkestan region | N 41°36′8.594″ E 69°21′58.013″ | 926.8 | 8 |
| Saryagash district, Zhemisti rural areas, Regional Branch, “Saryagash” LLC, “Kazakh Fruit and Vegetable Research Institute” (KazF&VRI) | Turkestan region | N 41°32′2.545″ E 69°21′36.069″ | 902.8 | 8 |
| Tulkubas district, Shakpak Baba rural areas, “Koktal” Peasant Farm | Turkestan region | N 41°29′2.69″ E 70°31′11.46″ | 945.3 | 8 |
| Merke district, Merke rural areas, “Merke experimental farm” LLP | Zhambyl region | N 42°48′.584″ E 73°10′.387″ | 925.2 | 3 |
| Yenbekshikazakh district, Baidibek bi rural areas, “Akkazy” Peasant Farm | Almaty region | N 43°39′.930″ E 77°86′.171″ | 902.8 | 8 |
| Yenbekshikazakh district, Baidibek bi rural areas, “Ermek” Peasant Farm | Almaty region | N 43°32′49.344″ E 77° 52′ 3.468″ | 906.9 | 8 |
| Yenbekshikazakh district, Koram rural areas, “Zhetysu Trade” | Almaty region | N 43°31′48.31″ E 78°11′48.09″ | 908.9 | 9 |
| Talgar District, Regional Branch, “Talgar” of KazF&VRI (Pomological Garden) | Almaty region | N 43°17′27″ E 77°12′15″ | 90.36 | 45 |
| Resistance Genes | Marker Name | Marker Type | Size of Allele (bp) | Primer Sequence (5′→3′) | References |
|---|---|---|---|---|---|
| Rvi6 (Vf) | AM19 | SCAR | 526 | F: CGTAGAACGGAATTTGACAGTG R: GACAAGGGCTTAAGTGCTCC | Bus et al. [17,18] |
| Rvi2, Rvi4, Rvi9, Rvi11 | CH05e03 | SSR | 172 | F: CGAATATTTTCACTCTGACTGGG R: CAAGTTGTTGTACTGCTCCGAC | Gygax et al. [46] Patocci et al. [27] |
| Rvi2, Rvi8 | OPL19 | SCAR | 433 | F: ACCTGCACTACAATCTTCACTAATC R: GACTCGTTTCCACTGAGGATATTTG | Bus et al. [17] Patocci et al. [27] |
| Rvi5 | Hi07f02 | SSR | 226 | F: ATTTGGGGTTTCAACAATGG R: GTTTCGGACATCAAACAAATGTGC | Silfverberg-Dilworth et al. [53] |
| Rvi6 | AL07 | SCAR | 466 | F: TGGAAGAGAGATCCAGAAAGTG R: CATCCCTCCACAAATGCC | Tartarini et al. [54] |
| Rvi11 | K08 | SCAR | 743 | F: GAACACTGGGCAAAGGAAAC R: TAAAAGCCACGTTCTCTCGC | Gygax et al. [46] |
| Rvi14 | HB09 | SSR | 210 | F: GCTCAAAATACTGAAGCCTTGC R: GGGGAAGCAGGATGGTTACT | Soufflet et al. [30] Patocchi et al. [27] |
| Rvi15 | CH02f06 | SSR | 147 | F: CCCTCTTCAGACCTGCATATG R: ACTGTTTCCAAGCGCTCAGG | Patocci et al. [48] Patocci et al. [27] |
| Turkestan Region | ||||||
|---|---|---|---|---|---|---|
| Akniyet Agro Orchards, Kazygurt District | Saryagash, Saryagash District | Koktal, Tulkubas District | ||||
| 2022 | 2023 | 2022 | 2023 | 2022 | 2023 | |
| Star Crimson | 8.75 | 0 | 18.75 | 9.37 | 0 | 0 |
| Idared | 20.88 | 0 | 0 | 0 | 0 | 0 |
| Gala | 5.62 | 0 | 0 | 0 | 0 | 0 |
| Fuji | 0 | 0 | 0 | 0 | 0 | 0 |
| Golden Delicious | 0 | 0 | 21.25 | 9.22 | 0 | 0 |
| Pink Lady | 0 | 0 | 0 | 0 | 0 | 0 |
| Landsberger Renette | 0 | 0 | 20.65 | 19.36 | 0 | 0 |
| Almaty Region | ||||||||
|---|---|---|---|---|---|---|---|---|
| Akkazy, Yenbekshikazakh District | Ermek, Yenbekshikazakh District | Zhetysu Trade, Yenbekshikazakh District | Talgar, Talgar District | |||||
| 2022 | 2023 | 2022 | 2023 | 2022 | 2023 | 2022 | 2023 | |
| Ainur | -* | - | - | - | - | - | 4.98 | 0 |
| Aigul | - | - | - | - | - | - | 4.69 | 0 |
| Maksat | - | - | - | - | - | - | 0 | 0 |
| Zaman | - | - | - | - | - | - | 5.06 | 0 |
| Kamila | - | - | - | - | - | - | 0 | 0 |
| Diana | - | - | - | - | - | - | 0 | 0 |
| Malus sieversii | - | - | - | - | - | - | 10.63 | 4.29 |
| Danalyk | - | - | - | - | - | - | 5.03 | 0 |
| Saltanat | - | - | - | - | - | - | 0 | 0 |
| Kandil Sinap | - | - | - | - | - | - | 4.38 | 5.63 |
| Golden Delicious | 21.88 | 10.63 | 0 | 0 | 9.36 | 4.38 | 48.75 | 28.75 |
| Granny Smith | - | - | - | - | - | - | 10.08 | 9.63 |
| Korey | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Mutsu | - | - | - | - | - | - | 0 | 0 |
| Landsberger Renette | - | - | - | - | - | - | 4.68 | 5.06 |
| Star Crimson | 19.48 | 4.38 | 0 | 0 | 0 | 0 | 0 | 9.68 |
| Stark’s Earliest | - | - | - | - | 9.79 | 9.88 | 5.06 | 9.38 |
| Talgarskoye | - | - | - | - | - | - | 0 | 0 |
| Tulpan | - | - | - | - | - | - | 0 | 0 |
| Williams Pride | - | - | - | - | - | - | 0 | 0 |
| Gala | 0 | 0 | 0 | 0 | 9.36 | 5.05 | 4.98 | 27.90 |
| Pestrushka | - | - | - | - | - | - | 4.38 | 0 |
| Idared | 0 | 0 | 0 | 0 | 0 | 0 | 5.03 | 9.65 |
| Pink Lady | - | - | - | - | - | - | 4.89 | 0 |
| Piros | - | - | - | - | - | - | 0 | 0 |
| Braeburn | - | - | - | - | - | - | 5.03 | 0 |
| Honeycrisp | - | - | - | - | - | - | 0 | 0 |
| Jeromine | - | - | - | - | - | - | 4.83 | 0 |
| SuperChief | - | - | - | - | - | - | 0 | 0 |
| Wilton’s Star | - | - | - | - | - | - | 4.78 | 0 |
| Red Topaz | - | - | - | - | - | - | 54.97 | 0 |
| SQ159 (Natyra) | - | - | - | - | - | - | 0 | 0 |
| Santana | - | - | - | - | - | - | 4.78 | 0 |
| Deljonca | - | - | - | - | - | - | 4.38 | 0 |
| Fuji | 0 | 0 | 0 | 0 | 0 | 0 | 4.78 | 9.48 |
| Pinova | - | - | - | - | - | - | 5.03 | |
| Nicola | - | - | - | - | - | - | 18.70 | 19.86 |
| Modi (Rvi6 control) | - | - | - | - | - | - | 0 | 0 |
| Quinte | 0 | 0 | 0 | 0 | 9.89 | 4.89 | 4.78 | 0 |
| Jonagold | - | - | - | - | - | - | 20.05 | 4.38 |
| Voskhod | - | - | - | - | - | - | 4.98 | 0 |
| Babuskino | - | - | - | - | - | - | 21.38 | 4.38 |
| Red Delicious | 0 | 0 | 0 | 0 | 4.68 | 4.48 | 49.50 | 0 |
| Golden Resistant | - | - | - | - | - | - | 0 | 0 |
| Prima (Rvi6 control) | - | - | - | - | - | - | 0 | 0 |
| Zhambyl Region | ||
|---|---|---|
| Merke Experimental Farm, Merke District | ||
| 2022 | 2023 | |
| Red Delicious | 29.75 | 4.63 |
| Star Crimson | 19.65 | 9.63 |
| Golden Delicious | 19.86 | 4.38 |
| Cultivar | Origin * | Rvi2/Rvi8 | Rvi2 | Rvi4 | Rvi9 | Rvi11 | Rvi5 | Rvi6 | Rvi11 | Rvi14 | Rvi15 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OPL19 | CH05e03 | Hi07f02 | AL07 | AM19 | K08 | HB09 | CH02f06 | |||||
| 433 bp | 163 bp | 172 bp | 169 bp | 160 bp | 220 bp | 466 bp | 526 bp | 743 bp | 210 bp | 135–158 bp | ||
| Ainur | KZ | + ** | + | − ** | − | − | − | − | − | − | − | + |
| Aigul | KZ | + | + | − | − | − | − | − | − | − | + | − |
| Maksat | KZ | − | − | − | − | − | − | − | − | − | − | + |
| Zaman | KZ | − | − | + | + | − | − | − | − | − | + | − |
| Kamila | KZ | + | − | − | − | − | − | − | − | + | + | + |
| Diana | RU | + | − | − | − | − | + | − | − | + | − | − |
| Malus sieversii | KZ | − | − | − | − | − | − | − | − | − | − | − |
| Danalyk | KZ | − | − | − | − | − | + | + | + | − | − | − |
| Saltanat | KZ | + | − | + | + | − | + | − | − | + | − | − |
| Kandil Sinap | UA | − | + | + | − | − | − | − | − | + | − | + |
| Golden Delicious | US | − | − | − | − | − | − | − | − | − | − | − |
| Granny Smith | AU | − | − | + | + | − | − | − | − | + | − | − |
| Korey | JP | + | − | − | − | − | − | − | − | + | + | − |
| Mutsu | JP | + | − | − | − | − | − | − | − | + | − | − |
| Landsberger Renette | DE | + | − | − | − | − | − | − | − | − | − | − |
| Star Crimson | US | + | − | − | + | − | − | − | − | + | − | − |
| Stark’s Earliest | US | + | + | + | + | − | + | − | − | + | + | + |
| Talgarskoye | KZ | + | − | − | − | − | − | − | − | + | + | + |
| Tulpan | KZ | − | − | − | − | − | − | − | − | − | − | − |
| Williams Pride | US | + | − | − | − | − | + | + | + | + | + | − |
| Gala | NZ | + | − | + | − | − | − | − | − | − | − | + |
| Pestrushka | RU | − | − | − | + | − | − | − | − | + | + | − |
| Idared | US | + | − | − | − | − | + | − | − | + | + | − |
| Pink Lady | AU | + | − | + | + | − | − | − | − | − | + | − |
| Piros | DE | − | − | − | + | − | − | − | − | − | + | + |
| Braeburn | NZ | + | − | + | + | − | − | − | − | − | − | + |
| Honeycrisp | US | − | − | + | + | − | − | − | − | − | − | − |
| Jeromine | US | + | − | + | − | − | + | − | − | + | + | − |
| SuperChief | US | + | − | + | − | − | + | − | − | + | + | − |
| Wilton’s Star | NL | − | − | − | − | − | − | − | − | − | − | + |
| Red Topaz | CZ | − | − | − | − | − | − | − | − | + | + | + |
| SQ159 (Natyra) | DE | + | − | − | − | − | − | + | + | + | − | + |
| Santana | NL | − | − | − | − | − | − | + | + | − | − | − |
| Deljonca | DE | − | − | − | − | − | + | − | − | − | − | + |
| Fuji | JP | + | + | + | + | − | − | − | − | − | − | − |
| Pinova | DE | + | + | + | − | − | − | − | − | + | − | + |
| Nicola | CA | − | − | − | − | − | − | − | − | − | − | − |
| Modi (Rvi6 control) | IT | + | − | + | + | + | − | + | + | − | − | − |
| Quinte | CA | − | − | − | + | − | − | − | − | − | − | − |
| Jonagold | US | − | − | − | − | − | − | − | − | − | − | − |
| Voskhod | KZ | + | − | − | − | − | − | − | − | + | − | − |
| Babuskino | RU | − | − | − | − | − | − | − | − | − | − | − |
| Red Delicious | US | − | − | − | − | − | − | − | − | + | − | − |
| Golden Resistant | US | − | − | − | − | − | − | − | − | − | − | − |
| Prima (Rvi6 control) | US | + | + | − | − | − | − | + | + | − | − | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Madenova, A.; Aitymbet, Z.; Bolat, M.; Kaldybayeva, D.; Galymbek, K.; Kuan, A.; Kabylbekova, B.; Irkitbay, A.; Yeszhanov, T.; Bakirov, S.; et al. Screening of Apple Cultivars for Scab Resistance in Kazakhstan. Horticulturae 2024, 10, 184. https://doi.org/10.3390/horticulturae10020184
Madenova A, Aitymbet Z, Bolat M, Kaldybayeva D, Galymbek K, Kuan A, Kabylbekova B, Irkitbay A, Yeszhanov T, Bakirov S, et al. Screening of Apple Cultivars for Scab Resistance in Kazakhstan. Horticulturae. 2024; 10(2):184. https://doi.org/10.3390/horticulturae10020184
Chicago/Turabian StyleMadenova, Aigul, Zhankeldy Aitymbet, Munira Bolat, Dinara Kaldybayeva, Kanat Galymbek, Angsagan Kuan, Balnur Kabylbekova, Azhargul Irkitbay, Tynyshbek Yeszhanov, Serik Bakirov, and et al. 2024. "Screening of Apple Cultivars for Scab Resistance in Kazakhstan" Horticulturae 10, no. 2: 184. https://doi.org/10.3390/horticulturae10020184
APA StyleMadenova, A., Aitymbet, Z., Bolat, M., Kaldybayeva, D., Galymbek, K., Kuan, A., Kabylbekova, B., Irkitbay, A., Yeszhanov, T., Bakirov, S., & Sapakhova, Z. (2024). Screening of Apple Cultivars for Scab Resistance in Kazakhstan. Horticulturae, 10(2), 184. https://doi.org/10.3390/horticulturae10020184





