Abstract
Since its domestication about a century ago in North America, highbush blueberry (Vaccinium corymbosum L.) has gained appreciation by consumers worldwide, and the demand for new blueberry varieties is increasing. Whole-genome resequencing can help plant breeders to decrease the time needed to create novel varieties by identifying novel genes linked to fruit-quality traits. The present study analyzed the genetic variability of eight V. corymbosum genotypes, seven Romanian varieties (‘Prod’, ‘Vital’, ‘Azur’, ‘Simultan’, ‘Delicia’, ‘Compact’, and ‘Safir’), and the American variety, ‘Bluecrop’. The analysis of the first ~10 Mb from scaffold 22, a hotspot of genomic variation, in the above-mentioned varieties revealed multiple differences in 11 upregulated and 50 downregulated genes involved in fruit growth and development. Of these differentially regulated genes, two upregulated and five downregulated genes were fully covered by at least 1× coverage depth by sequencing. The genes’ sequence analysis confirmed the high genetic variability of the region, with most of the genes presenting numerous SNPs and some InDels, and indicated that an attempted 10× medium-coverage depth of sequencing for V. corymbosum varieties yields useful preliminary data for use in breeding programs.
1. Introduction
Highbush blueberries (V. corymbosum), a perennial species belonging to the Ericaceae family, is native to North America and was domesticated at the end of 19th century in New England and Florida [1]. Over time, blueberry cultivation spread to the rest of the USA until the 1940s, to Europe in the 1970s, to Australia, New Zeeland and South America in the 1980s, and to China in the 2000s [2]. The species is now cultivated worldwide. In Romania, highbush blueberries were introduced in 1968 in Argeș county [3]. Subsequently, a breeding program was established in 1982 for highbush blueberries at the Research Institute for Fruit Growing Pitești, resulting in the creation of nine Romanian varieties: ‘Safir’ and ‘Azur’ (1998), ‘Augusta’ (1999), ‘Simultan’ and ‘Delicia’ (2001), ‘Lax’ and ‘Compact’ (2002), and ‘Vital’ and ‘Prod’ (2008) [4]. Moreover, after establishing a highbush blueberry collection and organizing yearly tasting sessions since 2016 with more than 60 blueberry varieties, the University of Agronomic Sciences and Veterinary Medicine of Bucharest started its own blueberry breeding program in 2021 [5,6].
One of the main reasons for cultivating highbush blueberries is that the fruits are considered a superfood, having nutraceutical qualities [7]. Blueberries contain high quantities of fiber, vitamins, minerals, secondary metabolites, and polyphenols such as anthocyanins and flavonols [8]. Polyphenols have been proven to have a positive role in the prevention of diseases like cancer, diabetes, cardiovascular disease, etc., [9,10,11,12].
In light of the growing demand for blueberries worldwide, the everchanging demand by consumers for different fruit colors, taste, flavors, together with climate change and the need for varieties resistant to biotic and abiotic stress, the pressure for creating new varieties resistant to environmental factors that will also satisfy the customers’ demands is ever-increasing [13,14,15,16]. One way to hasten the process of creating novel varieties is to employ molecular biology techniques such as the next-generation sequencing (NGS) of whole genomes [17,18,19,20]. Once a genome for a species has been fully sequenced and annotated, it serves as a reference genome. The resequencing of other genotypes from the same species leads to the detection of variations at the molecular level. The variations translate into phenotypic differences such as resistance to various environmental stress factors, and fruit-quality traits [21,22,23].
By 2012, Pasaniuc et al. [24] demonstrated that genome-wide associated studies (GWAS) with ultra-low coverage combined with genotype calling, using reference genomes from the 1000 Genomes Project [25], can use larger sample sizes at the same price as GWAS combined with SNP arrays, although the approach is limited to common variants [24]. Low-coverage resequencing has been used in crop species, such as wheat and passion fruit, and medicinal plants, such as milkweed, balloon flower and bonnet bellflower, not only to assess genetic diversity, and also to identify SNPs for use as markers for various traits of interest [26,27,28,29,30].
Although the price for whole-genome sequencing has decreased considerably, allowing for the sequencing of non-model plants [31,32,33], the question of which level of coverage depth is needed to obtain results useful for plant breeding for a particular species is still open.
The present study aimed to assess the genetic variability of eight V. corymbosum genotypes based on single-nucleotide polymorphism (SNP), insertion/deletion (InDel), structural variations (SV) and copy number variation (CNV) data from genome-level resequencing. It also aimed to determine if the whole-genome resequencing of V. corymbosum varieties with an average of 10× attempted coverage will yield enough data to breed new blueberry varieties with increased desirable fruit qualities. In this regard, the first ~10 Mb from scaffold 22, because it appears as a hotspot of genetic variation for both SNPs and InDels, was analyzed for the presence of genes differentially regulated throughout fruit growth and development.
2. Materials and Methods
2.1. Plant Material
In this study, seven Romanian blueberry varieties, ‘Prod’, ‘Vital’, ‘Azur’, ‘Simultan’, ‘Delicia’, ‘Compact’, and ‘Safir’, and a foreign variety, ‘Bluecrop’, were used. The blueberry varieties were cultivated in the orchard collection of the Horticulture Faculty, University of Agronomic Sciences and Veterinary Medicine of Bucharest, Romania. The origin and several characteristics of the Romanian varieties are presented in Table 1 [34].

Table 1.
Characteristics of Romanian blueberry varieties.
2.2. DNA Extraction
Genomic DNA was extracted from young leaves using the Innupure C16 extraction system (Analitik Jena GmbH, Jena, Germany) and the InnuPREP Plant DNA I kit (Analitik Jena GmbH, Jena, Germany), following the manufacturer’s instructions. Briefly, in a preliminary external lysis step, after grinding it to powder using liquid nitrogen, the plant material was homogenized with the SLS lysis solution, proteinase K, and RNase A solution. Thereafter, the extraction proceeded with the automated DNA extraction. DNA was quantified using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE, USA).
2.3. Sequencing, Computational Data Processing, and Sequencing Analysis
Whole-genome resequencing (WGRS), mapping and genomic variations detection and annotation were performed by Novogene Co., Ltd., Cambridge, UK. Sequencing was conducted using an Illumina platform (NGS). The filtered reads were mapped onto the V. corymbosum ‘Draper’ as a reference genome, downloaded from http://parrot.genomics.cn/gigadb/pub/10.5524/100001_101000/100537/V_corymbosum_genome_v1.0.fasta, accessed on 30 May 2022, using BWA software [35].
SNP and InDel variations were detected with SAMtools software [36] with the ‘mpileup -m 2 -F 0.002 -d 1000’ parameter; SVs were detected using the BreakDancer software [37]; CNVs were detected using the CNVnator software [38] with the ‘-call 100’ parameter. All variations were annotated using ANNOVAR software [39]. To reduce the error rate in the SNP and InDel detection, the results were filtered with the following filter conditions: first, the number of support reads for each SNP/InDel was higher than 4; second, the mapping quality of each SNP/InDel was higher than 20. Structural variations were filtered by removing those with less than 2 supporting pair-end reads.
The SNPs and InDels were located in the following regions within the genomes: upstream (within 1 kb upstream away from transcription start site of the gene), exonic (in the exonic region), intronic (in the intronic region), splicing (in the splicing site, within a 2 bp range of the intron/exon boundary), downstream (within 1 kb downstream away from the transcription termination site of the gene region), upstream/downstream (within the less than 2 kb intergenic region, which is in 1 kb downstream or upstream of the genes), intergenic (within the more than 2 kb intergenic region), and others (in other regions). The exonic regions were further defined as nonsynonymous (single-nucleotide mutation with changes in the amino acid sequence), synonymous (single-nucleotide mutation without changing the amino acid sequence), stop gain (a nonsynonymous SNP that leads to the introduction of a stop codon at the variant site), and stop loss (a nonsynonymous SNP that leads to the removal of the stop codon at the variant site).
2.4. Scaffold 22 Sequence Analysis
The comparison of Draper genome’s scaffold 22 with the V. myrtillus isolate NK2018 v1.0 genome sequence was conducted with the Synteny Viewer tool on the GDV website that used the Tripal Synteny Viewer developed by the Fei Bioinformatics Lab from the Boyce Thompson Institute at Cornell University, and MCScanX software [40].
Expression data from the Draper v1.0 scaffold 22, the first ~10 Mb, were obtained with the Expression Heatmap tool on the GDV website (vaccinium.org), accessed on 11 September 2023, based on transcriptome data published by Colle et al. [41].
Gene location, InterPro, and Go term data were acquired with the ‘Gene and transcript search’ tool for each of the differentially regulated genes in the first ~10 Mb of the Draper v1.0 scaffold 22 sequence.
The analysis of the differentially regulated genes located in the first ~10 Mb of the scaffold 22 sequences of the eight genomes wholly sequenced in the present study was conducted using Genome workbench software, version 3.9.0 [42].
3. Results
3.1. Genome Sequencing Data Analysis
3.1.1. Sequencing Data Quality Control
Seven Romanian blueberry cultivars (‘Prod’, ‘Vital’, ‘Azur’, ‘Simultan’, ‘Delicia’, ‘Compact’, ‘Safir’), and one foreign cultivar (‘Bluecrop’) were sequenced at the genomic level using Illumina technology. For all varieties sequenced, Q30 was over 88%, and the effective rate (ratio of clean data to raw data) was above 99% (Supplementary Table S1).
3.1.2. Mapping with the Reference Genome
The mapping rates, average depth, and percentage of 1 x and 4 x coverage depth are presented in Table 2. The mapping rates, calculated as a ratio of the reference genome assembly mapped reads to the total sequenced clean reads, had values of ~97%, indicating high similarity between the genotypes under study and the reference genome. Average depths varied between 3.24 x for ‘Bluecrop’ and 3.98 x for ‘Vital’. Coverage at least 1× varied between 65.24% for ‘Bluecrop’ and 70.19% for ‘Vital’, whereas coverage at least 4× varied between 19.25% for ‘Simultan’ and 28.61% for ‘Vital’.

Table 2.
Statistics of mapping rate, average depth, and coverage.
3.1.3. SNP Distribution and Mutation Frequency
SNPs were distributed across the genomes, within all regions defined (upstream, exonic, intronic, splicing, downstream, upstream/downstream, intergenic, and others (Supplementary Table S2)). The number of SNPs ranged between 794,931 in ‘Delicia’ and 1,228,096 in ‘Vital’. The highest number of SNPs in all the defined genome regions was observed in ‘Vital’, whereas the lowest number was observed in ‘Delicia’, except for synonymous and non-synonymous exonic SNPs, where the lowest number of SNPs was present in ‘Bluecrop’. The ratio between the number of transitions and the number of transversions (ts/tv) was over 1.9 in all genotypes, with the lowest ratio being observed in ‘Delicia’, 1.956, and the highest, 1.981, in ‘Simultan’.
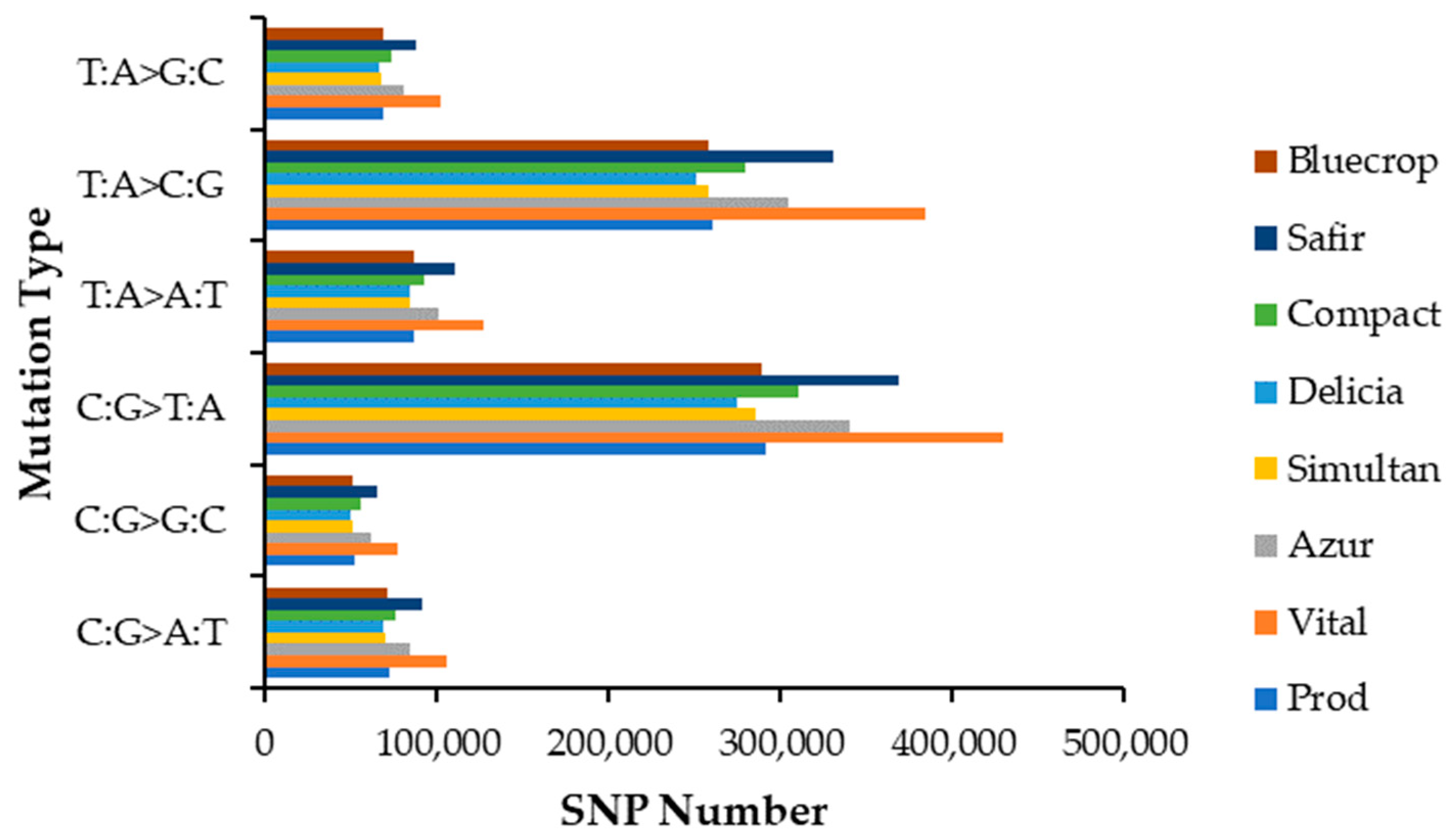
The distribution of SNP mutation types is depicted in Figure 1. When looking at the mutation spectrum, the SNP type C:G>T:A presented the highest mutation fraction for all genotypes studied, followed by T:A>C:G, T:A>A:T, C:G>A:T, T:A>G:C and C:G>G:C.
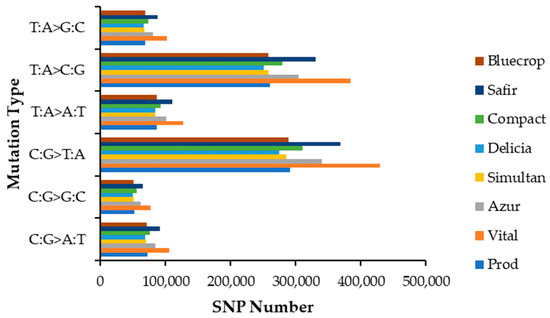
Figure 1.
SNP mutation type distribution.
3.1.4. Insertions/Deletions Distribution
Similar to the SNPs, InDels were distributed in all defined regions of the genomes (Supplementary Table S3). Like the SNPs, the highest number of InDels was observed in the ‘Vital’ variety for all defined regions. However, the lowest numbers of InDels varied among the genotypes, depending on the defined region. For instance, the ‘Bluecrop’ variety had the lowest number of InDels within the exonic regions, except for stop loss, where the lowest number of InDels was observed in the ‘Simultan’ variety. Outside of exonic regions, ‘Delicia’ presented the lowest number of InDels, except for the splicing region, where the lowest number of InDels was again observed in the ‘Simultan’ variety.
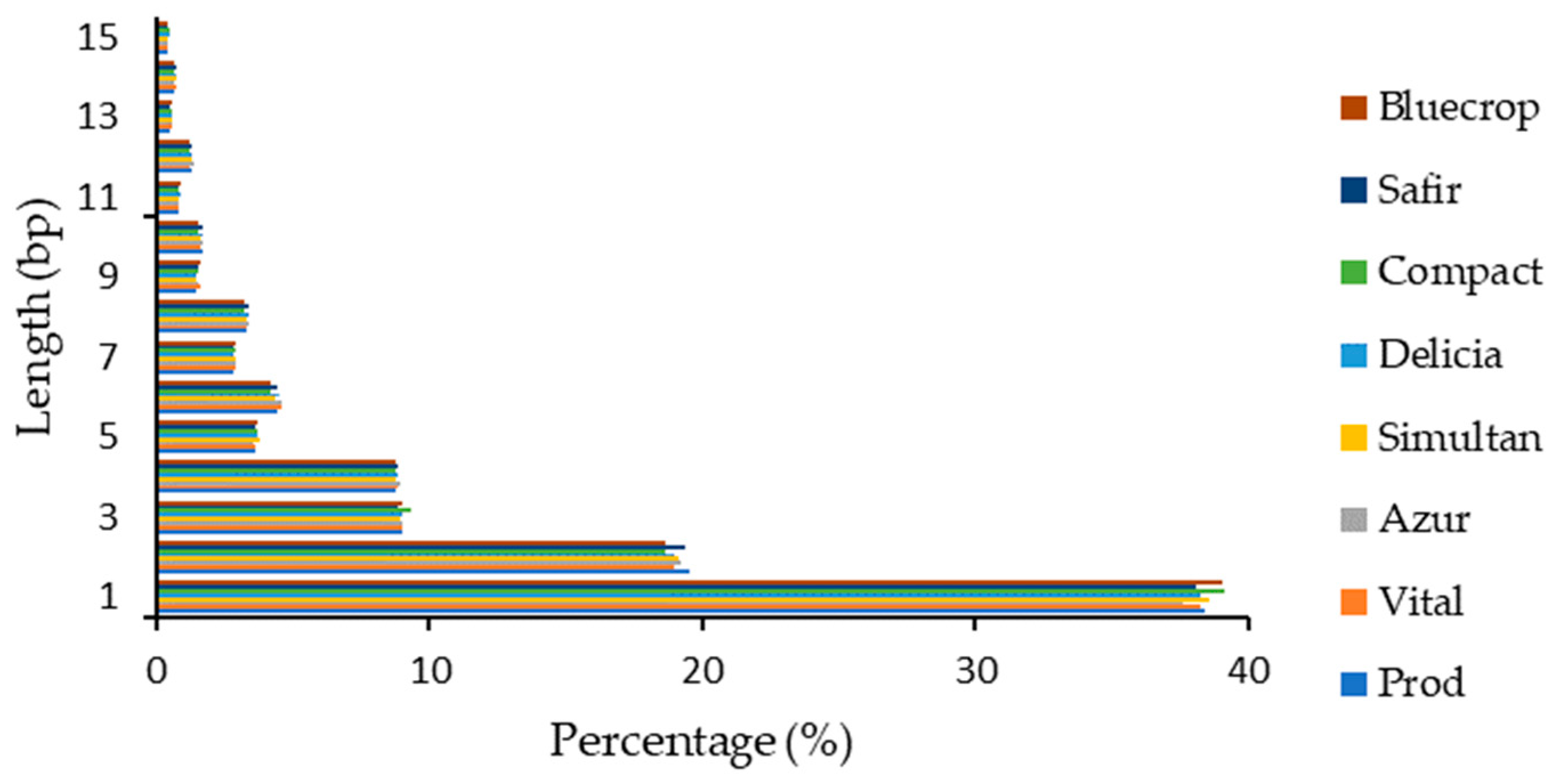
Within the coding sequences, the highest number of InDels was noted for the 1 bp insertion/deletion (~27%) and the decrease was inversely proportional with the InDel length (Figure 2). As expected, percentages of sequences with multiples of 3 bp were higher than the other sequences, since they do not cause frameshifts, and consequently, radically change the sequence of the translated polypeptide. The percentages of InDels with lengths above 15 bp were below 1%.
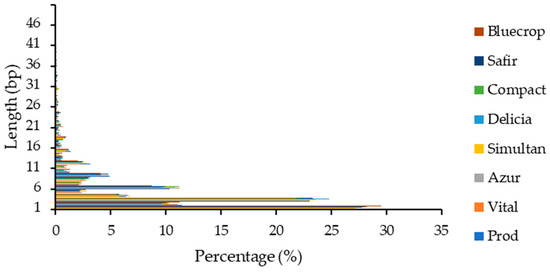
Figure 2.
InDel length distribution within coding sequences.
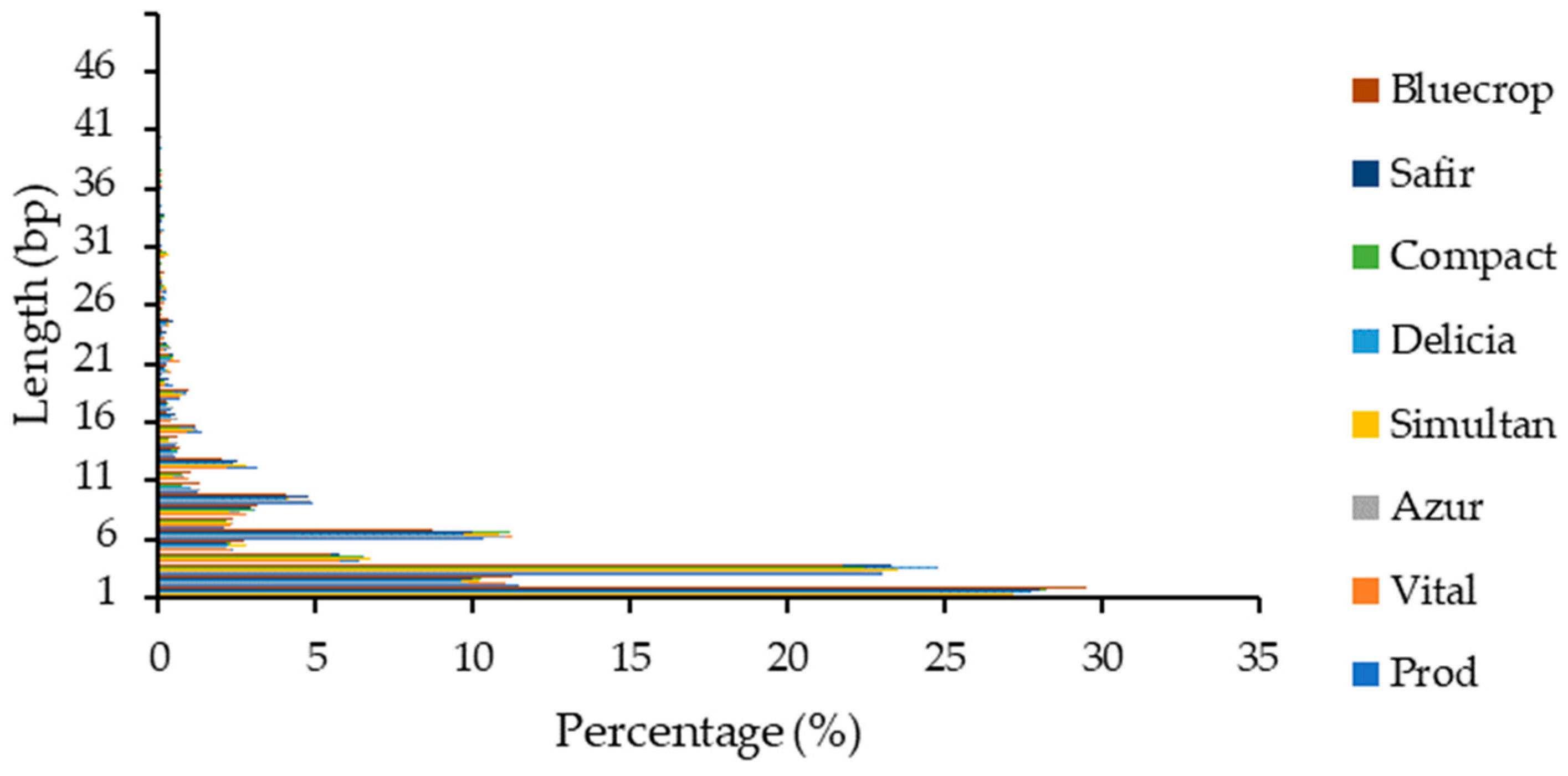
The highest number of InDels observed in the genomes was again noticed for the 1 bp insertion/deletion (~38%) and decreased similarly with the InDels within the coding sequence (Figure 3).
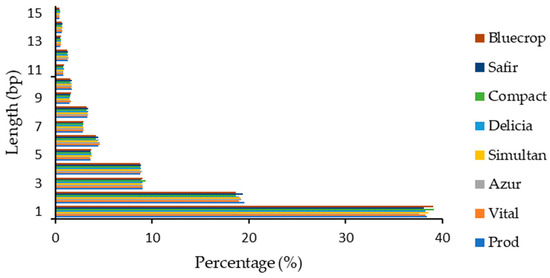
Figure 3.
InDel length distribution within the genomes.
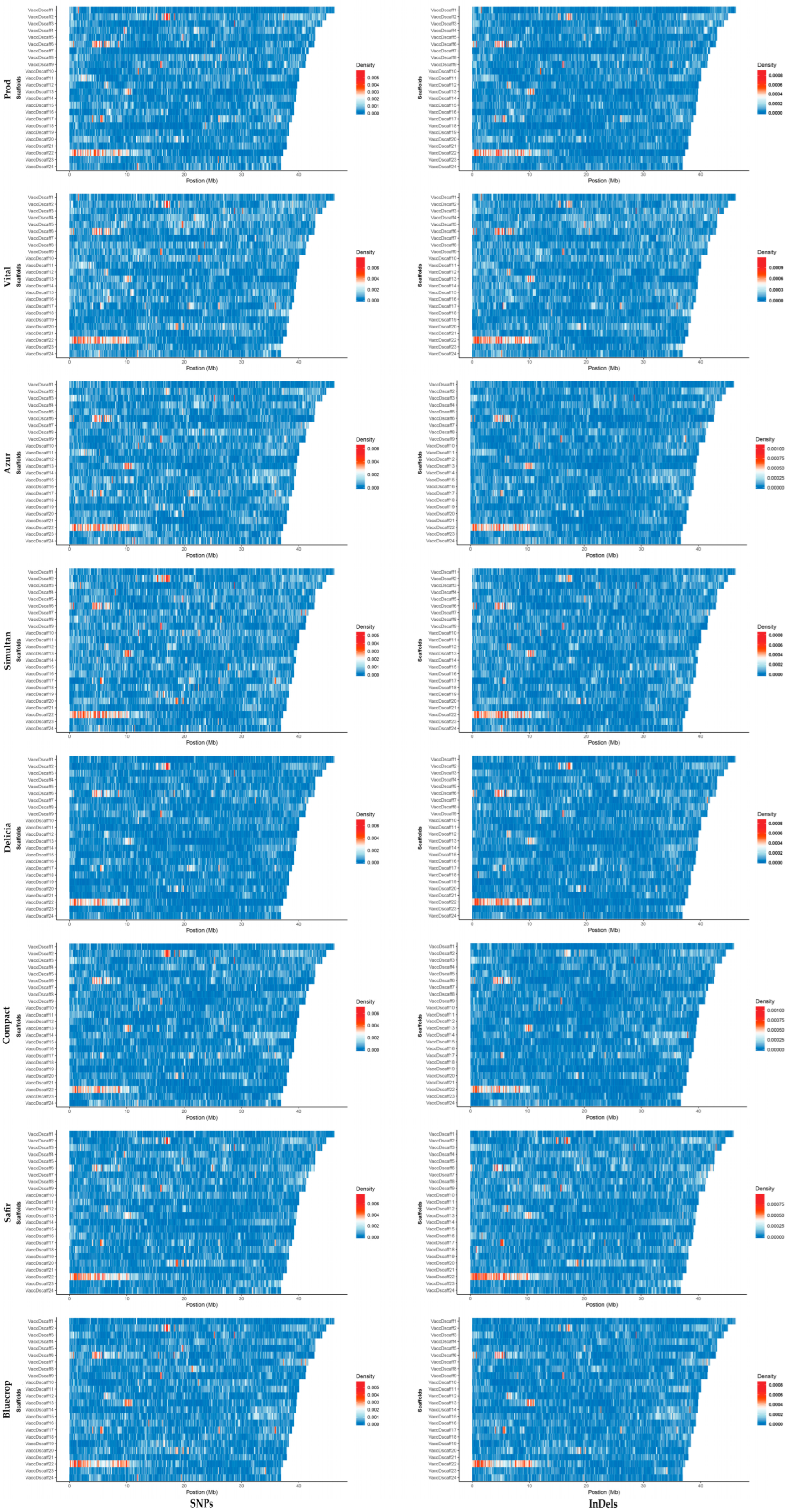
When looking at the SNP and InDel densities within the first 24 scaffolds, hotspots were observed within the same region of the scaffold for all genotypes studied (Figure 4). The widest hotspot region was observed in the first 10 Mb of the scaffold 22.
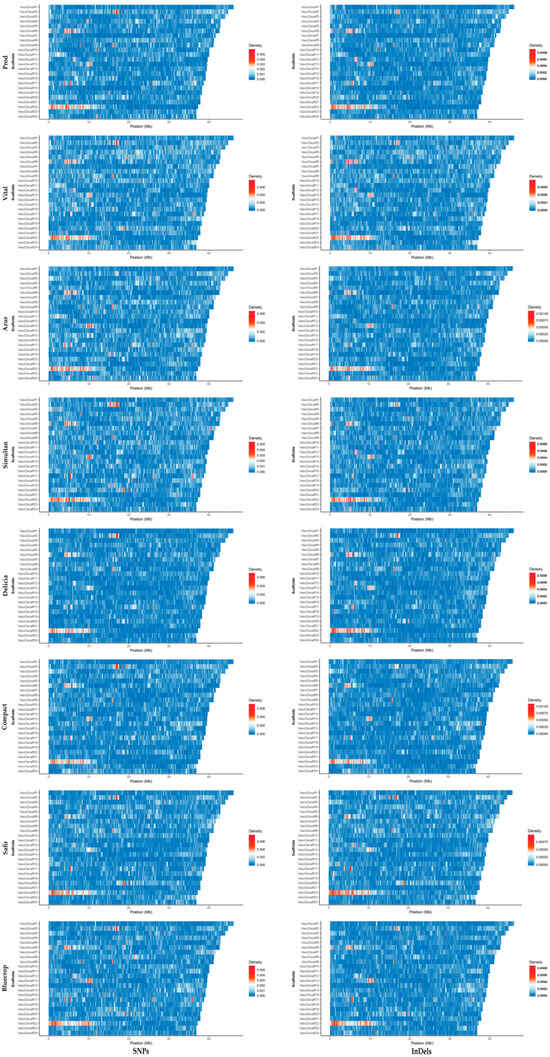
Figure 4.
SNP and InDel densities within the scaffolds 1–24 (VaccDscaff1-VaccDscaff24). For each variety, on the y axis the scaffolds are aligned in a size-decreasing order, with scaffold 1 on top, and scaffold 24 on bottom.
3.1.5. Structural Variations Detection and Annotation
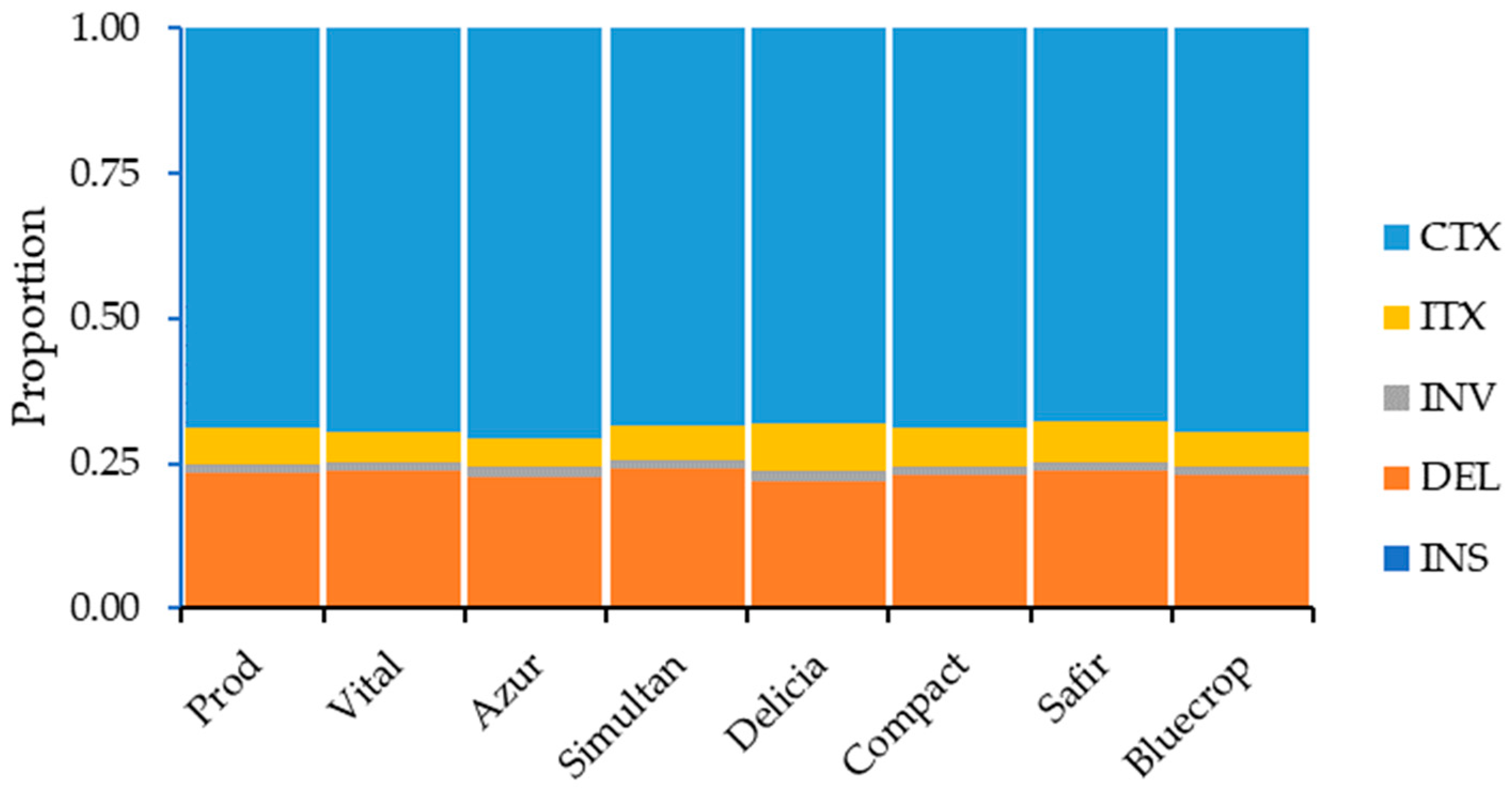
The distribution of the structural variations within the genomes is presented in Supplementary Table S4. Except for downstream, upstream/downstream, and intergenic regions (in the ‘Safir’ variety), the highest number of SVs in the genome regions was noted for the ‘Vital’ variety, whereas the lowest number of SVs was observed for the ‘Delicia’ variety. When considering SV types, the ‘Vital’ variety had the highest number of deletions, inversions, and inter-chromosomal translocations, while the highest number of intra-chromosomal translocations was observed for the ‘Safir’ variety. On the other hand, the lowest number of deletions, inversions, and inter-chromosomal translocations was noticed for the ‘Delicia’ variety and the lowest number of intra-chromosomal translocations was observed for the ‘Simultan’ variety. When looking at the SV types, the highest percentage of SVs was observed for the inter-chromosomal translocations, followed by deletions, inter-chromosomal translocations, inversions, and insertions (Figure 5).
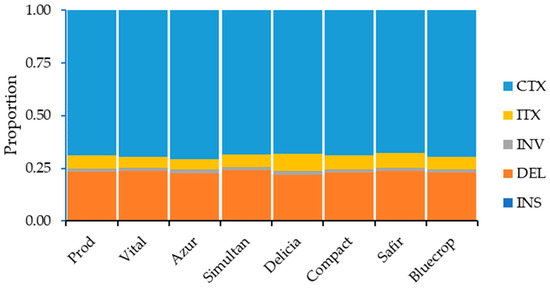
Figure 5.
SV type distribution within the genome.
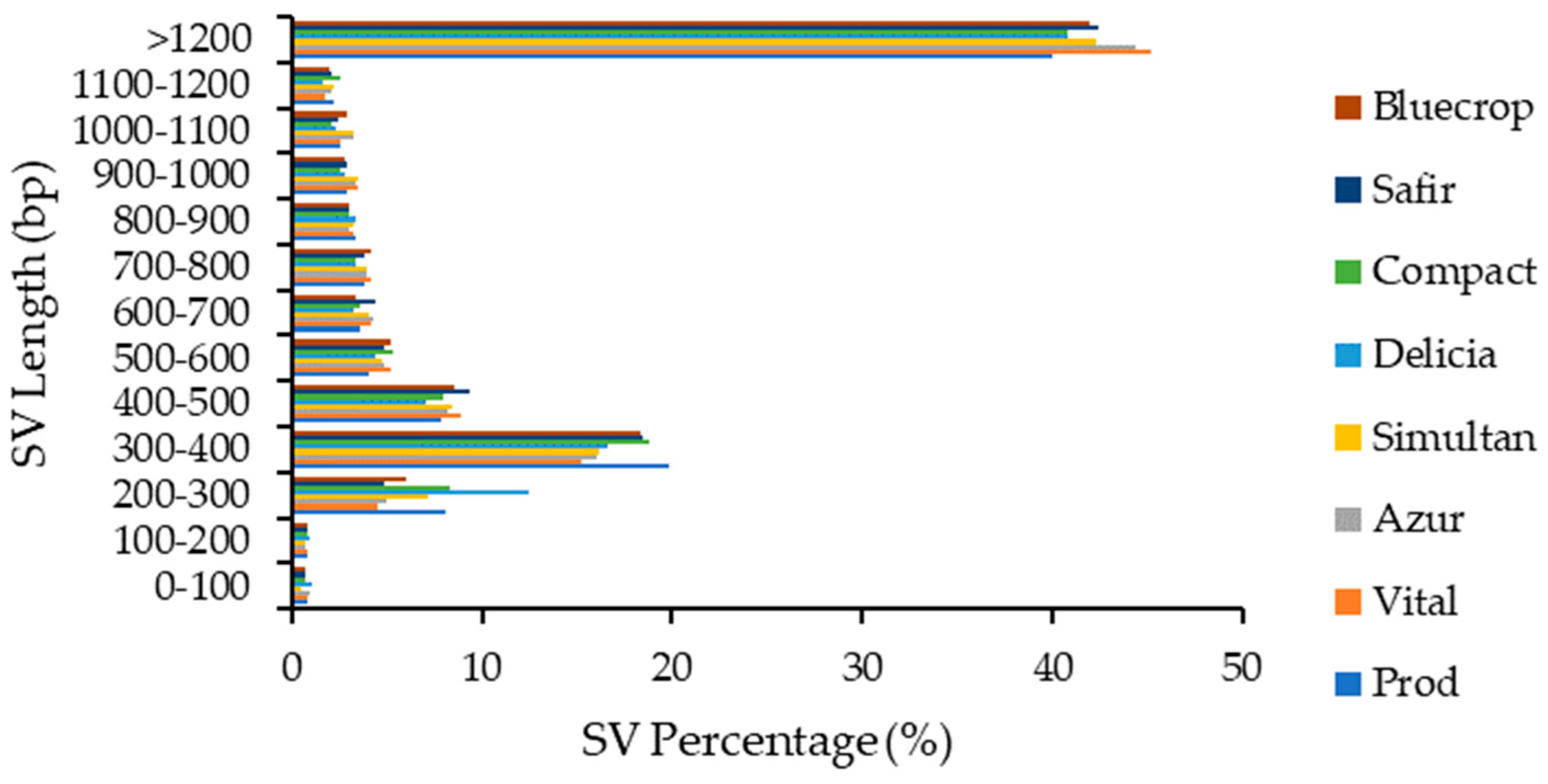
Structural variations with lengths above 1200 bp accounted for more than 40% of total SVs and were followed by SVs with lengths of 200–300 bp (~17%). The lowest percentages were observed for SVs with lengths of 0–200 bp (~0.6–1%) (Figure 6).
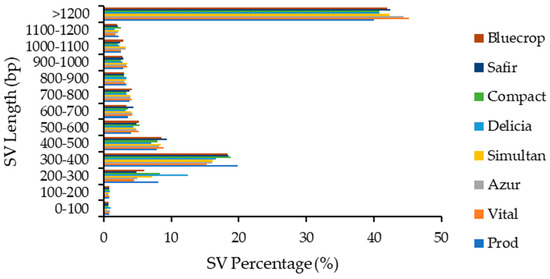
Figure 6.
Structural variations length distribution within the genomes.
3.1.6. Copy Number Variations Detection and Annotation
The highest number of CNVs was observed for the ‘Bluecrop’ variety in the intronic, downstream, and upstream/downstream genomic regions, for the ‘Vital’ variety in the exonic and intergenic regions of the genomes, and in the ‘Safir’ variety in the upstream genomic regions. The number of duplications, between 20,878 (‘Delicia’) and 52,280 (‘Vital’), was far higher than the number of deletions, with no deletions observed for the ‘Prod’, ‘Vital’, ‘Simultan’, ‘Safir’ and ‘Bluecrop’ varieties, and 9 deletions for ‘Compact’ variety, 16 for ‘Delicia’ variety and 29 for the ‘Azur’ variety (Supplementary Table S5).
3.2. Scaffold 22 Sequence Analysis
The block bivcdB0989 contains the scaffold 22 hotspot (V. corymbosum cv. Draper v1.0 genome sequence, VaccDscaff22_Vaccinium_corymbosum_Draper_v1: 7771–10,416,940) and matches the V. myrtillus isolate NK2018 v1.0 genome sequence Chr12_Vaccinium_myrtillus_NK2018_v1: 27,664–9,425,466. The genes present in both genomes within this block are shown in Supplementary Table S6, a number of 673 genes being present in the V. corymbosum sequence.
Using the Expression Heatmap tool on the GDV website (vaccinium.org) accessed on 11 September 2023, the expression data were obtained for these genes under the V. corymbosum cv. ‘Draper’ tissue during fruit development, FPKM analysis, with expression data available for 459 genes (Supplementary Table S7). Data available on the GDV website were submitted by Colle et al. [41] and were collected from the following tissue samples: flower bud, flower at anthesis, leaf day, leaf night, young shoot, leaves treated with methyl jasmonate for 1 h, 8 h and 24 h, fruit development (fruit at petal fall, green fruit, pink fruit, ripe fruit), and salt-treated and untreated roots.
For the fruit development data, out of the 459 genes, 11 were upregulated more than 10-fold during fruit development (Table 3, Supplementary Table S8) and 50 were downregulated more than 10-fold (Table 4, Supplementary Table S9).

Table 3.
Upregulated genes during fruit development.

Table 4.
Downregulated genes during fruit development.
When looking at the functions of the encoded upregulated genes, three have a protein-binding function, one has a heme-binding function, one has a nuclear-acid-binding function, and two have a serine-type carboxypeptidase activity. The rest of these genes do not have a GO (gene ontology) term.
Next, the sequences of these genes were analyzed in the sequenced blueberry varieties. Five of the genes, augustus-gene-4.24, processed-gene-12.3, augustus-gene-56.35, augustus-gene-89.30, and augustus-gene-89.31, were not completely covered by sequencing for any of the varieties analyzed. Augustus-gene-93.19 had full coverage for varieties ‘Vital’, ‘Azur’, ‘Delicia’, ‘Compact’, ‘Safir’, and ‘Bluecrop’, whereas augustus-gene-104.25 had full coverage only for the ‘Vital’ variety. Interestingly, the processed-gene-45.12, which was fully covered by sequencing for all varieties, had only one deletion for ‘Safir’ variety, a three-nucleotide sequence, GAC, at an SSR site: (GAC)3 instead of (GAC)4, as opposed to the rest of the genes, which had numerous SNPs, including ones not fully covered by sequencing. Processed-gene-0.20, which was also fully sequenced on all varieties, presented high variation within the first 60 nucleotides in the CDS sequence, between varieties, and within the variety at an SSR locus: (CCA)3 to (CCA)9, resulting in a variation in the number of histidine residues translated.
In the case of the downregulated genes, out of the 50, 21 genes were below 1 x coverage by sequencing, 5 genes had full coverage for all varieties studied, and the rest had full coverage for at least one variety. The putative functions of the downregulated genes varied from nucleic-acid-binding to protein-binding, ATP-binding, iron-ion-binding functions, etc., (Supplementary Table S9).
The following five downregulated genes were fully covered by sequencing for all varieties analyzed: VaccDscaff22-augustus-gene-23.30, VaccDscaff22-processed-gene-49.1, VaccDscaff22-processed-gene-63.2, VaccDscaff22-augustus-gene-71.34, and VaccDscaff22-augustus-gene-76.24.
VaccDscaff22-processed-gene-8.5 and VaccDscaff22-processed-gene-49.1 genes are both encoding proteins with glycosyl transferase activity. VaccDscaff22-augustus-gene-23.30 encodes a nucleic acid binding protein. VaccDscaff22-processed-gene-63.2 encodes a protein with a putative role in protein ubiquitination (RING/U-box/RING-Ubox_PUB-like protein). VaccDscaff22-augustus-gene-71.34 gene is encoding a putative protein-binding protein and contains a glutathione S-transferase (GST) domain. VaccDscaff22-augustus-gene-76.24 gene does not have a GO term, but InterPro predicts that it encodes a bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin with a structural domain consisting of 4-helices with a folded-leaf topology and forms a right-handed superhelix domain present in plant lipid-transfer proteins, proteinase/alpha-amylase inhibitors, and seed storage proteins [43]. In addition to the numerous SNPs present in this gene for all genotypes studied here, the ‘Delicia’ variety has a deletion of five nucleotides (As) within an intron.
4. Discussion
Next-generation sequencing techniques have made it possible to generate a tremendous amount of data from a single sequencing experiment that can be used in countless future studies. At the moment, the V. corymbosum genome has been sequenced at the scaffold level. The first to be wholly sequenced by shotgun sequencing was the diploid variety W8520, resulting in a draft genome sequence that was used to mine SSRs [44]. Thereafter, the highbush blueberry tetraploid variety, Draper, was sequenced at the scaffold level [41]. Lastly, data from the Vaccinium Pangenome Project were made available on the Genomic Database for the Vaccinium website (GDV, https://www.vaccinium.org/, accessed on 11 September 2023), on 22 V. corymbosum sequenced genomes at the scaffold level [45]. The present study offers an analysis of variability in eight V. corymbosum genomes, investigating SNPs, InDels, SVs, and CNVs. It also ‘zooms in’ to one of the SNP and InDel hotspots, the first ~10 Mb of the 22nd scaffold, to identify potential genes of interest involved in fruit development that present variability at the nucleotide level among the varieties presented here.
When looking at the SNP data, predictably, the highest number of SNP types were transitions (C:G>T:A and T:A>C:G), a phenomenon observed before [46], due to mutations resulting from the deamination of methylated cytosine residues [47]. Although the effective number of SNPs and InDels originate from just ~19–28% of the genome, it still indicates high numbers of mutations in regions of interest such as exonic regions. For instance, the number of non-synonymous SNPs varied between 29,881 for the ‘Bluecrop’ variety and 45,914 for the ‘Vital’ variety. Structural variations (SVs) varied between ~9700 and ~15,500 per genotype; however, it is possible that the number is higher, as high-throughput short-read sequencing does not characterize easily repetitive regions, and therefore might miss structural variations [48].
Both SNPs and InDels were detected in all defined genome regions, with hotspots with a higher density of SNPs and InDels, as observed before in another study [46]. These hotspots are usually located towards the ends of the chromosomes, most probably due to their higher recombination frequency [49,50].
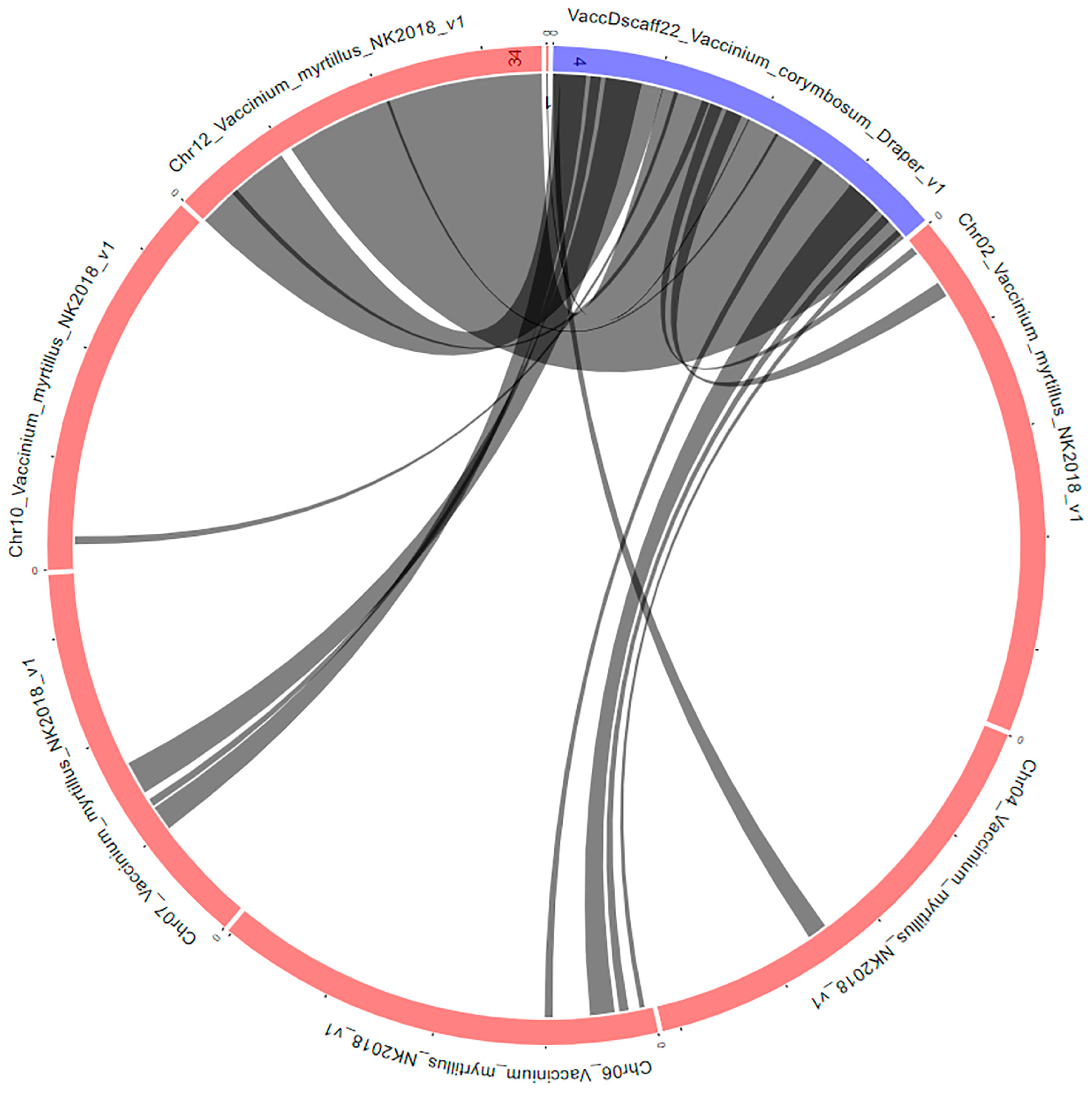
To date, V. corymbosum genome has been wholly sequenced at the scaffold level [44] and annotated by Gupta et al. [51]. In the present study, the large hotspot identified on scaffold 22, within the first 10 Mb (Figure 4), was chosen to be studied in more detail. Using the Synteny Viewer tool on the GDV website (vaccinium.org, accessed on 11 September 2023), the scaffold 22 from the Draper genome, sequenced in 2019 by Colle et al. [41], was compared to Vaccinium myrtillus L. isolate NK2018 v1.0 genome sequence. V. myrtillus has been wholly sequenced at the chromosome level [52], and the comparison yielded the location of scaffold 22 at the chromosome level for V. corymbosum (Figure 7). The comparison identified chromosome 12 as the putative location of this scaffold.
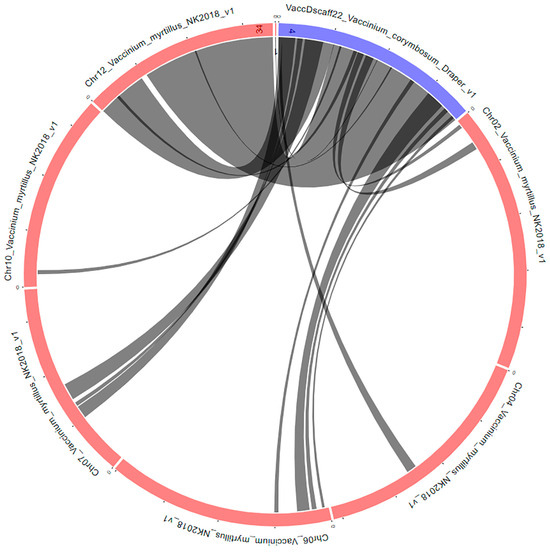
Figure 7.
Syntenic blocks between the V. corymbosum scaffold 22 and V. myrtillus genomes. The circular layout was obtained by using the Tripal Synteny Viewer from the GDV (https://www.vaccinium.org/ accessed on 11 September 2023) website.
The analysis of the hotspot region from the scaffold 22 revealed which of the differentially regulated genes during fruit growth and development presented differences in the varieties sequenced in the present study. These genes should be studied in the future to check if they could be linked to various traits related to fruit quality.
From the 11 upregulated genes, one gene wholly covered by sequencing for all varieties studied, VaccDscaff22-processed-gene-0.20, that encodes a S-adenosyl-l-methionine-dependent methyltransferase (SAM-MTase), contains SSR sites within its coding region. S-adenosyl-l-methionine-dependent methyltransferases catalyze various steps in ethylene and polyamine biosynthesis pathways, with both having roles in fruit ripening [53]. For instance, in a previous study looking for the genetic basis of several blueberry fruit traits, a gene encoding a SAM-MTase from the scaffold00012 (CUFF.1480.1) was linked to fruit firmness [54]. Another study characterized a SAM-MTase that catalyzes the biosynthesis of a key aroma compound in strawberry fruits [55]. If differences in this gene at the nucleotide level prove to translate into notable phenotypic differences in fruits, a molecular marker developed based on the SSR site present at the beginning of the coding sequence may be used to select for the desirable trait in breeding programs.
Of the 50 downregulated genes, 5 genes were covered by sequencing for all varieties studied here, and they encode two glycosyl transferases, a RING (Really Interesting New Gene) finger protein, a GST protein, and a bifunctional inhibitor/lipid-transfer protein/seed storage 2S albumin.
In plants, glycosyl transferases are involved in cell wall polysaccharides biosynthesis, in adding N-linked glycans to glycoproteins, but also in flavonoids biosynthesis [56]. It stands to reason that one or both genes encoding glycosyl transferases (VaccDscaff22-processed-gene-8.5 and VaccDscaff22-processed-gene-49.1) may have a role in anthocyanin biosynthesis during fruit ripening.
Plant RING finger proteins have been shown to play a role in stress resistance, both biotic and abiotic, signal transduction, and plant growth and development, including fruit development [57].
Some GST proteins have been proven to be involved in fruit development in multiple species, such as peach [58,59], pear [60,61], banana [62], apple [63], and strawberry [64].
Even though many of the genes differentially regulated throughout fruit growth and development were below at least 1× coverage by sequencing, two upregulated and five downregulated genes that vary at the nucleotide level were covered at least 1× by sequencing for all the genotypes.
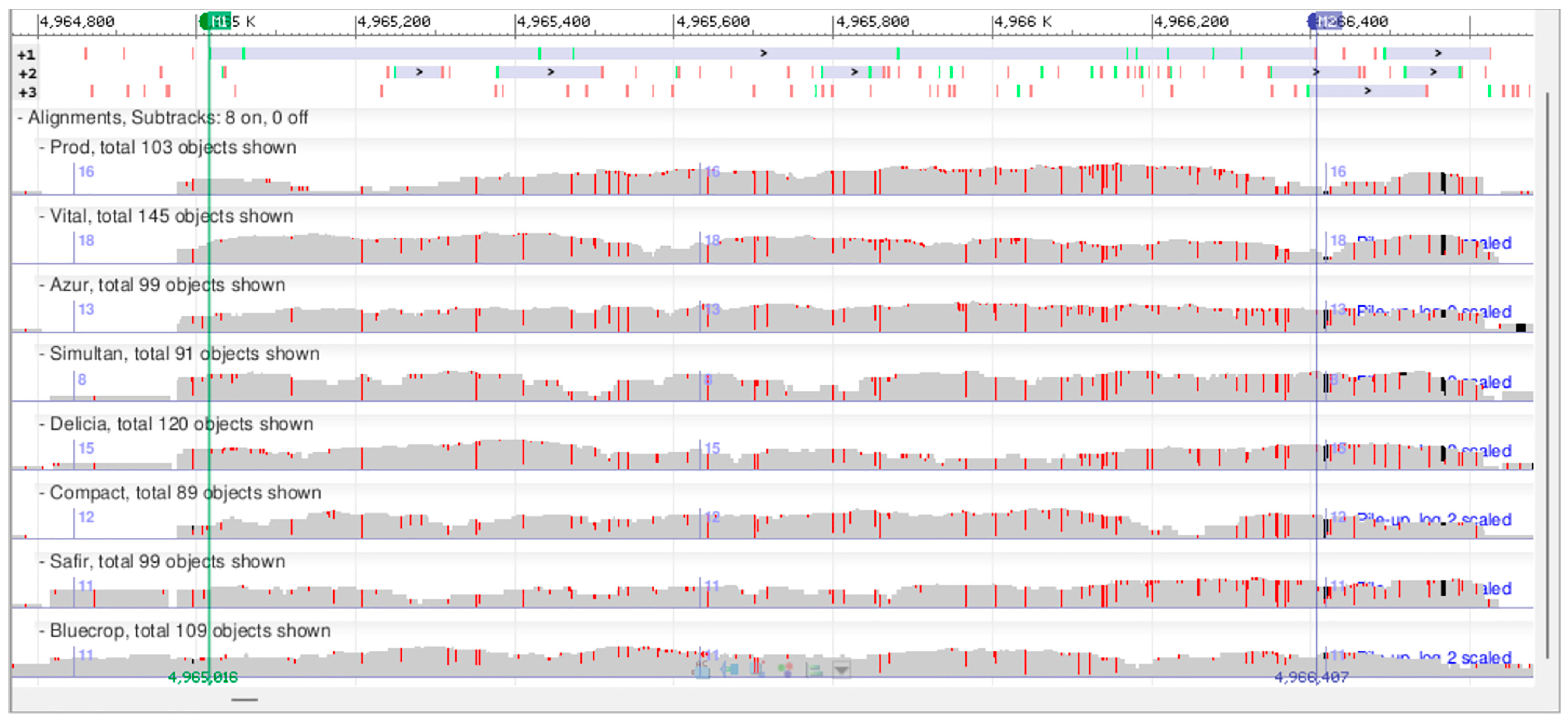
To further exemplify the potential of this type of preliminary low-coverage sequencing study, we chose one of the differentially expressed genes, VaccDscaff22-processed-gene-49.1, to be analyzed in further detail (Supplementary Table S10). This gene putatively encodes a glucosyl/glucuronosyl transferase. The minimum read depth, used as a filtering criterion for SNP detection, may be as low as three [65]. Here, a minimum of five supporting reads was used to detect an SNP. Within this gene’s coding region, 44 SNPs were identified, out of which 28 SNPs are silent and 16 SNPs (either homozygous or heterozygous) result in missense mutations (Figure 8). For each of the missense mutations, there are at least two varieties that are different at the SNP position, except for the SNP at the position 4,966,356, where only two varieties present heterozygous SNPs. In a subsequent study, the expression of this gene will be analyzed throughout fruit ripening in the genotypes studied here to see if there are any correlations between the DNA sequences, mRNA expression levels, and fruit traits of interest.
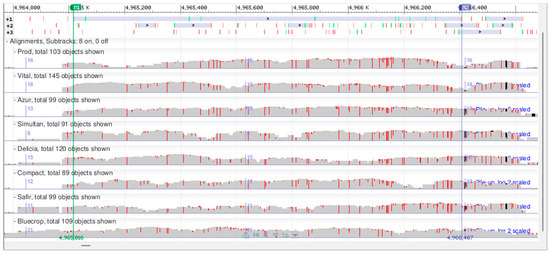
Figure 8.
Snapshot of alignment of the VaccDscaff22-processed-gene-49.1 gene DNA sequences for the eight V. corymbosum genotypes in the Genome Workbench software.
Furthermore, many of the genes analyzed were fully covered by sequencing for at least two genotypes, making a comparison at the nucleotide level possible, as well as the identification of SNPs and InDels. These genes are encoding for nucleic-acid-binding proteins (Zn finger protein, MYB-like DNA-binding protein, homeobox-leucine zipper protein, RNA-dependent RNA polymerase), protein-binding proteins (leucine-rich repeats (LRR) proteins, pentatricopeptide repeat protein, ARM repeat protein, iron-ion-binding proteins, polygalacturonase, hydrolase, and chloroplastic protein).
Zn finger proteins were shown to play a role in the regulation of fruit development in tomato [66,67], Chinese pear [68], cucumber [69], and banana [62], etc. MYB (myeloblastosis) transcription factors were also shown to play roles in fruit development in several species such as blueberry [70], tomato [71,72], Chinese pear [73], apple [74,75], and strawberry [76], etc. Likewise, leucine zipper transcription factors are associated with fruit morphogenesis in tomato, and with intense fruit cell division in grapes [77].
In addition to stress response, LRR proteins are also involved in fruit development in apple, strawberry [78], and peanuts [79]. Pentatricopeptide repeat (PPR) proteins play roles in fruit development in watermelon [80], kiwifruit [81], pepper [82,83], and maize [84], etc. The roles of polygalacturonases and hydrolases in fruit development began to be investigated more than two decades ago [85,86,87,88,89]. Therefore, full coverage for at least two genotypes for a differentially regulated gene revealed enough differences to choose candidate fruit development genes for further study. For genes that may prove to be of interest and have small gaps, Sanger sequencing using specific primers bordering the gaps may be an alternative solution to complete the missing data.
Therefore, all these data demonstrate that even at just 10× attempted average coverage by sequencing, this study offers a treasure trove of information, providing a plethora of preliminary data to be analysed and raising more questions to be answered in future studies.
5. Conclusions
Consumers preferences for blueberry fruit traits such as fruit color, size, firmness, taste, and flavor shape the goals of breeding programs. Whole-genome resequencing of the genotypes analyzed in the present study led to the identification of multiple genes putatively involved in fruit ripening that are showing high variability among the varieties, in one hotspot, and in one scaffold, and one criterion (fruit growth and development). If this criterion is applied in an in-depth study at the level of the whole genome, additional genes that vary among the genotypes are bound to be found. Moreover, additional criteria can be applied if looking for other traits of interest, such as resistance to stress, either biotic or abiotic. In this respect, despite the low–medium coverage depth of sequencing, the results presented in the current work indicate that a 10× attempted coverage depth is useful if sequencing a high number of genotypes of V. corymbosum. Thus, with a relatively low budget, this low-coverage sequencing offers reliable preliminary data for further use in subsequent studies engaged in discovering novel genes linked to useful traits. The genes identified here will be analyzed further in future studies for their effect on fruit-quality traits. If these genes are found to be linked to desirable fruit traits, these should definitely be selected in blueberry breeding programs.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae10020157/s1. Table S1: Quality of sequenced data; Table S2: SNP distribution across the genomes; Table S3: InDel distribution across the genomes; Table S4: SVs distribution across the genomes; Table S5: CNVs distribution across the genomes; Table S6. Genes present in both V. myrtillus isolate NK2018 v1.0 genome sequence Chr12_Vaccinium_myrtillus_NK2018_v1 and V. corymbosum VaccDscaff22_Vaccinium_corymbosum_Draper_v1: 7771—10416940); Table S7. Expression data for V. corymbosum cv. ‘Draper’ genes in various tissues during fruit development, FPKM analysis; Table S8. Upregulated genes in fruit development; Table S9. Downregulated genes in fruit development; Table S10. Sequence analysis of VaccDscaff22-processed-gene-49.1 gene.
Author Contributions
Conceptualization, C.A.M. and M.I.; methodology, C.A.M. and M.I.; software, M.I.; writing—original draft preparation, M.I.; writing—review and editing, C.A.M., M.I., L.B. and A.A.; project administration, C.A.M.; funding acquisition, C.A.M. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a grant of the University of Agronomic Sciences and Veterinary Medicine of Bucharest, project number 2021-0030, acronym BlueBerryGene, within IPC 2021.
Data Availability Statement
Data are contained within the article.
Acknowledgments
We thank Dan Popescu for taking care of and providing the plant material used in this study. We also thank Maria Ojog for helping with last minute changes in bioinformatics analysis.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Bassil, L.J.R.; James, F.; Hancock, N.V. Blueberry. In Genetics, Genomics and Breeding of Berries; CRC Press: Boca Raton, FL, USA, 2011; ISBN 978-0-429-06248-3. [Google Scholar]
- Lobos, G.A.; Hancock, J.F. Breeding Blueberries for a Changing Global Environment: A Review. Front. Plant Sci. 2015, 6, 782. [Google Scholar] [CrossRef]
- Asănică, A.; Delian, E.; Tudor, V.; Teodorescu, R.I. Physiological Activity of Some Blueberry Varieties in Protected and Outside Conditions. AgroLife Sci. J. 2017, 6, 31–39. [Google Scholar]
- Mladin, P.; Mladin, G.; Ancu, I.; Chitu, V. Results of the Blueberry Breeding at the Research Institute for Fruit Growing Pitești. Bull. UASVM Hortic. 2008, 65, 300–303. [Google Scholar]
- Popescu, D.; Asănică, A.; Tudor, V. Start of the Blueberry Breeding Program at the University of Agronomic Sciences and Veterinary Medicine of Bucharest. Sci. Pap. Ser. B Hortic. 2021, LXV, 50–55. [Google Scholar]
- Asănică, A. Sensorial Evaluation of 26 Highbush Blueberry Varieties in Romania. Sci. Pap. Ser. B Hortic. 2018, 62, 181–186. [Google Scholar]
- Williams, C. A Blueberry a Day…. New Sci. 2016, 231, 26–31. [Google Scholar] [CrossRef]
- Krishna, P.; Pandey, G.; Thomas, R.; Parks, S. Improving Blueberry Fruit Nutritional Quality through Physiological and Genetic Interventions: A Review of Current Research and Future Directions. Antioxidants 2023, 12, 810. [Google Scholar] [CrossRef]
- Bouyahya, A.; Omari, N.E.; EL Hachlafi, N.; Jemly, M.E.; Hakkour, M.; Balahbib, A.; El Menyiy, N.; Bakrim, S.; Naceiri Mrabti, H.; Khouchlaa, A.; et al. Chemical Compounds of Berry-Derived Polyphenols and Their Effects on Gut Microbiota, Inflammation, and Cancer. Molecules 2022, 27, 3286. [Google Scholar] [CrossRef]
- Wang, Y.; Gallegos, J.L.; Haskell-Ramsay, C.; Lodge, J.K. Effects of Chronic Consumption of Specific Fruit (Berries, Citrus and Cherries) on CVD Risk Factors: A Systematic Review and Meta-Analysis of Randomised Controlled Trials. Eur. J. Nutr. 2021, 60, 615–639. [Google Scholar] [CrossRef]
- Hameed, A.; Galli, M.; Adamska-Patruno, E.; Krętowski, A.; Ciborowski, M. Select Polyphenol-Rich Berry Consumption to Defer or Deter Diabetes and Diabetes-Related Complications. Nutrients 2020, 12, 2538. [Google Scholar] [CrossRef]
- Afrin, S.; Giampieri, F.; Gasparrini, M.; Forbes-Hernandez, T.Y.; Varela-López, A.; Quiles, J.L.; Mezzetti, B.; Battino, M. Chemopreventive and Therapeutic Effects of Edible Berries: A Focus on Colon Cancer Prevention and Treatment. Molecules 2016, 21, 169. [Google Scholar] [CrossRef] [PubMed]
- Tamada, T. Current Trends of Blueberry Culture in Japan. Acta Hortic. 2009, 810, 109–116. [Google Scholar] [CrossRef]
- Patel, N. Recent Trends in Australasian Blueberry Production. Acta Hortic. 1997, 446, 53–58. [Google Scholar] [CrossRef]
- Pliszka, K. Overview on Vaccinium Production in Europe. Acta Hortic. 1997, 446, 49–52. [Google Scholar] [CrossRef]
- Asănică, A.; Bădescu, A.; Bădescu, C. Blueberries in Romania: Past, Present and Future Perspective. Acta Hortic. 2017, 1180, 293–298. [Google Scholar] [CrossRef]
- Varshney, R.K.; Nayak, S.N.; May, G.D.; Jackson, S.A. Next-Generation Sequencing Technologies and Their Implications for Crop Genetics and Breeding. Trends Biotechnol. 2009, 27, 522–530. [Google Scholar] [CrossRef] [PubMed]
- Ray, S.; Satya, P. Next Generation Sequencing Technologies for next Generation Plant Breeding. Front. Plant Sci. 2014, 5, 367. [Google Scholar] [CrossRef]
- Barabaschi, D.; Tondelli, A.; Desiderio, F.; Volante, A.; Vaccino, P.; Valè, G.; Cattivelli, L. Next Generation Breeding. Plant Sci. 2016, 242, 3–13. [Google Scholar] [CrossRef]
- Hu, H.; Scheben, A.; Edwards, D. Advances in Integrating Genomics and Bioinformatics in the Plant Breeding Pipeline. Agriculture 2018, 8, 75. [Google Scholar] [CrossRef]
- Kilian, B.; Graner, A. NGS Technologies for Analyzing Germplasm Diversity in Genebanks. Brief. Funct. Genom. 2012, 11, 38–50. [Google Scholar] [CrossRef]
- Kumawat, S.; Raturi, G.; Dhiman, P.; Sudhakarn, S.; Rajora, N.; Thakral, V.; Yadav, H.; Padalkar, G.; Sharma, Y.; Rachappanavar, V.; et al. Opportunity and Challenges for Whole-Genome Resequencing-Based Genotyping in Plants. In Genotyping by Sequencing for Crop Improvement; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2022; pp. 38–51. ISBN 978-1-119-74568-6. [Google Scholar]
- Xu, X.; Bai, G. Whole-Genome Resequencing: Changing the Paradigms of SNP Detection, Molecular Mapping and Gene Discovery. Mol. Breed. 2015, 35, 33. [Google Scholar] [CrossRef]
- Pasaniuc, B.; Rohland, N.; McLaren, P.J.; Garimella, K.; Zaitlen, N.; Li, H.; Gupta, N.; Neale, B.M.; Daly, M.J.; Sklar, P.; et al. Extremely Low-Coverage Sequencing and Imputation Increases Power for Genome-Wide Association Studies. Nat. Genet. 2012, 44, 631–635. [Google Scholar] [CrossRef]
- Fairley, S.; Lowy-Gallego, E.; Perry, E.; Flicek, P. The International Genome Sample Resource (IGSR) Collection of Open Human Genomic Variation Resources. Nucleic Acids Res. 2020, 48, D941–D947. [Google Scholar] [CrossRef]
- Kersey, P.J. Plant Genome Sequences: Past, Present, Future. Curr. Opin. Plant Biol. 2019, 48, 1–8. [Google Scholar] [CrossRef]
- Straub, S.C.; Fishbein, M.; Livshultz, T.; Foster, Z.; Parks, M.; Weitemier, K.; Cronn, R.C.; Liston, A. Building a Model: Developing Genomic Resources for Common Milkweed (Asclepias syriaca) with Low Coverage Genome Sequencing. BMC Genom. 2011, 12, 211. [Google Scholar] [CrossRef]
- Lee, H.-O.; Choi, J.-W.; Baek, J.-H.; Oh, J.-H.; Lee, S.-C.; Kim, C.-K. Assembly of the Mitochondrial Genome in the Campanulaceae Family Using Illumina Low-Coverage Sequencing. Genes 2018, 9, 383. [Google Scholar] [CrossRef]
- Keilwagen, J.; Lehnert, H.; Badaeva, E.D.; Özkan, H.; Sharma, S.; Civáň, P.; Kilian, B. Finding Needles in a Haystack: Identification of Inter-Specific Introgressions in Wheat Genebank Collections Using Low-Coverage Sequencing Data. Front. Plant Sci. 2023, 14, 1166854. [Google Scholar] [CrossRef]
- Pamponét, V.C.C.; Souza, M.M.; Silva, G.S.; Micheli, F.; de Melo, C.A.F.; de Oliveira, S.G.; Costa, E.A.; Corrêa, R.X. Low Coverage Sequencing for Repetitive DNA Analysis in Passiflora Edulis Sims: Citogenomic Characterization of Transposable Elements and Satellite DNA. BMC Genom. 2019, 20, 262. [Google Scholar] [CrossRef]
- McCombie, W.R.; McPherson, J.D.; Mardis, E.R. Next-Generation Sequencing Technologies. Cold Spring Harb Perspect. Med. 2019, 9, a036798. [Google Scholar] [CrossRef]
- Unamba, C.I.N.; Nag, A.; Sharma, R.K. Next Generation Sequencing Technologies: The Doorway to the Unexplored Genomics of Non-Model Plants. Front. Plant Sci. 2015, 6, 1074. [Google Scholar] [CrossRef]
- Kim, K.D.; Kang, Y.; Kim, C. Application of Genomic Big Data in Plant Breeding: Past, Present, and Future. Plants 2020, 9, 1454. [Google Scholar] [CrossRef]
- Ștefan, N.; Glăman, G.; Braniște, N.; Stănică, F.; Duțu, I.; Coman, M. Pomologia României Vol. IX—Soiuri Noi de Măr, Păr, Gutui, Cireș, Vișin, Prun și Cais Create în România; CERES: Bucharest, Romania, 2018; ISBN 978-973-40-1125-4. [Google Scholar]
- Li, H.; Durbin, R. Fast and Accurate Short Read Alignment with Burrows–Wheeler Transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map Format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Chen, K.; Wallis, J.W.; McLellan, M.D.; Larson, D.E.; Kalicki, J.M.; Pohl, C.S.; McGrath, S.D.; Wendl, M.C.; Zhang, Q.; Locke, D.P.; et al. BreakDancer: An Algorithm for High-Resolution Mapping of Genomic Structural Variation. Nat. Methods 2009, 6, 677–681. [Google Scholar] [CrossRef]
- Abyzov, A.; Urban, A.E.; Snyder, M.; Gerstein, M. CNVnator: An Approach to Discover, Genotype, and Characterize Typical and Atypical CNVs from Family and Population Genome Sequencing. Genome Res. 2011, 21, 974–984. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional Annotation of Genetic Variants from High-Throughput Sequencing Data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A Toolkit for Detection and Evolutionary Analysis of Gene Synteny and Collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Colle, M.; Leisner, C.P.; Wai, C.M.; Ou, S.; Bird, K.A.; Wang, J.; Wisecaver, J.H.; Yocca, A.E.; Alger, E.I.; Tang, H.; et al. Haplotype-Phased Genome and Evolution of Phytonutrient Pathways of Tetraploid Blueberry. GigaScience 2019, 8, giz012. [Google Scholar] [CrossRef]
- Kuznetsov, A.; Bollin, C.J. NCBI Genome Workbench: Desktop Software for Comparative Genomics, Visualization, and GenBank Data Submission. In Multiple Sequence Alignment: Methods and Protocols; Katoh, K., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2021; pp. 261–295. ISBN 978-1-07-161036-7. [Google Scholar]
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427. [Google Scholar] [CrossRef]
- Bian, Y.; Ballington, J.; Raja, A.; Brouwer, C.; Reid, R.; Burke, M.; Wang, X.; Rowland, L.J.; Bassil, N.; Brown, A. Patterns of Simple Sequence Repeats in Cultivated Blueberries (Vaccinium Section Cyanococcus spp.) and Their Use in Revealing Genetic Diversity and Population Structure. Mol. Breed. 2014, 34, 675–689. [Google Scholar] [CrossRef]
- Yocca, A.E.; Platts, A.; Alger, E.; Teresi, S.; Mengist, M.F.; Benevenuto, J.; Ferrão, L.F.V.; Jacobs, M.; Babinski, M.; Magallanes-Lundback, M.; et al. Blueberry and Cranberry Pangenomes as a Resource for Future Genetic Studies and Breeding Efforts. Hortic. Res. 2023, 10, uhad202. [Google Scholar] [CrossRef]
- Udriște, A.-A.; Iordachescu, M.; Ciceoi, R.; Bădulescu, L. Next-Generation Sequencing of Local Romanian Tomato Varieties and Bioinformatics Analysis of the Ve Locus. Int. J. Mol. Sci. 2022, 23, 9750. [Google Scholar] [CrossRef]
- Edwards, D.; Forster, J.W.; Chagné, D.; Batley, J. What Are SNPs? In Association Mapping in Plants; Oraguzie, N.C., Rikkerink, E.H.A., Gardiner, S.E., De Silva, H.N., Eds.; Springer: New York, NY, USA, 2007; pp. 41–52. ISBN 978-0-387-36011-9. [Google Scholar]
- Yuan, Y.; Bayer, P.E.; Batley, J.; Edwards, D. Current Status of Structural Variation Studies in Plants. Plant Biotechnol. J. 2021, 19, 2153–2163. [Google Scholar] [CrossRef]
- Sim, S.-C.; Durstewitz, G.; Plieske, J.; Wieseke, R.; Ganal, M.W.; Deynze, A.V.; Hamilton, J.P.; Buell, C.R.; Causse, M.; Wijeratne, S.; et al. Development of a Large SNP Genotyping Array and Generation of High-Density Genetic Maps in Tomato. PLoS ONE 2012, 7, e40563. [Google Scholar] [CrossRef]
- Aguilar, M.; Prieto, P. Telomeres and Subtelomeres Dynamics in the Context of Early Chromosome Interactions During Meiosis and Their Implications in Plant Breeding. Front. Plant Sci. 2021, 12, 672489. [Google Scholar] [CrossRef]
- Gupta, V.; Estrada, A.D.; Blakley, I.; Reid, R.; Patel, K.; Meyer, M.D.; Andersen, S.U.; Brown, A.F.; Lila, M.A.; Loraine, A.E. RNA-Seq Analysis and Annotation of a Draft Blueberry Genome Assembly Identifies Candidate Genes Involved in Fruit Ripening, Biosynthesis of Bioactive Compounds, and Stage-Specific Alternative Splicing. GigaScience 2015, 4, 5. [Google Scholar] [CrossRef]
- Wu, C.; Deng, C.; Hilario, E.; Albert, N.W.; Lafferty, D.; Grierson, E.R.P.; Plunkett, B.J.; Elborough, C.; Saei, A.; Günther, C.S.; et al. A Chromosome-Scale Assembly of the Bilberry Genome Identifies a Complex Locus Controlling Berry Anthocyanin Composition. Mol. Ecol. Resour. 2022, 22, 345–360. [Google Scholar] [CrossRef]
- Cappai, F.; Benevenuto, J.; Ferrão, L.F.V.; Munoz, P. Molecular and Genetic Bases of Fruit Firmness Variation in Blueberry—A Review. Agronomy 2018, 8, 174. [Google Scholar] [CrossRef]
- Ferrão, L.F.V.; Johnson, T.S.; Benevenuto, J.; Edger, P.P.; Colquhoun, T.A.; Munoz, P.R. Genome-Wide Association of Volatiles Reveals Candidate Loci for Blueberry Flavor. New Phytol. 2020, 226, 1725–1737. [Google Scholar] [CrossRef]
- Wein, M.; Lavid, N.; Lunkenbein, S.; Lewinsohn, E.; Schwab, W.; Kaldenhoff, R. Isolation, Cloning and Expression of a Multifunctional O-Methyltransferase Capable of Forming 2,5-Dimethyl-4-Methoxy-3(2H)-Furanone, One of the Key Aroma Compounds in Strawberry Fruits. Plant J. 2002, 31, 755–765. [Google Scholar] [CrossRef]
- Keegstra, K.; Raikhel, N. Plant Glycosyltransferases. Curr. Opin. Plant Biol. 2001, 4, 219–224. [Google Scholar] [CrossRef]
- Sun, J.; Sun, Y.; Ahmed, R.I.; Ren, A.; Xie, M. Research Progress on Plant RING-Finger Proteins. Genes 2019, 10, 973. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Cao, H.; Pan, L.; Niu, L.; Wei, B.; Cui, G.; Wang, L.; Yao, J.-L.; Zeng, W.; Wang, Z. Two Loss-of-Function Alleles of the Glutathione S-Transferase (GST) Gene Cause Anthocyanin Deficiency in Flower and Fruit Skin of Peach (Prunus persica). Plant J. 2021, 107, 1320–1331. [Google Scholar] [CrossRef]
- Zhao, Y.; Dong, W.; Zhu, Y.; Allan, A.C.; Lin-Wang, K.; Xu, C. PpGST1, an Anthocyanin-Related Glutathione S-Transferase Gene, Is Essential for Fruit Coloration in Peach. Plant Biotechnol. J. 2020, 18, 1284–1295. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.-Y.; Li, Z.-H.; Zhang, Y.-X.; Chen, L.; Xiang, D.-Y.; Zhang, Y.-F. Two Pear Glutathione S-Transferases Genes Are Regulated during Fruit Development and Involved in Response to Salicylic Acid, Auxin, and Glucose Signaling. PLoS ONE 2014, 9, e89926. [Google Scholar] [CrossRef]
- Wang, L.; Qian, M.; Wang, R.; Wang, L.; Zhang, S. Characterization of the Glutathione S-Transferase (GST) Gene Family in Pyrus Bretschneideri and Their Expression Pattern upon Superficial Scald Development. Plant Growth Regul. 2018, 86, 211–222. [Google Scholar] [CrossRef]
- Han, Y.; Fu, C.; Kuang, J.; Chen, J.; Lu, W. Two Banana Fruit Ripening-Related C2H2 Zinc Finger Proteins Are Transcriptional Repressors of Ethylene Biosynthetic Genes. Postharvest Biol. Technol. 2016, 116, 8–15. [Google Scholar] [CrossRef]
- Zhao, Y.-W.; Wang, C.-K.; Huang, X.-Y.; Hu, D.-G. Genome-Wide Analysis of the Glutathione S-Transferase (GST) Genes and Functional Identification of MdGSTU12 Reveals the Involvement in the Regulation of Anthocyanin Accumulation in Apple. Genes 2021, 12, 1733. [Google Scholar] [CrossRef]
- Lin, Y.; Zhang, L.; Zhang, J.; Zhang, Y.; Wang, Y.; Chen, Q.; Luo, Y.; Zhang, Y.; Li, M.; Wang, X.; et al. Identification of Anthocyanins-Related Glutathione S-Transferase (GST) Genes in the Genome of Cultivated Strawberry (Fragaria × Ananassa). Int. J. Mol. Sci. 2020, 21, 8708. [Google Scholar] [CrossRef]
- Kumar, S.; Banks, T.W.; Cloutier, S. SNP Discovery through Next-Generation Sequencing and Its Applications. Int. J. Plant Genom. 2012, 2012, 831460. [Google Scholar] [CrossRef]
- Weng, L.; Zhao, F.; Li, R.; Xu, C.; Chen, K.; Xiao, H. The Zinc Finger Transcription Factor SlZFP2 Negatively Regulates Abscisic Acid Biosynthesis and Fruit Ripening in Tomato. Plant Physiol. 2015, 167, 931–949. [Google Scholar] [CrossRef] [PubMed]
- Sicard, A.; Petit, J.; Mouras, A.; Chevalier, C.; Hernould, M. Meristem Activity during Flower and Ovule Development in Tomato Is Controlled by the Mini Zinc Finger Gene INHIBITOR OF MERISTEM ACTIVITY. Plant J. 2008, 55, 415–427. [Google Scholar] [CrossRef]
- Cao, Y.; Han, Y.; Meng, D.; Abdullah, M.; Li, D.; Jin, Q.; Lin, Y.; Cai, Y. Systematic Analysis and Comparison of the PHD-Finger Gene Family in Chinese Pear (Pyrus bretschneideri) and Its Role in Fruit Development. Funct. Integr. Genom. 2018, 18, 519–531. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, W.; He, H.; Nie, J.; Bie, B.; Zhao, J.; Ren, G.; Li, Y.; Zhang, D.; Pan, J.; et al. Tuberculate Fruit Gene Tu Encodes a C2H2 Zinc Finger Protein That Is Required for the Warty Fruit Phenotype in Cucumber (Cucumis sativus L.). Plant J. 2014, 78, 1034–1046. [Google Scholar] [CrossRef]
- Zhao, M.; Li, J.; Zhu, L.; Chang, P.; Li, L.; Zhang, L. Identification and Characterization of MYB-bHLH-WD40 Regulatory Complex Members Controlling Anthocyanidin Biosynthesis in Blueberry Fruits Development. Genes 2019, 10, 496. [Google Scholar] [CrossRef]
- Machemer, K.; Shaiman, O.; Salts, Y.; Shabtai, S.; Sobolev, I.; Belausov, E.; Grotewold, E.; Barg, R. Interplay of MYB Factors in Differential Cell Expansion, and Consequences for Tomato Fruit Development. Plant J. 2011, 68, 337–350. [Google Scholar] [CrossRef]
- Hassanin, A.A.; Eldomiaty, A.S.; Ujjan, J.A.; Al-Mushhin, A.A.M.; ALrashidi, A.A.; Saad, A.M.; Sakit ALHaithloul, H.A.; El-Saadony, M.T.; Awad, M.F.; Sitohy, M.Z. Assessment of the R2R3 MYB Gene Expression Profile during Tomato Fruit Development Using in Silico Analysis, Quantitative and Semi-Quantitative RT-PCR. Saudi J. Biol. Sci. 2022; in press. [Google Scholar] [CrossRef]
- Cao, Y.; Han, Y.; Li, D.; Lin, Y.; Cai, Y. MYB Transcription Factors in Chinese Pear (Pyrus bretschneideri Rehd.): Genome-Wide Identification, Classification, and Expression Profiling during Fruit Development. Front. Plant Sci. 2016, 7, 577. [Google Scholar] [CrossRef]
- Espley, R.V.; Hellens, R.P.; Putterill, J.; Stevenson, D.E.; Kutty-Amma, S.; Allan, A.C. Red Colouration in Apple Fruit Is Due to the Activity of the MYB Transcription Factor, MdMYB10. Plant J. 2007, 49, 414–427. [Google Scholar] [CrossRef]
- Vimolmangkang, S.; Han, Y.; Wei, G.; Korban, S.S. An Apple MYB Transcription Factor, MdMYB3, Is Involved in Regulation of Anthocyanin Biosynthesis and Flower Development. BMC Plant Biol. 2013, 13, 176. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, H.; Yang, Y.; Li, M.; Zhang, Y.; Liu, J.; Dong, J.; Li, J.; Butelli, E.; Xue, Z.; et al. The Control of Red Colour by a Family of MYB Transcription Factors in Octoploid Strawberry (Fragaria × ananassa) Fruits. Plant Biotechnol. J. 2020, 18, 1169–1184. [Google Scholar] [CrossRef]
- Ribone, P.A.; Capella, M.; Arce, A.L.; Chan, R.L. Chapter 22—What Do We Know about Homeodomain–Leucine Zipper I Transcription Factors? Functional and Biotechnological Considerations. In Plant Transcription Factors; Gonzalez, D.H., Ed.; Academic Press: Boston, MA, USA, 2016; pp. 343–356. ISBN 978-0-12-800854-6. [Google Scholar]
- Sun, J.; Li, L.; Wang, P.; Zhang, S.; Wu, J. Genome-Wide Characterization, Evolution, and Expression Analysis of the Leucine-Rich Repeat Receptor-like Protein Kinase (LRR-RLK) Gene Family in Rosaceae Genomes. BMC Genom. 2017, 18, 763. [Google Scholar] [CrossRef]
- Zhao, K.; Wang, L.; Qiu, D.; Cao, Z.; Wang, K.; Li, Z.; Wang, X.; Wang, J.; Ma, Q.; Cao, D.; et al. PSW1, an LRR Receptor Kinase, Regulates Pod Size in Peanut. Plant Biotechnol. J. 2023, 21, 2113–2124. [Google Scholar] [CrossRef]
- Subburaj, S.; Tu, L.; Lee, K.; Park, G.-S.; Lee, H.; Chun, J.-P.; Lim, Y.-P.; Park, M.-W.; McGregor, C.; Lee, G.-J. A Genome-Wide Analysis of the Pentatricopeptide Repeat (PPR) Gene Family and PPR-Derived Markers for Flesh Color in Watermelon (Citrullus lanatus). Genes 2020, 11, 1125. [Google Scholar] [CrossRef]
- Zhang, A.; Xiong, Y.; Liu, F.; Zhang, X. A Genome-Wide Analysis of the Pentatricopeptide Repeat Protein Gene Family in Two Kiwifruit Species with an Emphasis on the Role of RNA Editing in Pathogen Stress. Int. J. Mol. Sci. 2023, 24, 13700. [Google Scholar] [CrossRef]
- Barchenger, D.W.; Said, J.I.; Zhang, Y.; Song, M.; Ortega, F.A.; Ha, Y.; Kang, B.-C.; Bosland, P.W. Genome-Wide Identification of Chile Pepper Pentatricopeptide Repeat Domains Provides Insight into Fertility Restoration. J. Am. Soc. Hortic. Sci. 2018, 143, 418–429. [Google Scholar] [CrossRef]
- Jo, Y.D.; Ha, Y.; Lee, J.-H.; Park, M.; Bergsma, A.C.; Choi, H.-I.; Goritschnig, S.; Kloosterman, B.; van Dijk, P.J.; Choi, D.; et al. Fine Mapping of Restorer-of-Fertility in Pepper (Capsicum annuum L.) Identified a Candidate Gene Encoding a Pentatricopeptide Repeat (PPR)-Containing Protein. Theor. Appl. Genet. 2016, 129, 2003–2017. [Google Scholar] [CrossRef]
- Ren, R.C.; Lu, X.; Zhao, Y.J.; Wei, Y.M.; Wang, L.L.; Zhang, L.; Zhang, W.T.; Zhang, C.; Zhang, X.S.; Zhao, X.Y. Pentatricopeptide Repeat Protein DEK40 Is Required for Mitochondrial Function and Kernel Development in Maize. J. Exp. Bot. 2019, 70, 6163–6179. [Google Scholar] [CrossRef]
- Hadfield, K.A.; Bennett, A.B. Polygalacturonases: Many Genes in Search of a Function1. Plant Physiol. 1998, 117, 337–343. [Google Scholar] [CrossRef]
- Prasanna, V.; Prabha, T.N.; Tharanathan, R.N. Fruit Ripening Phenomena—An Overview. Crit. Rev. Food Sci. Nutr. 2007, 47, 1–19. [Google Scholar] [CrossRef]
- Visser, J.A.E.B. Jaap Polygalacturonases. In Handbook of Food Enzymology; CRC Press: Boca Raton, FL, USA, 2002; ISBN 978-0-429-22254-2. [Google Scholar]
- Lang, C.; Dörnenburg, H. Perspectives in the Biological Function and the Technological Application of Polygalacturonases. Appl. Microbiol. Biotechnol. 2000, 53, 366–375. [Google Scholar] [CrossRef]
- Fischer, R.L.; Bennett, A.B. Role of Cell Wall Hydrolases in Fruit Ripening. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1991, 42, 675–703. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).