Evaluation of the Genome Size and Ploidy Level of Pears (Pyrus spp.) in Relation to Their Morphological Traits
Abstract
1. Introduction
2. Materials and Methods
2.1. Flow Cytometry
2.2. Morphological Research
3. Results and Discussion
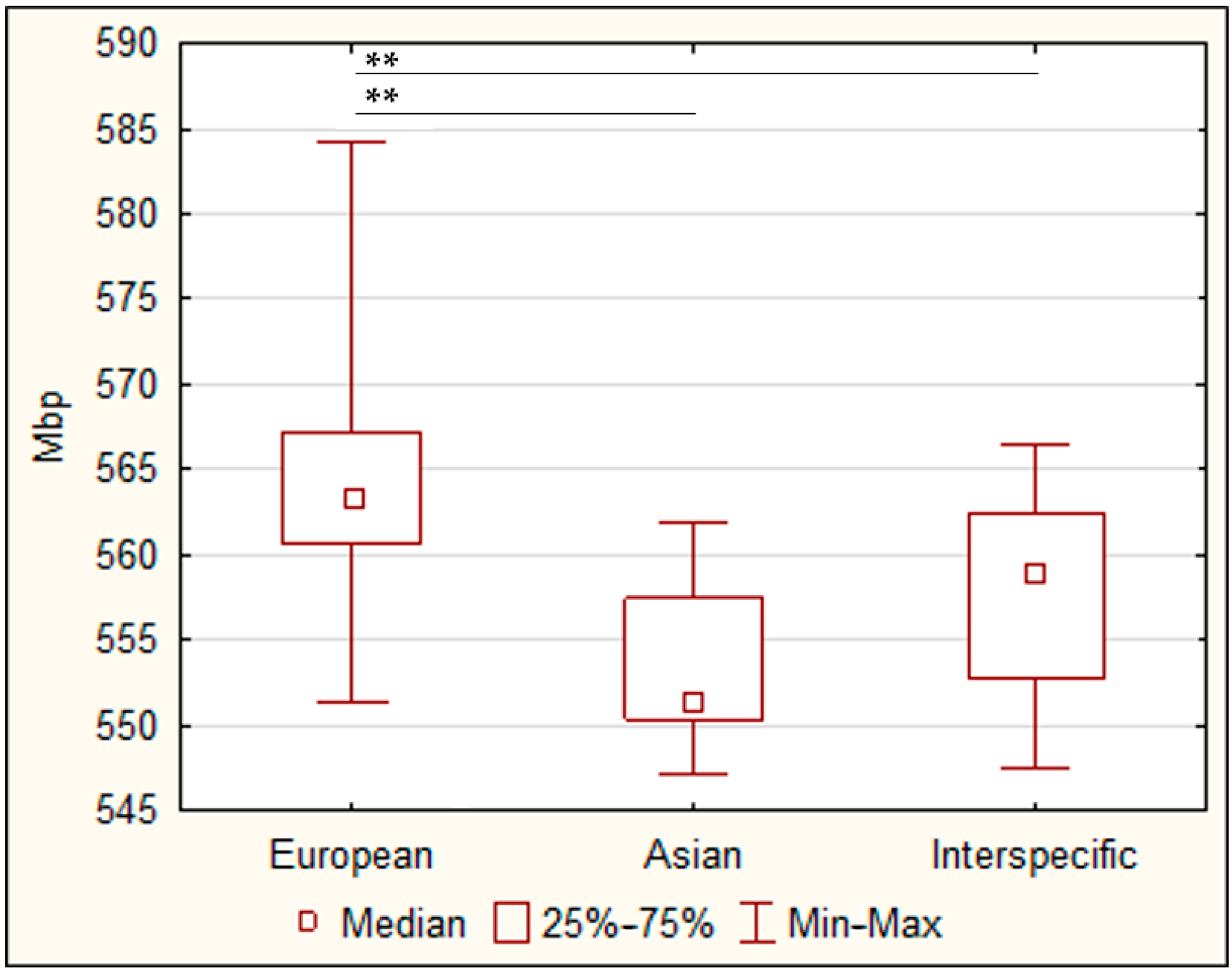
3.1. The Relative Genome Size
3.2. Selected Morphological Characteristics
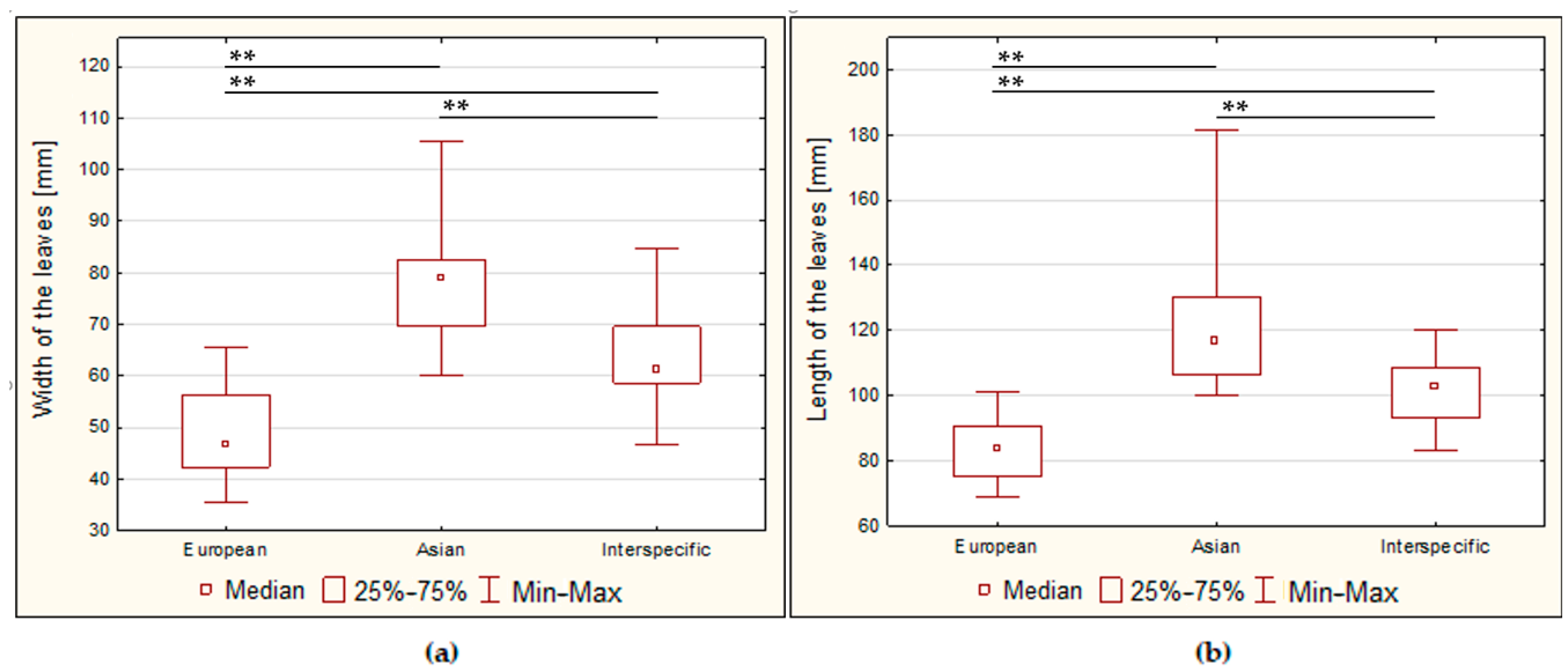
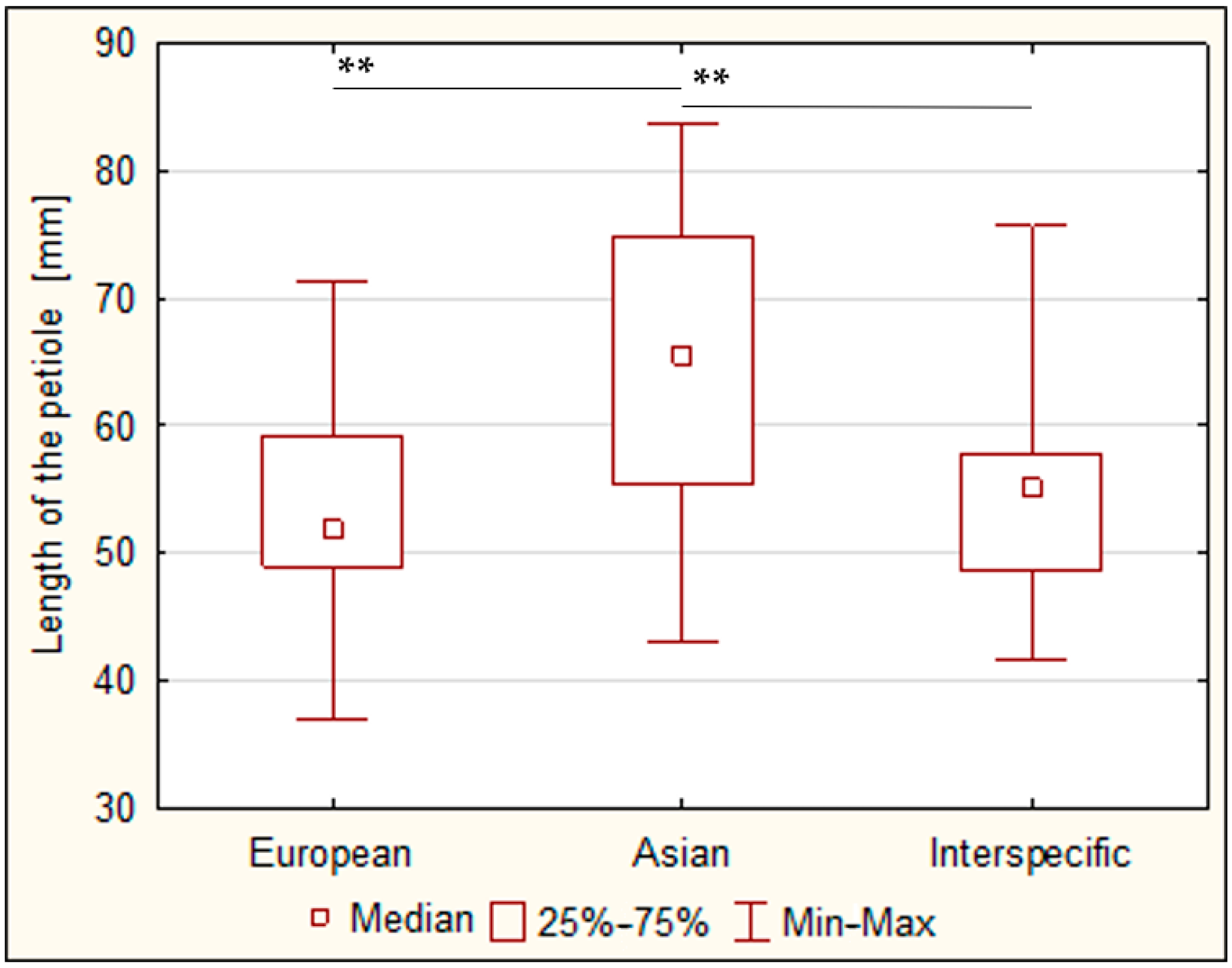
3.2.1. Size of the Leaves
3.2.2. Diameter of the Flowers
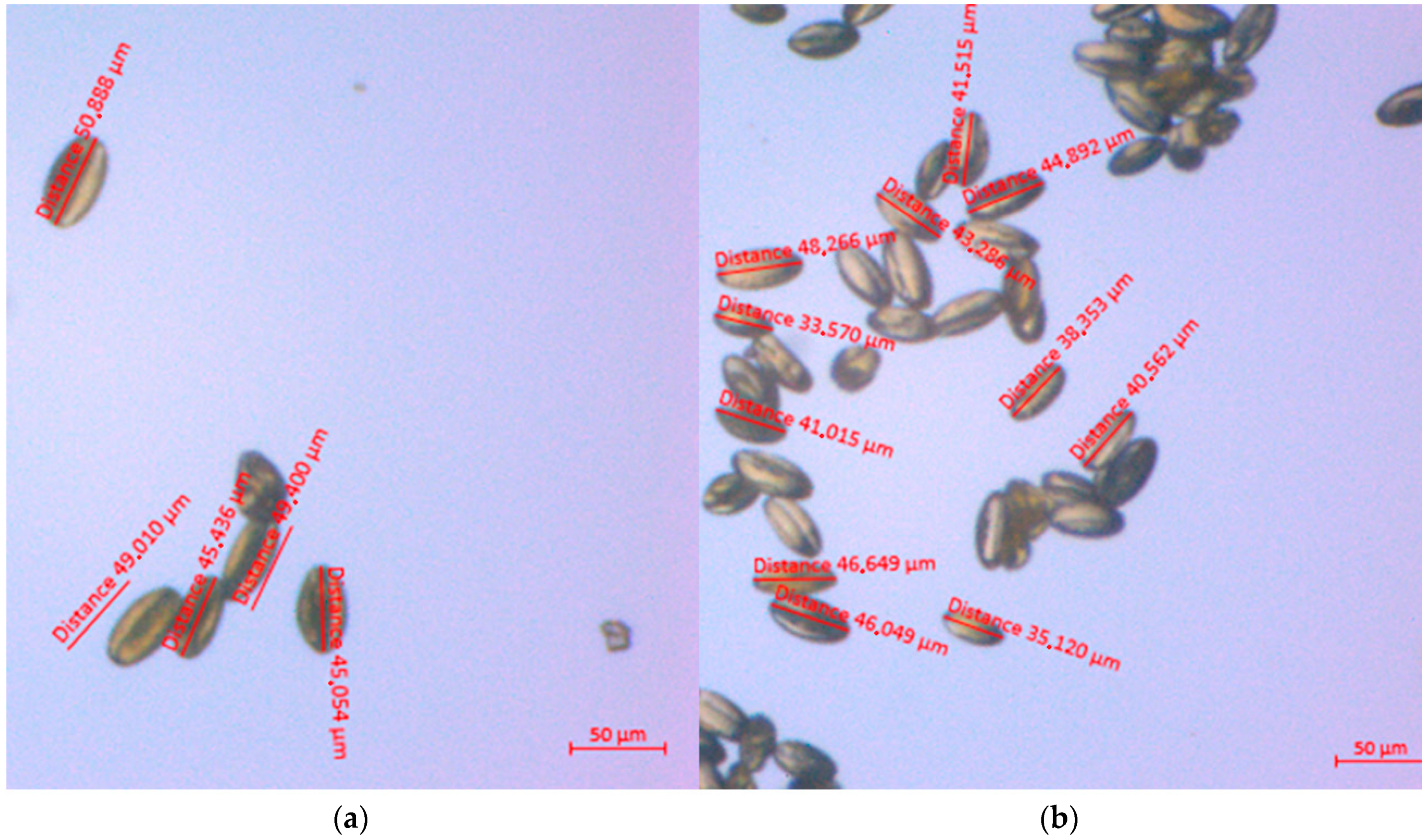
3.2.3. Length of the Pollen Grains
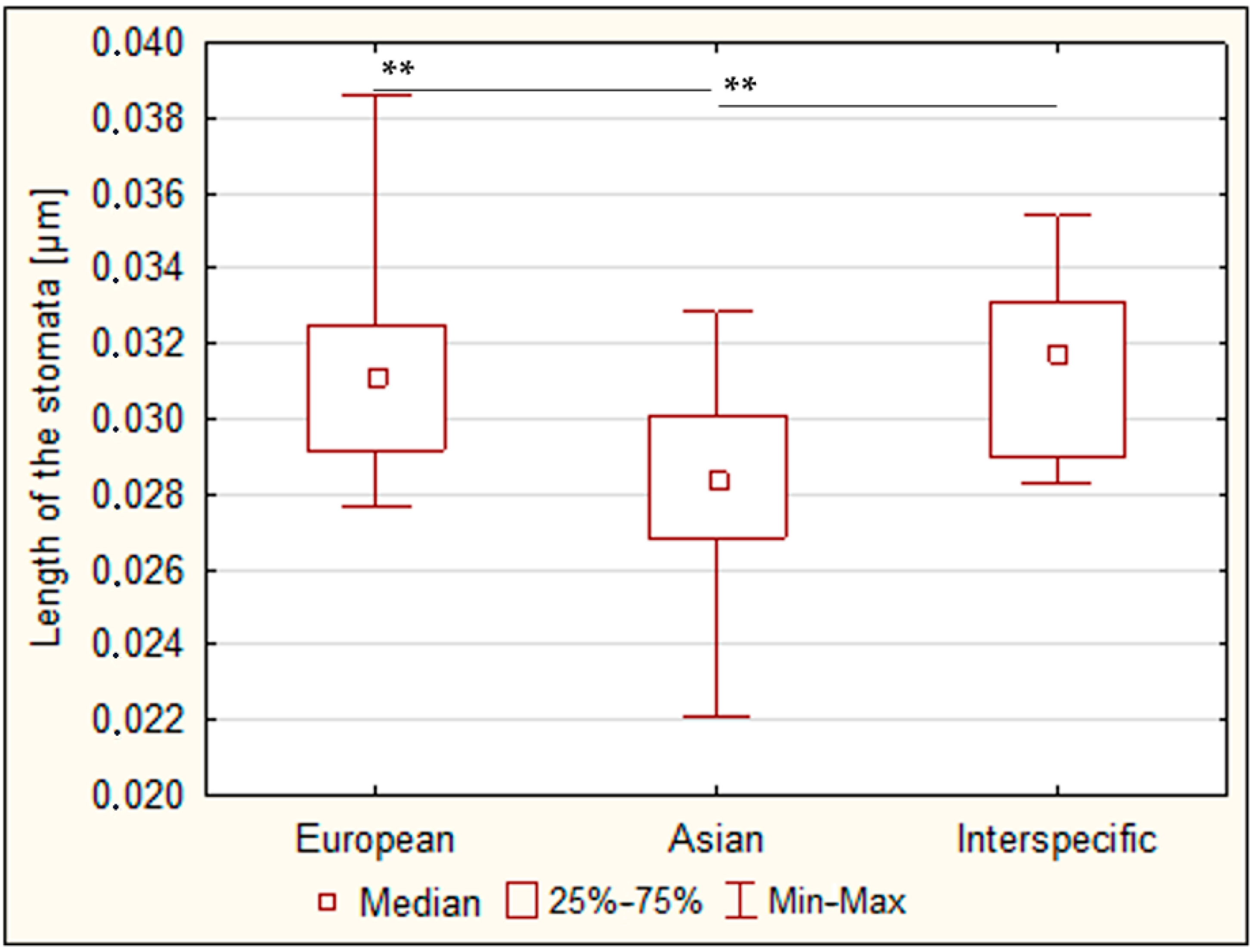
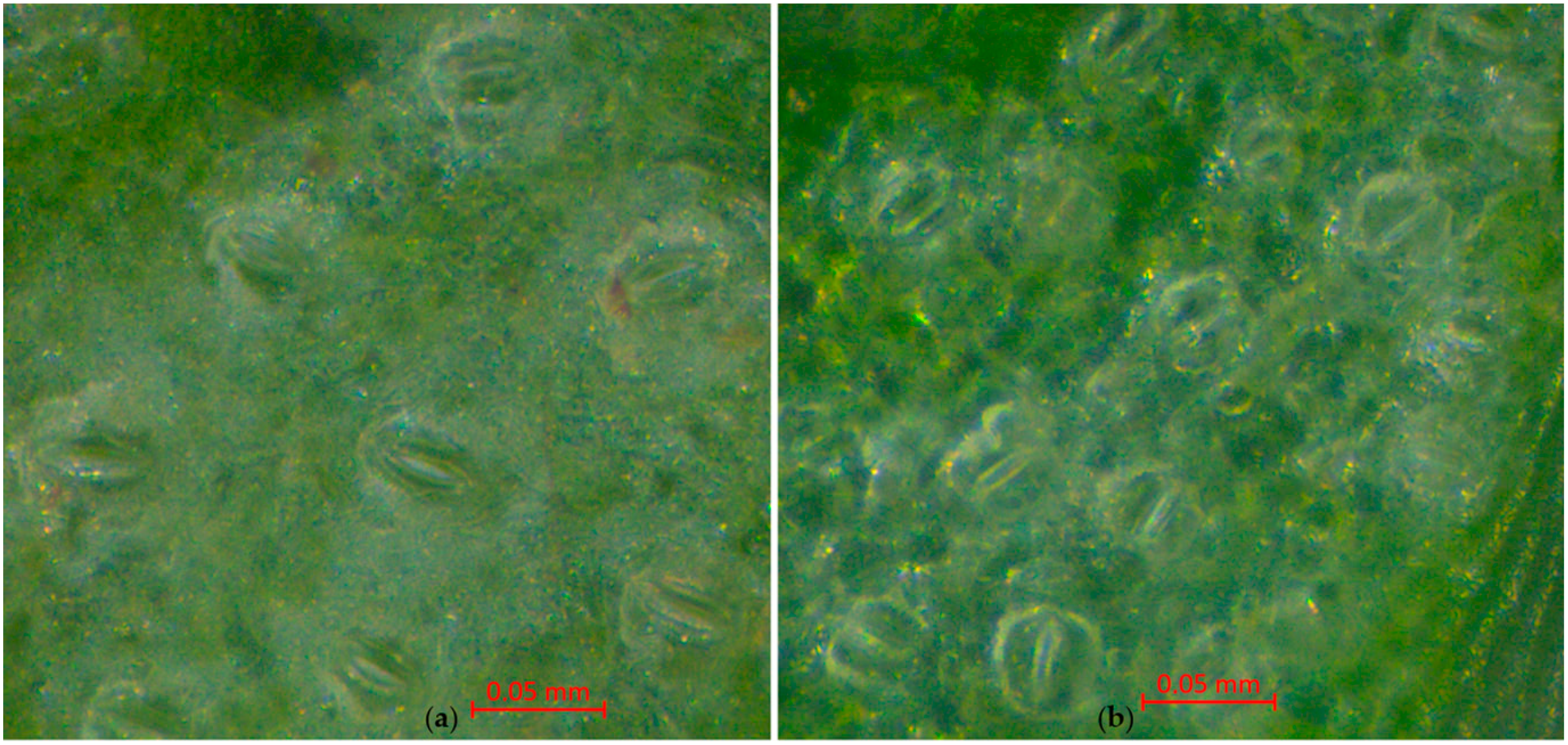
3.2.4. Length of the Stomata
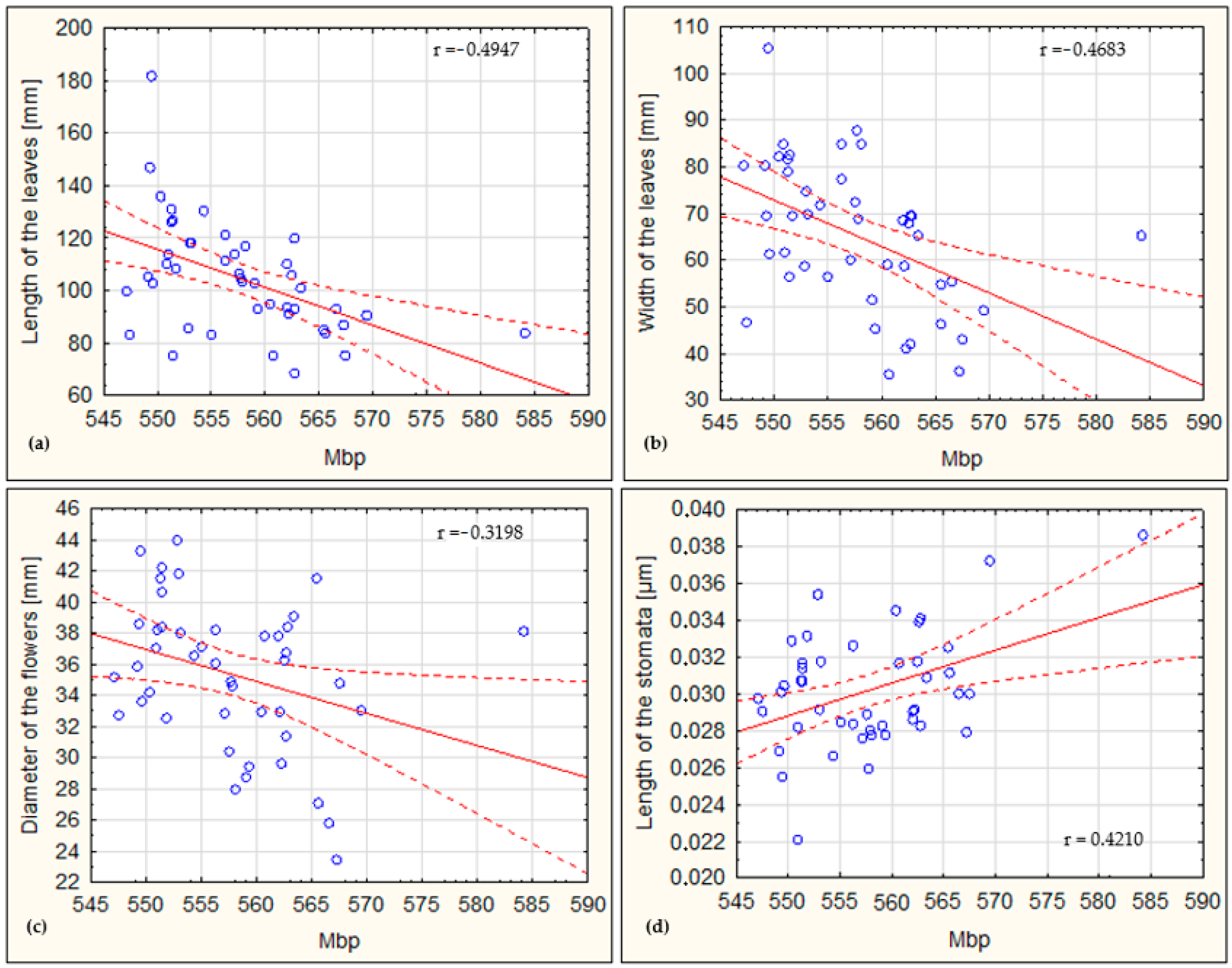
3.3. Correlations of the Genome Size and Morphological Traits
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kadkhodaei, S.; Arzani, K.; Yadollahi, A.; Karimzadeh, G.; Abdollahi, H. Genetic Diversity and Similarity of Asian and European Pears (Pyrus spp.) Revealed by Genome Size and Morphological Traits Prediction. Int. J. Fruit Sci. 2021, 21, 619–633. [Google Scholar] [CrossRef]
- Nečas, T.; Balík, J.; Wolf, J.; Goliáš, J.; Láčík, J.; Šillerová, J.; Kiss, T.; Ondrášek, I.; Horák, M.; Kožíšková, J.; et al. ASIJSKÉ HRUŠNĚ V PODMÍNKÁCH ČESKÉ REPUBLIKY, Pěstování, Pomologie, Skladování a Choroby, 1st ed.; Ediční středisko Mendelovy Univerzity: Brno, Czech Republic, 2018; 184p, ISBN 978-80-7509-557-2. [Google Scholar]
- Rampáčková, E.; Mrázová, M.; Čížková, J.; Nečas, T. Pomological Traits and Genome Size of Prunus armeniaca L. Considering to Geographical Origin. Horticulturae 2022, 8, 199. [Google Scholar] [CrossRef]
- Sakhanokho, H.F.; Timothy Rinehart, A.; Stringer, S.J.; Islam-Faridi, M.N.; Pounders, C.T. Variation in nuclear DNA content and chromosome numbers in blueberry. Sci. Hortic. 2018, 233, 108–113. [Google Scholar] [CrossRef]
- Kolano, B.; Siwinska, D.; Gomez Pando, L.; Szymanowska-Pulka, J.; Maluszynska, J. Genome size variation in Chenopodium quinoa (Chenopodiaceae). Plant Syst. Evol. 2012, 298, 251–255. [Google Scholar] [CrossRef][Green Version]
- Phillips, W.D.; Ranney, T.G.; Touchell, D.H.; Eaker, T.A. Fertility and reproductive pathways of triploid flowering pears (Pyrus sp.). HortScience 2016, 51, 968–971. [Google Scholar] [CrossRef]
- Niu, Y.; Zhou, W.; Chen, X.; Fan, G.; Zhang, S.; Liao, K. Genome size and chromosome ploidy identification in pear germplasm represented by Asian pears—Local pear varieties. Sci. Hortic. 2020, 265, 109202. [Google Scholar] [CrossRef]
- Baccichet, I.; Foria, S.; Messina, R.; Peccol, E.; Losa, A.; Fabro, M.; Gori, G.; Zandigiacomo, P.; Cipriani, G.; Testolin, R. Genetic and ploidy diversity of Pear (Pyrus spp.) germplasm of Friuli Venezia Giulia, Italy. Genet. Resour. Crop Evol. 2019, 67, 83–96. [Google Scholar] [CrossRef]
- Postman, J.; Bassil, N.; Bell, R. Ploidy of USDA World Pear Germplasm Collection determined by flow cytometry. Acta Hortic. 2014, 1094, 75–81. [Google Scholar] [CrossRef]
- Doležel, J.; Sgorbatti, S.; Lucretti, S. Comparison of three DNA fluorochromes for flow cytometric estimation of nuclear DNA content in plants. Physiol. Plant. 1992, 85, 625–631. [Google Scholar] [CrossRef]
- Doležel, J.; Greihuber, J.; Suda, J. Flow Cytometry with Plant Cells. Analysis of Genes, Chromosomes and Genomes; WILEY-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2007; ISBN 978-3-527-31487-4. [Google Scholar]
- Doležel, J.; Bartoš, J.A.N. Plant DNA flow cytometry and estimation of nuclear genome size. Ann. Bot. 2005, 95, 99–110. [Google Scholar] [CrossRef]
- Dickson, E.E.; Arumuganathan, K.; Kresovich, S.; Doyle, J.J. Nuclear DNA content variation within the Rosaceae. Am. J. Bot. 1992, 79, 1081–1086. [Google Scholar] [CrossRef]
- Pustahija, F.; Brown, S.C.; Bogunić, F.; Bašić, N.; Muratović, E.; Ollier, S.; Hidalgo, O.; Bourge, M.; Stevanović, V.; Siljak-Yakovlev, S. Small genomes dominate in plants growing on serpentine soils in West Balkans, an exhaustive study of 8 habitats covering 308 taxa. Plant Soil 2013, 373, 427–453. [Google Scholar] [CrossRef]
- Wu, J.; Wang, Z.; Shi, Z.; Zhang, S.; Ming, R.; Zhu, S.; Khan, M.A.; Tao, S.; Korban, S.S.; Wang, H.; et al. The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res. 2012, 23, 396–408. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; He, J.; Zhao, T.-H.; Cao, Y.; Wang, G.; Sun, B.; Yan, X.; Guo, W.; Li, M.-H. The smaller the leaf is, the faster the leaf water loses in a temperate forest. Front. Plant Sci. 2019, 10, 58. [Google Scholar] [CrossRef]
- Antkowiak, W.; Czarna, A.; Wawrzyniak, M. Pyrus x myloslavensis (P. communis L. x P. salicifolia Pall.)—A new spontaneous pear hybrid. Dendrobiology 2008, 60, 45–49. [Google Scholar]
- Dalalbashi, A.A.; Alhatem, J.Y. Morphological and anatomical study of some Italian pear leaves Pyrus communis L. cultivated in Iraq. In Proceedings of the 2nd International Conference on Materials Engineering & Science (IConMEAS 2019), Baghdad, Iraq, 25–26 September 2019. [Google Scholar] [CrossRef]
- Farkas, A.; Orosz-Kovács, Z.S. Primary and secondary attractants of flowers in Pear Pyrus betulifolia. Acta Hortic. 2004, 636, 317–324. [Google Scholar] [CrossRef]
- Singh, S.K.; Bist, L.D.; Patel, V.B. Flowering and floral biology studies in low-chill pear cultivars. Indian J. Hortic. 2004, 61, 94–96. [Google Scholar]
- Motyleva, S.; Brindza, J.; Kulikov, I. The morphology of pollen grains of the some species of Rosaceae Juss. family. Agrobiodivers. Improv. Nutr. Health Life Qual. 2017, 1, 338–341. [Google Scholar] [CrossRef][Green Version]
- Westwood, M.N.; Challice, J.S. Morphology and surface topography of pollen and anthers of Pyrus species. J. Am. Soc. Hortic. Sci. 1978, 103, 28–37. [Google Scholar] [CrossRef]
- Matsuta, N.; Omura, M.; Akihama, T. Difference in micromorphological pattern on pollen surface of Japanese pear cultivars. Jpn. J. Breed. 1982, 32, 123–128. [Google Scholar] [CrossRef]
- Babosha, A.; Kumachova, T.; Ryabchenko, A.; Komarova, G. Microrelief of the leaf epidermis and stomatal polymorphism of malus orientalis, Pyrus caucasica and Mespilus germanica in mountains and Plains. Flora 2022, 291, 152074. [Google Scholar] [CrossRef]
- Ameri, A.; Davarynejad, G.; Moshtaghi, N.; Tehranifar, A. SEMICOMPACT canopy form in mixoploid plants differentiated from the endosperm of Pyrus communis cv. Natanzi: Evidence from flow cytometric analysis and anatomical and morphological traits. Ann. Appl. Biol. 2020, 177, 385–394. [Google Scholar] [CrossRef]
- Wang, X.; Wang, H.; Shi, C.; Zhang, X.; Duan, K.; Luo, J. Morphological, cytological and fertility consequences of a spontaneous tetraploid of the diploid pear (Pyrus pyrifolia Nakai) cultivar ‘cuiguan’. Sci. Hortic. 2015, 189, 59–65. [Google Scholar] [CrossRef]
- Beaulieu, J.M.; Leitch, I.J.; Patel, S.; Pendharkar, A.; Knight, C.A. Genome size is a strong predictor of cell size and stomatal density in angiosperms. New Phytol. 2008, 179, 975–986. [Google Scholar] [CrossRef]
- Basak, S.; Sun, X.; Wang, G.; Yang, Y. Genome size unaffected by variation in morphological traits, temperature, and precipitation in Turnip. Appl. Sci. 2019, 9, 253. [Google Scholar] [CrossRef]
- Dušková, E.; Kolář, F.; Sklenář, P.; Rauchová, J.; Kubešová, M.; Fér, T.; Suda, J.; Marhold, K. Genome size correlates with growth form, habitat and phylogeny in the Andean genus Lasiocephalus (Asteraceae). Preslia 2010, 82, 127–148. [Google Scholar]
- Savaş Tuna, G.; Bbaşer, İ.; Tuna, M. Genome size variation among natural populations of Brachypodium distachyon and B. hybridum collected from different regions of Turkey. Turk. J. Bot. 2019, 43, 196–207. [Google Scholar] [CrossRef]
- Greilhuber, J. Intraspecific variation in genome size in angiosperms: Identifying its existence. Ann. Bot. 2005, 95, 91–98. [Google Scholar] [CrossRef]
- Šmarda, P.; Bureš, P. Understanding intraspecific variation in genome size in plants. Preslia 2010, 82, 41–61. [Google Scholar]
- Wolf, J.; Kiss, T.; Nečas, T. The use of SSRS for the identification of unknown Asian pear cultivars. Acta Hortic. 2021, 1307, 205–212. [Google Scholar] [CrossRef]
- Fiala, J.; Nečas, T. Asijské a mezidruhové odrůdy hrušní pro podmínky měnícího se klimatu v čr. Zahrad. = Záhradníctvo Měsíčník Pro Prof. Zahradníky. Odb. Recenzovaný Časopis 2024, XXIII, 38–43. [Google Scholar]
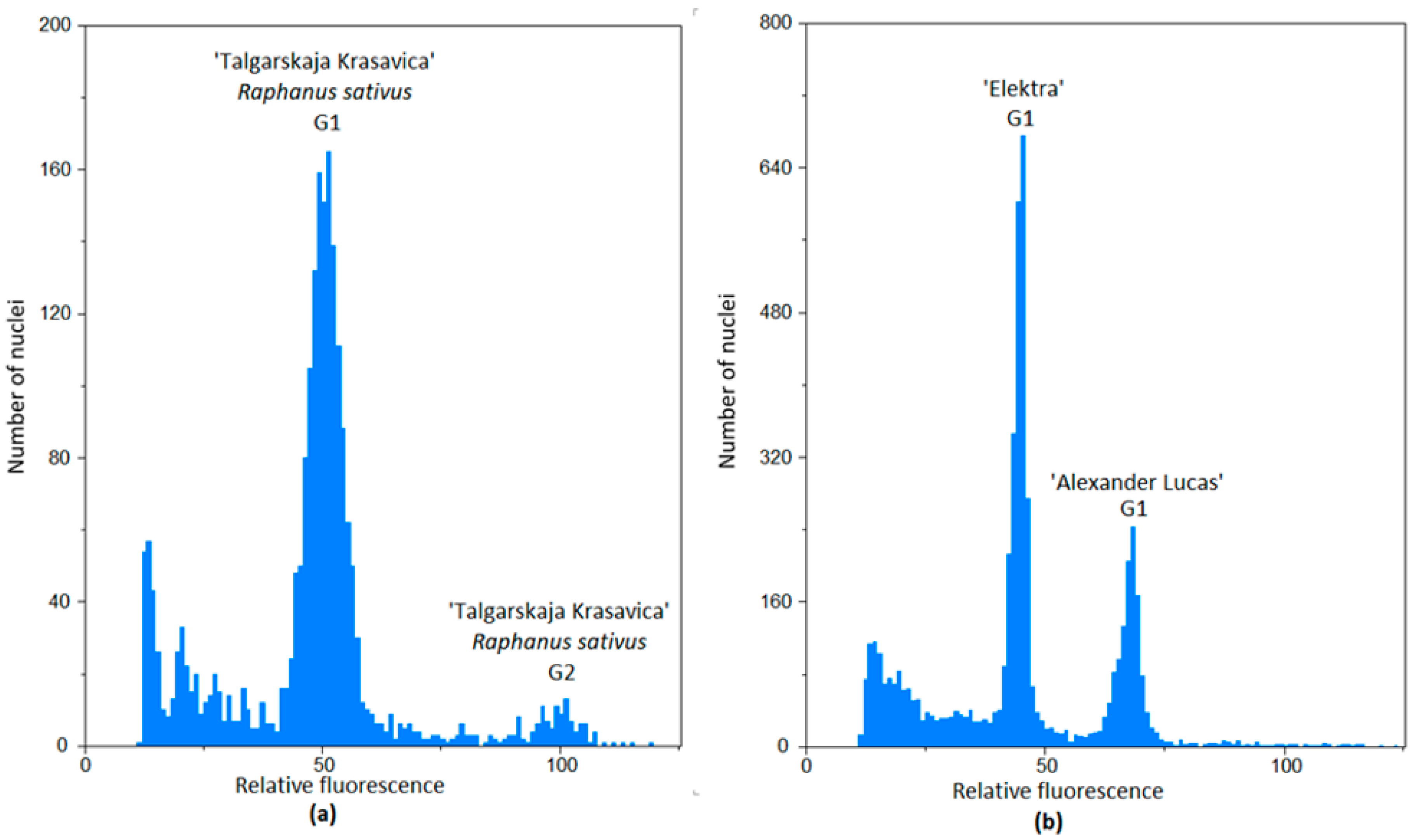
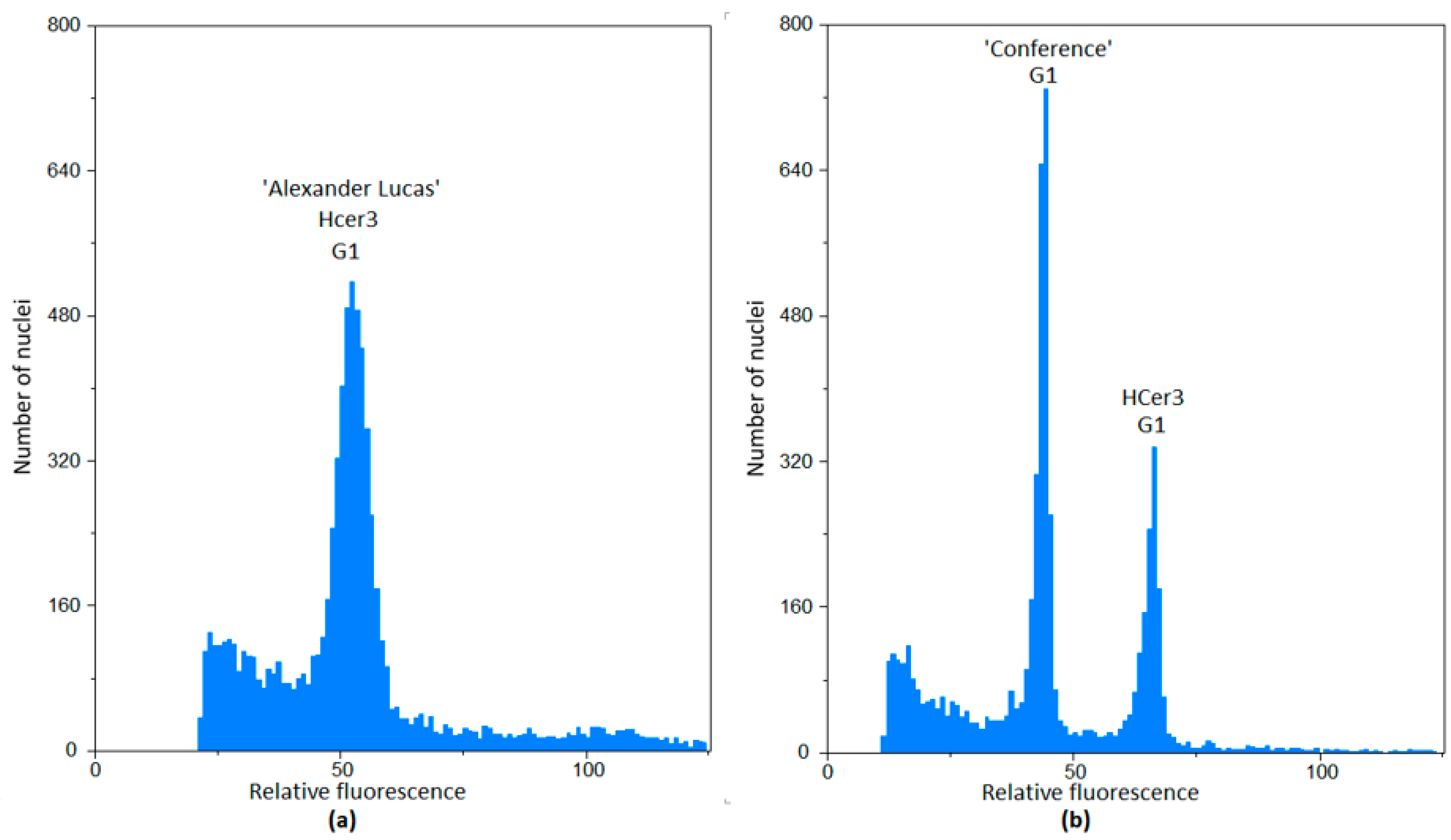


| Asian Varieties | European Varieties | Interspecific Varieties | European Hybrids |
|---|---|---|---|
| Anhuixueli | Alexander Lucas | Benita | Hcer1 |
| Baoshy | Bosc | Hood | Hcer2 |
| Cangxixueli | Conference | Kieffer | Hcer3 |
| Dangshanshuli | Elektra | Wujiuxiang | Hcer4 |
| Dong Guo | Karina | ||
| Huang Hua | Kirgizskaja Zimnaja | Interspecific Hybrids | Botanical Species |
| Chang Ba | Kulovita | H2 | P. betulifolia |
| Chili | Oharkula | H4 | P. calleryana |
| Jin Hua | Talgarskaja krasavica | H7 | P. caucasica |
| Jing Bai | Williams Red | H8 | P. communis |
| Mut Chen | H12 | P. pyrifolia | |
| Nanguoli | H13 | P. ussuriensis | |
| Nutika | H15 | P. bretschneiderii | |
| Shali | H19 | ||
| Shon Shu | H24 | ||
| Snow Flower | |||
| Xin Gao | |||
| Yali | |||
| Zaosuli |
| 1 Cx (Mbp) | Width of the Leaves (mm) | Length of the Leaves (mm) | Length of the Petiole (mm) | Diameter of the Flowers (mm) | Length of the Pollen Grains (μm) | Length of the Stomata (μm) | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| European | 564.1 | ± | 2.2 | 49.0 | ± | 2.8 | 84.1 | ± | 2.5 | 53.4 | ± | 2.8 | 34.3 | ± | 1.5 | 44.5 | ± | 0.5 | 31.6 | ± | 0.9 |
| Asian | 553.4 | ± | 0.9 | 77.6 | ± | 2.4 | 121.1 | ± | 4.4 | 64.9 | ± | 2.7 | 36.8 | ± | 0.9 | 45.9 | ± | 1.3 | 28.4 | ± | 0.6 |
| Interspecific | 557.5 | ± | 1.7 | 63.3 | ± | 2.7 | 101.0 | ± | 3.2 | 54.7 | ± | 2.4 | 34.3 | ± | 1.3 | 43.5 | ± | 1.2 | 31.4 | ± | 0.7 |
| P. betulifolia | 571.7 | ± | 0.9 | 56.4 | ± | 0.7 | 83.2 | ± | 4.7 | 45.8 | ± | 4.2 | 25.8 | ± | 0.6 | N | 28.0 | ± | 0.8 | ||
| P. calleryana | 554.2 | ± | 1.5 | 48.4 | ± | 1.7 | 84.2 | ± | 2.1 | 52.8 | ± | 2.3 | 23.3 | ± | 0.6 | 41.5 | ± | 0.5 | 28.0 | ± | 1.3 |
| P. caucasica | 542.5 | ± | 0.6 | 54.8 | ± | 2.1 | 66.0 | ± | 2.5 | 56.2 | ± | 3.6 | 35.0 | ± | 0.5 | 43.6 | ± | 0.7 | 31.1 | ± | 0.5 |
| P. communis | 559.1 | ± | 1.5 | 43.6 | ± | 0.9 | 55.6 | ± | 1.9 | 44.8 | ± | 1.6 | 28.6 | ± | 0.8 | 46.9 | ± | 1.0 | 34.2 | ± | 1.2 |
| P. pyrifolia | 553.1 | ± | 2.2 | 67.2 | ± | 2.0 | 104.6 | ± | 3.4 | 49.8 | ± | 2.4 | 31.0 | ± | 0.7 | N | 26.0 | ± | 0.9 | ||
| P. ussuriensis | 560.7 | ± | 1.1 | 67.0 | ± | 1.1 | 89.0 | ± | 0.7 | 61.6 | ± | 1.3 | 31.7 | ± | 0.9 | N | 35.1 | ± | 1.1 | ||
| P. bretschneiderii | 558.1 | ± | 3.5 | 34.8 | ± | 1.7 | 85.8 | ± | 1.8 | 45.0 | ± | 1.5 | 29.6 | ± | 0.4 | N | 28.5 | ± | 0.8 | ||
| Origin ** | CV (s) | CV (ref) | 2C (pg DNA) | 1Cx (Mbp) | Width of the Leaves (mm) | Length of the Leaves (mm) | Length of the Petiole (mm) | Diameter of the Flowers (mm) | Length of the Pollen Grains (µm) | Length of the Stomata (µm) | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Alexander Lucas * | E | 5.1 | 4.9 | 1.75 | ± | 0.009 | 569.4 | ± | 2.9 | 49.2 | ± | 1.1 | 90.6 | ± | 3.8 | 51.6 | ± | 2.6 | 33.0 | ± | 1.0 | 46.6 | ± | 0.9 | 37.2 | ± | 1.2 |
| Anhuixueli | A | 4.3 | 3.2 | 1.14 | ± | 0.002 | 557.1 | ± | 1.1 | 60.2 | ± | 1.4 | 114.0 | ± | 2.8 | 62.0 | ± | 2.6 | 32.8 | ± | 0.4 | 38.6 | ± | 1.9 | 27.6 | ± | 0.8 |
| Baoshy | A | 3.9 | 3.1 | 1.13 | ± | 0.002 | 551.0 | ± | 1.2 | 61.8 | ± | 1.8 | 114.0 | ± | 6.3 | 49.0 | ± | 3.1 | 38.2 | ± | 1.0 | 48.5 | ± | 1.1 | 28.2 | ± | 1.2 |
| Benita | I | 4.4 | 3.4 | 1.12 | ± | 0.001 | 547.5 | ± | 0.4 | 46.8 | ± | 1.5 | 83.0 | ± | 2.5 | 60.8 | ± | 2.2 | 32.7 | ± | 0.6 | 43.8 | ± | 1.2 | 29.0 | ± | 1.1 |
| Bosc | E | 3.6 | 3.2 | 1.16 | ± | 0.002 | 565.4 | ± | 1.0 | 54.8 | ± | 1.9 | 85.0 | ± | 1.5 | 51.8 | ± | 4.0 | 41.5 | ± | 0.7 | 44.9 | ± | 2.3 | 32.5 | ± | 1.0 |
| Cangxixueli | A | 3.8 | 2.8 | 1.12 | ± | 0.003 | 549.3 | ± | 1.6 | 69.6 | ± | 2.1 | 146.6 | ± | 4.8 | 83.6 | ± | 4.8 | 38.6 | ± | 0.8 | 38.1 | ± | 0.6 | 30.1 | ± | 0.6 |
| Conference | E | 2.8 | 2.1 | 1.16 | ± | 0.004 | 565.6 | ± | 1.9 | 46.4 | ± | 2.5 | 83.6 | ± | 2.7 | 52.8 | ± | 5.2 | 27.1 | ± | 0.6 | 42.3 | ± | 1.0 | 31.1 | ± | 1.6 |
| Dangshanshuli | A | 3.9 | 2.9 | 1.14 | ± | 0.001 | 557.7 | ± | 0.4 | 87.8 | ± | 0.5 | 104.4 | ± | 2.9 | 78.4 | ± | 2.9 | 34.9 | ± | 0.7 | 48.8 | ± | 1.4 | 26.0 | ± | 1.0 |
| Dong Guo | A | 4.8 | 4.1 | 1.13 | ± | 0.003 | 551.3 | ± | 1.6 | 79.0 | ± | 1.6 | 126.0 | ± | 4.4 | 55.4 | ± | 7.4 | 42.2 | ± | 1.0 | 48.2 | ± | 1.1 | 30.7 | ± | 0.8 |
| Elektra | E | 3.6 | 3.6 | 1.15 | ± | 0.001 | 562.7 | ± | 0.2 | 42.2 | ± | 1.2 | 68.8 | ± | 0.7 | 37.0 | ± | 2.3 | 36.7 | ± | 0.7 | 43.6 | ± | 1.2 | 34.0 | ± | 0.6 |
| Hood | I | 3.3 | 2.9 | 1.14 | ± | 0.001 | 559.0 | ± | 0.3 | 51.6 | ± | 2.6 | 102.6 | ± | 2.9 | 57.0 | ± | 2.3 | 28.7 | ± | 0.6 | 39.2 | ± | 0.7 | 28.3 | ± | 1.1 |
| Huang Hua | A | 3.4 | 2.8 | 1.13 | ± | 0.004 | 550.3 | ± | 1.8 | 82.2 | ± | 2.1 | 135.8 | ± | 1.7 | 63.8 | ± | 1.2 | 34.2 | ± | 0.3 | 47.3 | ± | 0.9 | 32.9 | ± | 1.0 |
| Chang Ba | A | 3.2 | 3.0 | 1.13 | ± | 0.002 | 552.9 | ± | 0.7 | 74.6 | ± | 1.5 | 118.2 | ± | 2.5 | 76.2 | ± | 2.4 | 41.8 | ± | 0.6 | 48.0 | ± | 0.7 | 29.1 | ± | 0.8 |
| Chili | A | 4.1 | 2.9 | 1.13 | ± | 0.002 | 551.4 | ± | 1.0 | 82.6 | ± | 2.9 | 126.4 | ± | 5.2 | 72.6 | ± | 5.0 | 40.6 | ± | 0.5 | 37.7 | ± | 1.3 | 31.4 | ± | 1.4 |
| Jin Hua | A | 4.3 | 3.4 | 1.12 | ± | 0.001 | 549.5 | ± | 0.7 | 105.6 | ± | 6.7 | 181.6 | ± | 8.2 | 74.8 | ± | 3.3 | 43.3 | ± | 0.8 | 47.7 | ± | 0.6 | 25.5 | ± | 0.7 |
| Jing Bai | A | 4.5 | 3.4 | 1.14 | ± | 0.003 | 557.5 | ± | 1.5 | 72.6 | ± | 0.9 | 106.2 | ± | 3.5 | 77.8 | ± | 3.8 | 30.4 | ± | 0.7 | 39.0 | ± | 1.6 | 28.8 | ± | 1.3 |
| Karina | E | 4.9 | 4.1 | 1.16 | ± | 0.002 | 567.2 | ± | 1.0 | 36.2 | ± | 1.3 | 87.2 | ± | 3.0 | 41.8 | ± | 6.1 | 23.5 | ± | 0.5 | 45.3 | ± | 0.8 | 27.9 | ± | 1.4 |
| Kieffer | I | 3.1 | 2.7 | 1.15 | ± | 0.002 | 562.1 | ± | 1.2 | 58.6 | ± | 1.7 | 93.8 | ± | 2.3 | 41.6 | ± | 1.2 | 32.9 | ± | 0.5 | 41.8 | ± | 0.9 | 28.6 | ± | 1.1 |
| Kirgizskaja Zimnaja | E | 3.5 | 3.2 | 1.14 | ± | 0.001 | 559.3 | ± | 0.6 | 45.2 | ± | 0.2 | 93.0 | ± | 1.5 | 59.2 | ± | 2.7 | 29.4 | ± | 0.5 | N | 27.7 | ± | 1.6 | ||
| Kulovita | E | 3.2 | 2.7 | 1.15 | ± | 0.002 | 560.7 | ± | 0.9 | 35.6 | ± | 0.9 | 75.0 | ± | 1.6 | 51.8 | ± | 6.0 | 37.8 | ± | 1.0 | 47.6 | ± | 1.2 | 31.6 | ± | 1.1 |
| Mut Chen | A | 4.4 | 3.3 | 1.14 | ± | 0.000 | 557.9 | ± | 0.2 | 68.8 | ± | 0.9 | 103.2 | ± | 4.1 | 60.2 | ± | 2.7 | 34.6 | ± | 0.6 | 41.0 | ± | 1.2 | 28.0 | ± | 1.3 |
| Nanguoli | A | 4.1 | 3.5 | 1.15 | ± | 0.002 | 562.0 | ± | 1.2 | 68.6 | ± | 2.4 | 110.4 | ± | 2.4 | 70.0 | ± | 1.8 | 37.8 | ± | 1.3 | 42.7 | ± | 0.7 | 29.0 | ± | 0.8 |
| Nutika | A | 3.7 | 2.7 | 1.12 | ± | 0.002 | 549.2 | ± | 0.8 | 80.4 | ± | 1.4 | 105.2 | ± | 3.9 | 49.0 | ± | 3.4 | 35.9 | ± | 0.7 | N | 26.9 | ± | 1.1 | ||
| Oharkula | E | 4.4 | 3.2 | 1.13 | ± | 0.002 | 555.0 | ± | 1.2 | 56.4 | ± | 1.0 | 83.0 | ± | 2.8 | 68.0 | ± | 3.8 | 37.1 | ± | 1.5 | 43.0 | ± | 0.8 | 28.4 | ± | 1.4 |
| Shali | A | 4.8 | 4.9 | 1.14 | ± | 0.001 | 556.3 | ± | 0.6 | 77.4 | ± | 1.8 | 121.2 | ± | 2.6 | 58.0 | ± | 4.3 | 36.1 | ± | 1.0 | 49.2 | ± | 0.9 | 28.3 | ± | 0.8 |
| Shon Shu | A | 3.9 | 2.8 | 1.14 | ± | 0.001 | 558.1 | ± | 0.7 | 85.0 | ± | 3.5 | 117.0 | ± | 4.0 | 74.2 | ± | 7.3 | 28.0 | ± | 0.4 | 47.9 | ± | 1.2 | 27.7 | ± | 1.4 |
| Snow Flower | A | 4.8 | 3.5 | 1.13 | ± | 0.002 | 550.9 | ± | 1.1 | 85.0 | ± | 2.8 | 110.4 | ± | 1.5 | 68.8 | ± | 5.2 | 37.0 | ± | 0.4 | 49.8 | ± | 1.0 | 22.1 | ± | 0.7 |
| Talgarskaja krasavica | E | 4.3 | 3.2 | 1.15 | ± | 0.002 | 563.3 | ± | 1.0 | 65.4 | ± | 1.2 | 101.0 | ± | 3.5 | 49.0 | ± | 2.4 | 39.1 | ± | 0.8 | 41.3 | ± | 1.5 | 30.9 | ± | 1.1 |
| Williams Red | E | 2.9 | 2.5 | 1.15 | ± | 0.002 | 562.2 | ± | 1.2 | 41.2 | ± | 3.0 | 91.2 | ± | 2.3 | 40.6 | ± | 4.4 | 29.6 | ± | 0.7 | 44.4 | ± | 0.5 | 29.2 | ± | 1.4 |
| Wujiuxiang | I | 3.5 | 2.6 | 1.15 | ± | 0.001 | 562.7 | ± | 0.4 | 69.6 | ± | 2.9 | 120.0 | ± | 11.7 | 49.6 | ± | 5.1 | 31.4 | ± | 0.6 | 46.3 | ± | 1.3 | 28.3 | ± | 1.2 |
| Xin Gao | A | 3.8 | 3.2 | 1.12 | ± | 0.003 | 547.1 | ± | 1.7 | 80.2 | ± | 1.5 | 100.0 | ± | 2.5 | 43.0 | ± | 1.7 | 35.2 | ± | 0.6 | 59.0 | ± | 0.8 | 29.7 | ± | 0.6 |
| Yali | A | 4.2 | 3.2 | 1.13 | ± | 0.001 | 551.3 | ± | 0.3 | 81.6 | ± | 2.2 | 131.0 | ± | 2.7 | 65.6 | ± | 3.1 | 41.5 | ± | 1.3 | 48.2 | ± | 0.9 | 30.7 | ± | 1.2 |
| Zaosuli | A | 4.5 | 3.8 | 1.13 | ± | 0.001 | 554.3 | ± | 0.6 | 71.8 | ± | 2.0 | 130.2 | ± | 3.5 | 51.2 | ± | 3.6 | 36.5 | ± | 0.5 | 45.8 | ± | 1.1 | 26.6 | ± | 0.8 |
| Hcer1 | E | 3.2 | 2.6 | 1.13 | ± | 0.003 | 550.8 | ± | 1.4 | 48.2 | ± | 1.9 | 82.2 | ± | 3.2 | 53.4 | ± | 1.7 | 35.0 | ± | 0.7 | 45.0 | ± | 1.0 | 26.9 | ± | 0.7 |
| Hcer2 | E | 4.1 | 3.9 | 1.13 | ± | 0.002 | 551.4 | ± | 1.0 | 56.4 | ± | 1.2 | 75.4 | ± | 3.6 | 57.2 | ± | 3.4 | 38.4 | ± | 0.5 | 42.0 | ± | 1.2 | 31.7 | ± | 1.6 |
| Hcer3* | E | 2.0 | 2.7 | 1.79 | ± | 0.009 | 584.2 | ± | 3.0 | 65.2 | ± | 1.5 | 83.6 | ± | 1.9 | 71.4 | ± | 3.7 | 38.1 | ± | 0.9 | 46.1 | ± | 1.4 | 38.6 | ± | 0.7 |
| Hcer4 | E | 4.7 | 3.5 | 1.16 | ± | 0.001 | 567.5 | ± | 0.4 | 43.2 | ± | 1.1 | 75.4 | ± | 1.6 | 62.4 | ± | 2.0 | 34.8 | ± | 0.8 | 45.8 | ± | 0.7 | 30.0 | ± | 1.0 |
| H2 | I | 3.3 | 2.8 | 1.15 | ± | 0.002 | 560.5 | ± | 0.9 | 59.2 | ± | 2.2 | 94.8 | ± | 1.2 | 46.4 | ± | 3.5 | 32.9 | ± | 0.7 | N | 34.5 | ± | 1.4 | ||
| H4 | I | 3.7 | 2.6 | 1.13 | ± | 0.003 | 552.8 | ± | 1.5 | 58.8 | ± | 2.2 | 85.8 | ± | 4.4 | 46.4 | ± | 4.9 | 44.0 | ± | 0.7 | 40.6 | ± | 1.2 | 35.4 | ± | 1.4 |
| H7 | I | 3.8 | 2.7 | 1.14 | ± | 0.003 | 556.3 | ± | 1.6 | 84.8 | ± | 1.5 | 111.6 | ± | 2.2 | 54.0 | ± | 3.0 | 38.2 | ± | 0.6 | 47.5 | ± | 0.7 | 32.6 | ± | 1.0 |
| H8 | I | 4.2 | 3.1 | 1.13 | ± | 0.001 | 551.8 | ± | 0.4 | 69.6 | ± | 1.7 | 108.4 | ± | 2.0 | 60.2 | ± | 3.6 | 32.5 | ± | 0.5 | N | 33.1 | ± | 1.3 | ||
| H12 | I | 3.8 | 2.9 | 1.13 | ± | 0.001 | 553.1 | ± | 0.7 | 70.0 | ± | 2.3 | 118.0 | ± | 5.8 | 55.2 | ± | 4.1 | 38.0 | ± | 0.6 | 48.3 | ± | 1.1 | 31.7 | ± | 1.0 |
| H13 | I | 3.0 | 2.6 | 1.15 | ± | 0.002 | 562.5 | ± | 1.1 | 67.8 | ± | 2.0 | 105.6 | ± | 3.4 | 57.8 | ± | 1.6 | 36.2 | ± | 0.5 | 40.7 | ± | 0.8 | 31.7 | ± | 1.4 |
| H19 | I | 5.1 | 4.8 | 1.15 | ± | 0.001 | 562.8 | ± | 0.5 | 69.4 | ± | 2.9 | 93.2 | ± | 3.7 | 57.8 | ± | 4.0 | 38.4 | ± | 0.7 | N | 34.1 | ± | 1.9 | ||
| H24 | I | 4.2 | 3.0 | 1.16 | ± | 0.001 | 566.5 | ± | 0.7 | 55.6 | ± | 2.0 | 93.2 | ± | 3.2 | 75.8 | ± | 3.7 | 25.8 | ± | 0.8 | N | 30.0 | ± | 1.0 | ||
| Mbp | Width of the Leaves | Length of the Leaves | Length of the Petiole | Diameter of the Flowers | Length of the Pollen Grains | Length of the Stomata | |
|---|---|---|---|---|---|---|---|
| p | 0.00004 | <10−8 | <10−7 | 0.005 | 0.2 | 0.7 | 0.002 |
| Width of the Leaves (mm) | Length of the Leaves (mm) | Length of the Petiole (mm) | Diameter of the Flowers (mm) | Length of the Stomata (µm) | |
|---|---|---|---|---|---|
| Mbp | −0.4683 | −0.4947 | −0.1029 | −0.3198 | 0.4210 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fiala, J.; Zezulová, E.; Nečas, T. Evaluation of the Genome Size and Ploidy Level of Pears (Pyrus spp.) in Relation to Their Morphological Traits. Horticulturae 2024, 10, 1241. https://doi.org/10.3390/horticulturae10121241
Fiala J, Zezulová E, Nečas T. Evaluation of the Genome Size and Ploidy Level of Pears (Pyrus spp.) in Relation to Their Morphological Traits. Horticulturae. 2024; 10(12):1241. https://doi.org/10.3390/horticulturae10121241
Chicago/Turabian StyleFiala, Jonáš, Eliška Zezulová, and Tomáš Nečas. 2024. "Evaluation of the Genome Size and Ploidy Level of Pears (Pyrus spp.) in Relation to Their Morphological Traits" Horticulturae 10, no. 12: 1241. https://doi.org/10.3390/horticulturae10121241
APA StyleFiala, J., Zezulová, E., & Nečas, T. (2024). Evaluation of the Genome Size and Ploidy Level of Pears (Pyrus spp.) in Relation to Their Morphological Traits. Horticulturae, 10(12), 1241. https://doi.org/10.3390/horticulturae10121241







