The GCN4 Transcription Factor: A Review of Its Functional Progress in Fungi
Abstract
:1. Introduction
2. Research Progress on the GCN4-Mediated General Amino Acid Control (GAAC) Signaling Pathway
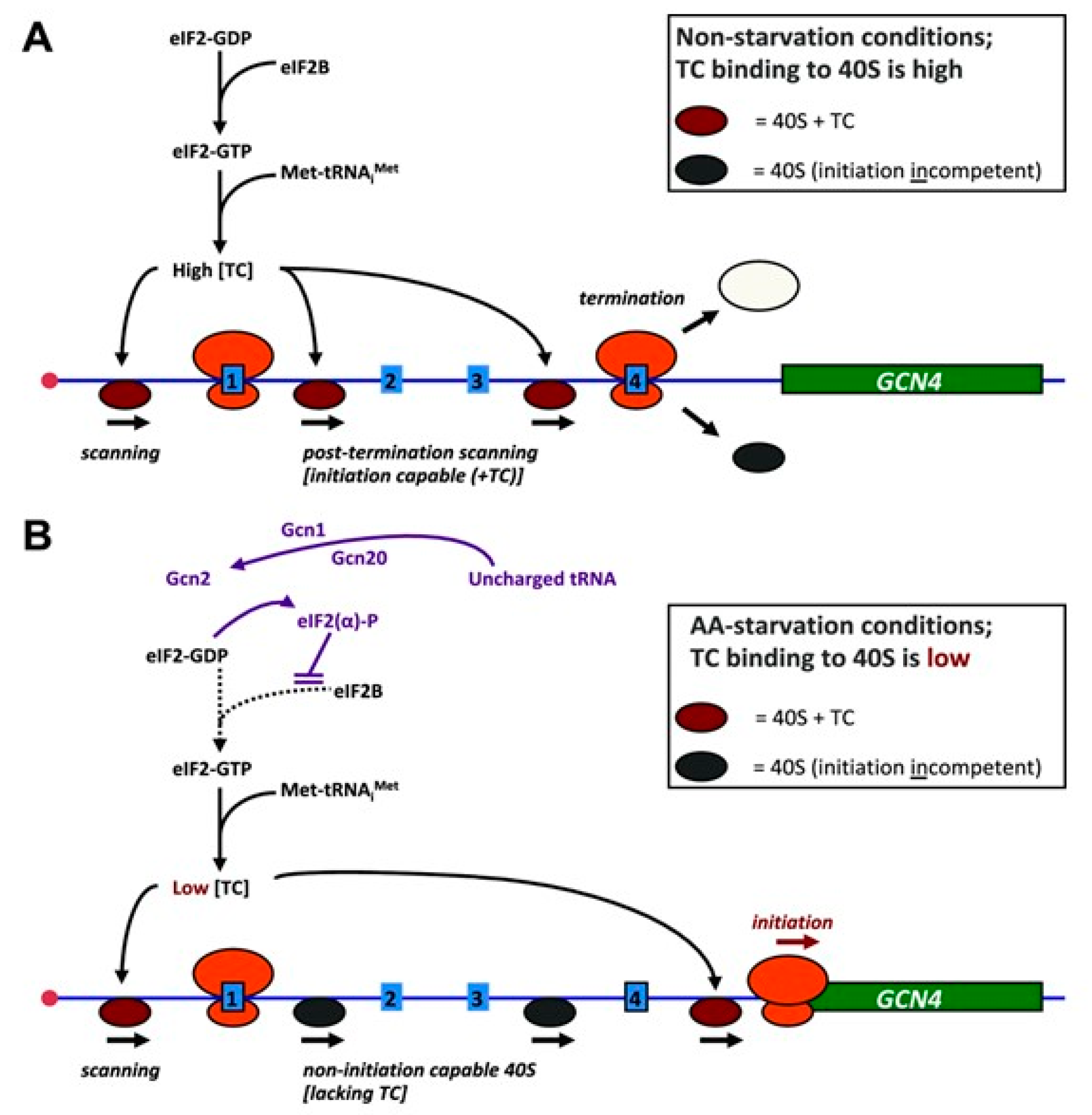
2.1. The Translation Mechanism of GCN4
2.2. The Stability of the GCN4 Protein
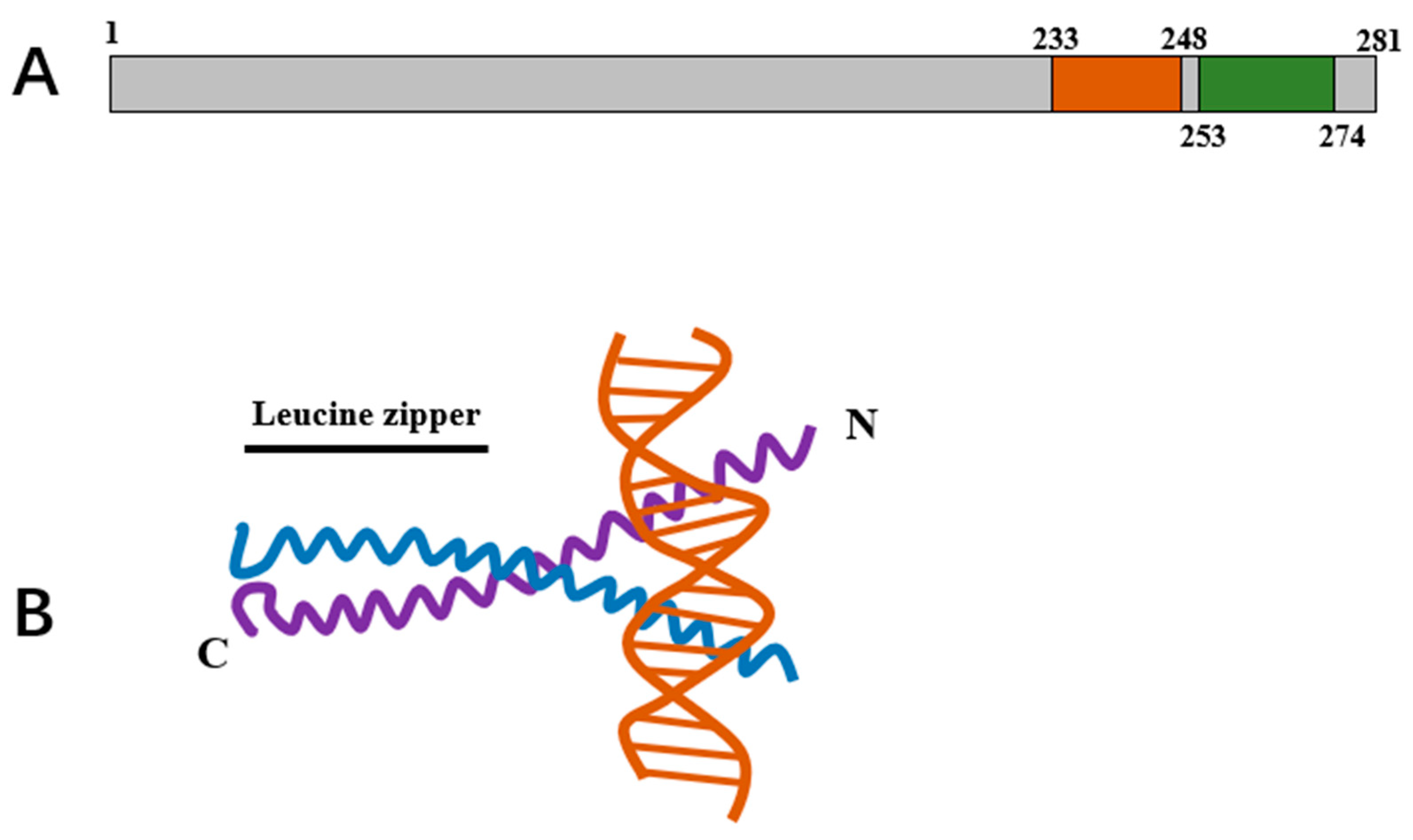
3. The Characteristics of Conserved Structural Domain in the Transcription Factor GCN4
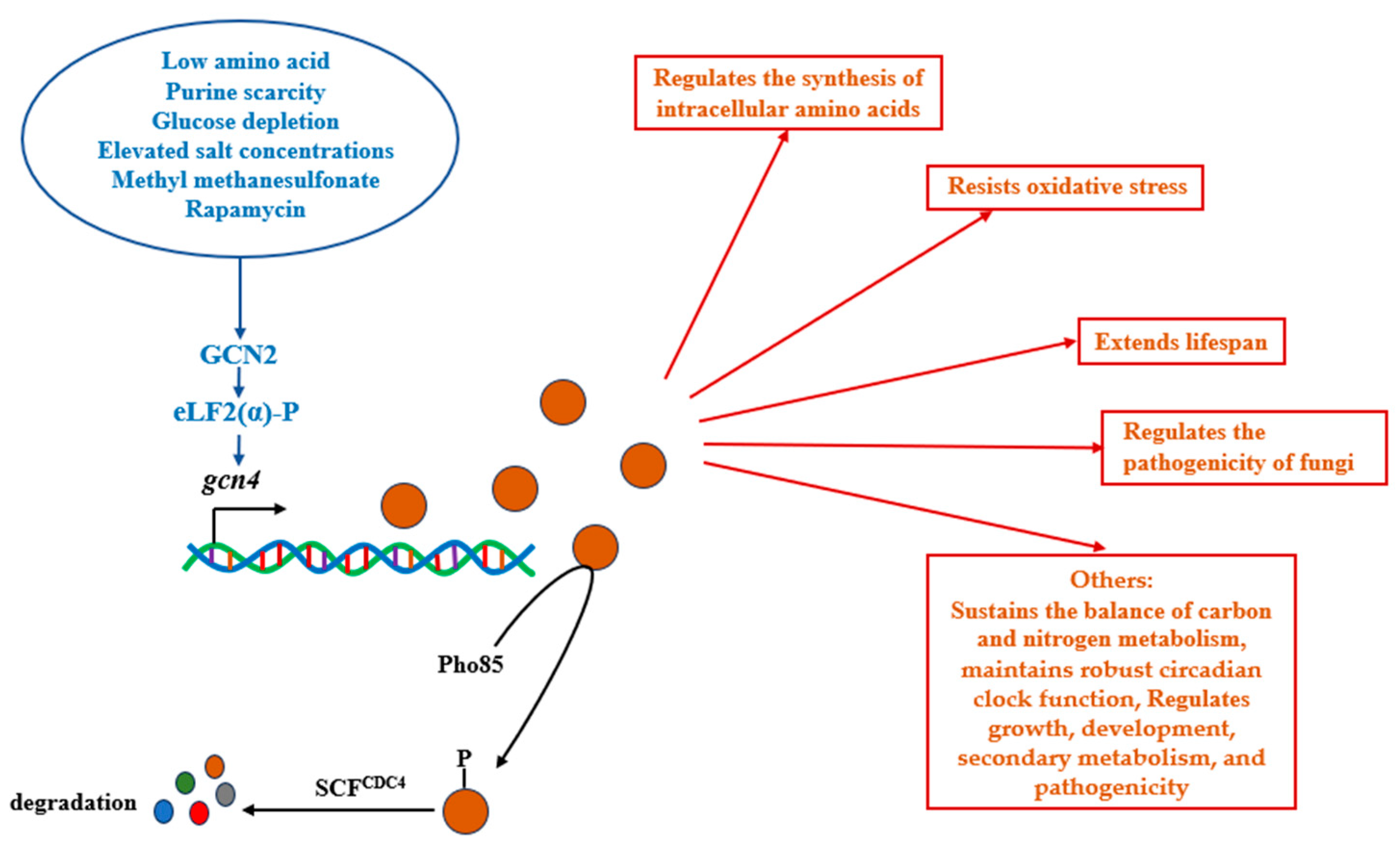
4. The Transcription Factor GCN4 Regulates the Physiological Processes of Fungi by Activating the Expression of Downstream Target Genes
4.1. The Transcription Factor GCN4 Responds to Nitrogen Starvation and Regulates the Synthesis of Intracellular Amino Acids
4.2. The Transcription Factor GCN4 Extends Lifespan
4.3. The Transcription Factor GCN4 Regulates Oxidative Stress
4.4. Transcription Factor GCN4 Regulates the Pathogenicity of Fungi
4.5. The Other Regulatory Functions of the Transcription Factor GCN4
5. The Research Progress on the Connection Between the Transcription Factor GCN4 and Other Signaling Pathways
6. Conclusions and Prospects
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Tudzynski, B. Nitrogen regulation of fungal secondary metabolism in fungi. Front. Microbiol. 2014, 5, 656. [Google Scholar] [CrossRef]
- Hinnebusch, A.G. The general control of amino acid biosynthetic genes in the Saccharomyces cerevisiae. CRC Crit. Rev. Biochem. 1986, 21, 277–317. [Google Scholar] [CrossRef]
- Niederberger, P.; Miozzari, G.; Hütter, R. Biological role of the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 1981, 1, 584–593. [Google Scholar] [CrossRef]
- Hoffmann, B.; Valerius, O.; Andermann, M.; Braus, G.H. Transcriptional autoregulation and inhibition of mRNA translation of amino acid regulator gene cpcA of filamentous fungus Aspergillus nidulans. Mol. Biol. Cell 2001, 12, 2846–2857. [Google Scholar] [CrossRef]
- Krappmann, S.; Bignell, E.M.; Reichard, U.; Rogers, T.; Haynes, K.; Braus, G.H. The Aspergillus fumigatus transcriptional activator CpcA contributes significantly to the virulence of this fungal pathogen. Mol. Microbiol. 2004, 52, 785–799. [Google Scholar] [CrossRef]
- Tian, C.; Kasuga, T.; Sachs, M.S.; Glass, N.L. Transcriptional profiling of cross pathway control in Neurospora crassa and comparative analysis of the Gcn4 and CPC1 regulons. Eukaryot. Cell 2007, 6, 1018–1029. [Google Scholar] [CrossRef]
- Liu, H.; Tang, X.; Zhang, N.; Li, S.; Si, H. Role of bZIP Transcription Factors in Plant Salt Stress. Int. J. Mol. Sci. 2023, 24, 7893. [Google Scholar] [CrossRef]
- Landschulz, W.H.; Johnson, P.F.; McKnight, S.L. The leucine zipper: A hypothetical structure common to a new class of DNA binding proteins. Science 1988, 240, 1759–1764. [Google Scholar] [CrossRef]
- Meng, X.; Reed, A.; Lai, S.; Szavits-Nossan, J.; McCarthy JE, G. Stochastic scanning events on the GCN4 mRNA 5′ untranslated region generate cell-to-cell heterogeneity in the Saccharomyces cerevisiae nutritional stress response. Nucleic Acids Res. 2023, 51, 6609–6621. [Google Scholar] [CrossRef] [PubMed]
- Williams, N.P.; Hinnebusch, A.G.; Donahue, T.F. Mutations in the structural genes for eukaryotic initiation factors 2 alpha and 2 beta of Saccharomyces cerevisiae disrupt translational control of GCN4 mRNA. Proc. Natl. Acad. Sci. USA 1989, 86, 7515–7519. [Google Scholar] [CrossRef] [PubMed]
- Mariner, B.L.; Felker, D.P.; Cantergiani, R.J.; Peterson, J.; McCormick, M.A. Multiomics of GCN4-Dependent Replicative Lifespan Extension Models Reveals Gcn4 as a Regulator of Protein Turnover in Saccharomyces cerevisiae. Int. J. Mol. Sci. 2023, 24, 16163. [Google Scholar] [CrossRef] [PubMed]
- Mascarenhas, C.; Edwards-Ingram, L.C.; Zeef, L.; Shenton, D.; Ashe, M.P.; Grant, C.M. Gcn4 is required for the response to peroxide stress in the Saccharomyces cerevisiae. Mol. Biol. Cell 2008, 19, 2995–3007. [Google Scholar] [CrossRef] [PubMed]
- Ebbole, D.J.; Paluh, J.L.; Plamann, M.; Sachs, M.S.; Yanofsky, C. cpc-1, the general regulatory gene for genes of amino acid biosynthesis in Neurospora crassa, is differentially expressed during the asexual life cycle. Mol. Cell. Biol. 1991, 11, 928–934. [Google Scholar] [PubMed]
- Tripathi, G.; Wiltshire, C.; Macaskill, S.; Tournu, H.; Budge, S.; Brown, A.J. Gcn4 co-ordinates morphogenetic and metabolic responses to amino acid starvation in Candida albicans. EMBO J. 2002, 21, 5448–5456. [Google Scholar] [CrossRef]
- Pan, Y.; Gao, L.; Zhang, X.; Qin, Y.; Liu, G.; Qu, Y. The Role of Cross-Pathway Control Regulator CpcA in the Growth and Extracellular Enzyme Production of Penicillium oxalicum. Curr. Microbiol. 2020, 77, 49–54. [Google Scholar] [CrossRef]
- Busch, S.; Bode, H.B.; Brakhage, A.A.; Braus, G.H. Impact of the cross-pathway control on the regulation of lysine and penicillin biosynthesis in Aspergillus nidulans. Curr. Genet. 2003, 42, 209–219. [Google Scholar] [CrossRef]
- Dever, T.E.; Feng, L.; Wek, R.C.; Cigan, A.M.; Donahue, T.F.; Hinnebusch, A.G. Phosphorylation of initiation factor 2 alpha by protein kinase GCN2 mediates gene-specific translational control of GCN4 in Saccharomyces cerevisiae. Cell 1992, 68, 585–596. [Google Scholar] [CrossRef]
- Dever, T.E.; Ivanov, I.P.; Hinnebusch, A.G. Translational regulation by uORFs and start codon selection stringency. Genes Dev. 2023, 37, 474–489. [Google Scholar] [CrossRef]
- Mueller, P.P.; Hinnebusch, A.G. Multiple upstream AUG codons mediate translational control of GCN4. Cell 1986, 45, 201–207. [Google Scholar] [CrossRef]
- Grant, C.M.; Miller, P.F.; Hinnebusch, A.G. Requirements for intercistronic distance and level of eukaryotic initiation factor 2 activity in reinitiation on GCN4 mRNA vary with the downstream cistron. Mol. Cell. Biol. 1994, 14, 2616–2628. [Google Scholar]
- Hinnebusch, A.G. Translational regulation of GCN4 and the general amino acid control of Saccharomyces cerevisiae. Annu. Rev. Microbiol. 2005, 59, 407–450. [Google Scholar] [CrossRef] [PubMed]
- Dey, M.; Velyvis, A.; Li, J.J.; Chiu, E.; Chiovitti, D.; Kay, L.E.; Sicheri, F.; Dever, T.E. Requirement for kinase-induced conformational change in eukaryotic initiation factor 2alpha (eIF2alpha) restricts phosphorylation of Ser51. Proc. Natl. Acad. Sci. USA 2011, 108, 4316–4321. [Google Scholar] [CrossRef]
- Albrecht, G.; Mösch, H.U.; Hoffmann, B.; Reusser, U.; Braus, G.H. Monitoring the Gcn4 protein-mediated response in the Saccharomyces cerevisiae. J. Biol. Chem. 1998, 273, 12696–12702. [Google Scholar] [CrossRef]
- Sundaram, A.; Grant, C.M. A single inhibitory upstream open reading frame (uORF) is sufficient to regulate Candida albicans GCN4 translation in response to amino acid starvation conditions. RNA 2014, 20, 559–567. [Google Scholar] [CrossRef]
- Garcia-Barrio, M.T.; Naranda, T.; Vazquez de Aldana, C.R.; Cuesta, R.; Hinnebusch, A.G.; Hershey, J.W.; Tamame, M. GCD10, a translational repressor of GCN4, is the RNA-binding subunit of eukaryotic translation initiation factor-3. Genes Dev. 1995, 9, 1781–1796. [Google Scholar] [CrossRef]
- Abastado, J.P.; Miller, P.F.; Jackson, B.M.; Hinnebusch, A.G. Suppression of ribosomal reinitiation at upstream open reading frames in amino acid-starved cells forms the basis for GCN4 translational control. Mol. Cell. Biol. 1991, 11, 486–496. [Google Scholar]
- Dver, T.E.; Yang, W.; Aström, S.; Byström, A.S.; Hinnebusch, A.G. Modulation of tRNA(iMet), eIF-2, and eIF-2B expression shows that GCN4 translation is inversely coupled to the level of eIF-2.GTP.Met-tRNA(iMet) ternary complexes. Mol. Cell. Biol. 1995, 15, 6351–6363. [Google Scholar] [CrossRef]
- Rolfes, R.J.; Hinnebusch, A.G. Translation of the Saccharomyces cerevisiae transcriptional activator GCN4 is stimulated by purine limitation: Implications for activation of the protein kinase GCN2. Mol. Cell. Biol. 1993, 13, 5099–5111. [Google Scholar]
- Marbach, I.; Licht, R.; Frohnmeyer, H.; Engelberg, D. Gcn2 mediates Gcn4 activation in response to glucose stimulation or UV radiation not via GCN4 translation. J. Biol. Chem. 2001, 276, 16944–16951. [Google Scholar] [CrossRef]
- Goossens, A.; Dever, T.E.; Pascual-Ahuir, A.; Serrano, R. The protein kinase Gcn2p mediates sodium toxicity in Saccharomyces cerevisiae. J. Biol. Chem. 2001, 276, 30753–30760. [Google Scholar] [CrossRef]
- Natarajan, K.; Meyer, M.R.; Jackson, B.M.; Slade, D.; Roberts, C.; Hinnebusch, A.G.; Marton, M.J. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001, 21, 4347–4368. [Google Scholar] [CrossRef]
- Valenzuela, L.; Aranda, C.; González, A. TOR modulates GCN4-dependent expression of genes turned on by nitrogen limitation. J. Bacteriol. 2001, 183, 2331–2334. [Google Scholar] [CrossRef]
- Gottesman, S.; Maurizi, M.R. Regulation by proteolysis: Energy-dependent proteases and their targets. Microbiol. Rev. 1992, 56, 592–621. [Google Scholar] [CrossRef]
- Schimke, R.T. Regulation of enzyme levels in animal tissues: The role of enzyme degradation. Birth Defects Orig. Artic. Ser. 1973, 9, 1–8. [Google Scholar]
- Kornitzer, D.; Raboy, B.; Kulka, R.G.; Fink, G.R. Regulated degradation of the transcription factor Gcn4. EMBO J. 1994, 13, 6021–6030. [Google Scholar] [CrossRef]
- Meimoun, A.; Holtzman, T.; Weissman, Z.; McBride, H.J.; Stillman, D.J.; Fink, G.R.; Kornitzer, D. Degradation of the transcription factor Gcn4 requires the kinase Pho85 and the SCF(CDC4) ubiquitin-ligase complex. Mol. Biol. Cell 2000, 11, 915–927. [Google Scholar] [CrossRef]
- Rawal, Y.; Qiu, H.; Hinnebusch, A.G. Accumulation of a threonine biosynthetic intermediate attenuates general amino acid control by accelerating degradation of Gcn4 via Pho85 and Cdk8. PLoS Genet. 2014, 10, e1004534. [Google Scholar] [CrossRef]
- Kyrpides, N.; Tavernarakis, N.; Papamatheakis, J.; Thireos, G. A transient GCN4 mRNA destabilization follows GCN4 translational derepression. J. Biol. Chem. 1995, 270, 17317–17320. [Google Scholar] [CrossRef]
- Jung, Y.; Seong, K.M.; Baek, J.H.; Kim, J. Ssb2 is a novel factor in regulating synthesis and degradation of Gcn4 in Saccharomyces cerevisiae. Mol. Microbiol. 2018, 110, 728–740. [Google Scholar] [CrossRef] [PubMed]
- Walvekar, A.S.; Kadamur, G.; Sreedharan, S.; Gupta, R.; Srinivasan, R.; Laxman, S. Methylated PP2A stabilizes Gcn4 to enable a methionine-induced anabolic program. J. Biol. Chem. 2020, 295, 18390–18405. [Google Scholar] [CrossRef] [PubMed]
- Váchová, L.; Plocek, V.; Maršíková, J.; Rešetárová, S.; Hatáková, L.; Palková, Z. Differential stability of Gcn4p controls its cell-specific activity in differentiated Saccharomyces cerevisiae colonies. mBio 2024, 15, e0068924. [Google Scholar] [CrossRef]
- Kouzarides, T.; Ziff, E. Leucine zippers of fos, jun and GCN4 dictate dimerization specificity and thereby control DNA binding. Nature 1989, 340, 568–571. [Google Scholar] [CrossRef]
- Vinson, C.R.; Sigler, P.B.; McKnight, S.L. Scissors-grip model for DNA recognition by a family of leucine zipper proteins. Science 1989, 246, 911–916. [Google Scholar] [CrossRef]
- O’Neil, K.T.; Hoess, R.H.; DeGrado, W.F. Design of DNA-binding peptides based on the leucine zipper motif. Science 1990, 249, 774–778. [Google Scholar] [CrossRef]
- Dahiyat, B.I.; Mayo, S.L. Protein design automation. Protein Sci. A Publ. Protein Soc. 1996, 5, 895–903. [Google Scholar] [CrossRef]
- O’Donoghue, S.I.; Junius, F.K.; King, G.F. Determination of the structure of symmetric coiled-coil proteins from NMR data: Application of the leucine zipper proteins Jun and GCN4. Protein Eng. 1993, 6, 557–564. [Google Scholar] [CrossRef]
- Cranz, S.; Berger, C.; Baici, A.; Jelesarov, I.; Bosshard, H.R. Monomeric and dimeric bZIP transcription factor GCN4 bind at the same rate to their target DNA site. Biochemistry 2004, 43, 718–727. [Google Scholar] [CrossRef]
- Metallo, S.J.; Schepartz, A. Certain bZIP peptides bind DNA sequentially as monomers and dimerize on the DNA. Nat. Struct. Biol. 1997, 4, 115–117. [Google Scholar] [CrossRef]
- Berger, C.; Piubelli, L.; Haditsch, U.; Bosshard, H.R. Diffusion-controlled DNA recognition by an unfolded, monomeric bZIP transcription factor. FEBS Lett. 1998, 425, 14–18. [Google Scholar] [CrossRef]
- Hollenbeck, J.J.; McClain, D.L.; Oakley, M.G. The role of helix stabilizing residues in GCN4 basic region folding and DNA binding. Protein Sci. A Publ. Protein Soc. 2002, 11, 2740–2747. [Google Scholar] [CrossRef]
- Rawal, Y.; Chereji, R.V.; Valabhoju, V.; Qiu, H.; Ocampo, J.; Clark, D.J.; Hinnebusch, A.G. Gcn4 Binding in Coding Regions Can Activate Internal and Canonical 5′ Promoters in Yeast. Mol. Cell 2018, 70, 297–311. [Google Scholar] [CrossRef]
- Coey, C.T.; Clark, D.J. A systematic genome-wide account of binding sites for the model transcription factor Gcn4. Genome Res. 2022, 32, 367–377. [Google Scholar] [CrossRef]
- Yuan, W.; Guo, S.; Gao, J.; Zhong, M.; Yan, G.; Wu, W.; Chao, Y.; Jiang, Y. General Control Nonderepressible 2 (GCN2) Kinase Inhibits Target of Rapamycin Complex 1 in Response to Amino Acid Starvation in Saccharomyces cerevisiae. J. Biol. Chem. 2017, 292, 2660–2669. [Google Scholar] [CrossRef]
- Vlahakis, A.; Graef, M.; Nunnari, J.; Powers, T. TOR complex 2-Ypk1 signaling is an essential positive regulator of the general amino acid control response and autophagy. Proc. Natl. Acad. Sci. USA 2014, 111, 10586–10591. [Google Scholar] [CrossRef]
- Ljungdahl, P.O.; Daignan-Fornier, B. Regulation of amino acid, nucleotide, and phosphate metabolism in Saccharomyces cerevisiae. Genetics 2012, 190, 885–929. [Google Scholar] [CrossRef]
- Ballester-Tomás, L.; Prieto, J.A.; Alepuz, P.; González, A.; Garre, E.; Randez-Gil, F. Inappropriate translation inhibition and P-body formation cause cold-sensitivity in tryptophan-auxotroph Saccharomyces cerevisiae mutants. Biochim. Et Biophys. Acta. Mol. Cell Res. 2017, 1864, 314–323. [Google Scholar] [CrossRef]
- Hinnebusch, A.G.; Natarajan, K. Gcn4p, a master regulator of gene expression, is controlled at multiple levels by diverse signals of starvation and stress. Eukaryot. Cell 2002, 1, 22–32. [Google Scholar] [CrossRef]
- Lian, L.; Qiao, J.; Guo, X.; Xing, Z.; Ren, A.; Zhao, M.; Zhu, J. The transcription factor GCN4 contributes to maintaining intracellular amino acid contents under nitrogen-limiting conditions in the mushroom Ganoderma lucidum. Microb. Cell Factories 2023, 22, 205. [Google Scholar] [CrossRef]
- Lian, L.; Shi, L.; Zhu, J.; Shi, L.; Ren, A.; You, H.; Liu, R.; Zhao, M. GCN4 Enhances the Transcriptional Regulation of AreA by Interacting with SKO1 To Mediate Nitrogen Utilization in Ganoderma lucidum. Appl. Environ. Microbiol. 2022, 88, e0132222. [Google Scholar] [CrossRef]
- Eckert, S.E.; Kübler, E.; Hoffmann, B.; Braus, G.H. The tryptophan synthase-encoding trpB gene of Aspergillus nidulans is regulated by the cross-pathway control system. Mol. Gen. Genet. MGG 2000, 263, 867–876. [Google Scholar] [CrossRef]
- Tavernarakis, N. Ageing and the regulation of protein synthesis: A balancing act? Trends Cell Biol. 2008, 18, 228–235. [Google Scholar] [CrossRef]
- Syntichaki, P.; Troulinaki, K.; Tavernarakis, N. Protein synthesis is a novel determinant of aging in Caenorhabditis elegans. Ann. N. Y. Acad. Sci. 2007, 1119, 289–295. [Google Scholar] [CrossRef]
- Hu, Z.; Xia, B.; Postnikoff, S.D.; Shen, Z.J.; Tomoiaga, A.S.; Harkness, T.A.; Seol, J.H.; Li, W.; Chen, K.; Tyler, J.K. Ssd1 and Gcn2 suppress global translation efficiency in replicatively aged Saccharomyces cerevisiae while their activation extends lifespan. eLife 2018, 7, e35551. [Google Scholar] [CrossRef]
- Mittal, N.; Guimaraes, J.C.; Gross, T.; Schmidt, A.; Vina-Vilaseca, A.; Nedialkova, D.D.; Aeschimann, F.; Leidel, S.A.; Spang, A.; Zavolan, M. The Gcn4 transcription factor reduces protein synthesis capacity and extends Saccharomyces cerevisiae lifespan. Nat. Commun. 2017, 8, 457. [Google Scholar] [CrossRef]
- Lei, Y.; Huang, T.; Jiang, G.; Zhang, Y.; Liu, S.; Li, H.; Lu, K. Transcriptional regulation of autophagy by RNA polymerase II. Autophagy 2023, 19, 1867–1868. [Google Scholar] [CrossRef]
- Kruegel, U.; Robison, B.; Dange, T.; Kahlert, G.; Delaney, J.R.; Kotireddy, S.; Tsuchiya, M.; Tsuchiyama, S.; Murakami, C.J.; Schleit, J.; et al. Elevated proteasome capacity extends replicative lifespan in Saccharomyces cerevisiae. PLoS Genet. 2011, 7, e1002253. [Google Scholar] [CrossRef]
- Steffen, K.K.; MacKay, V.L.; Kerr, E.O.; Tsuchiya, M.; Hu, D.; Fox, L.A.; Dang, N.; Johnston, E.D.; Oakes, J.A.; Tchao, B.N.; et al. Saccharomyces cerevisiae life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell 2008, 133, 292–302. [Google Scholar] [CrossRef]
- Delaney, J.R.; Ahmed, U.; Chou, A.; Sim, S.; Carr, D.; Murakami, C.J.; Schleit, J.; Sutphin, G.L.; An, E.H.; Castanza, A.; et al. Stress profiling of longevity mutants identifies Afg3 as a mitochondrial determinant of cytoplasmic mRNA translation and aging. Aging Cell 2013, 12, 156–166. [Google Scholar] [CrossRef]
- McCormick, M.A.; Delaney, J.R.; Tsuchiya, M.; Tsuchiyama, S.; Shemorry, A.; Sim, S.; Chou, A.C.; Ahmed, U.; Carr, D.; Murakami, C.J.; et al. A Comprehensive Analysis of Replicative Lifespan in 4698 Single-Gene Deletion Strains Uncovers Conserved Mechanisms of Aging. Cell Metab. 2015, 22, 895–906. [Google Scholar] [CrossRef]
- Robbins, C.E.; Patel, B.; Sawyer, D.L.; Wilkinson, B.; Kennedy, B.K.; McCormick, M.A. Cytosolic and mitochondrial tRNA synthetase inhibitors increase lifespan in a GCN4/atf-4-dependent manner. iScience 2022, 25, 105410. [Google Scholar] [CrossRef]
- Gulias, J.F.; Niesi, F.; Arán, M.; Correa-García, S.; Bermúdez-Moretti, M. Gcn4 impacts metabolic fluxes to promote Saccharomyces cerevisiae chronological lifespan. PLoS ONE 2023, 18, e0292949. [Google Scholar] [CrossRef]
- Scandalios, J.G. The rise of ROS. Trends Biochem. Sci. 2002, 27, 483–486. [Google Scholar] [CrossRef]
- Hansberg, W.; Salas-Lizana, R.; Domínguez, L. Fungal catalases: Function, phylogenetic origin and structure. Arch. Biochem. Biophys. 2012, 525, 170–180. [Google Scholar] [CrossRef]
- Zheng, Q.; Qiu, H.; Zhang, H.; Hinnebusch, A.G. Differential requirements for Gcn5 and NuA4 HAT activities in the starvation-induced versus basal transcriptomes. Nucleic Acids Res. 2023, 51, 3696–3721. [Google Scholar] [CrossRef]
- Kuo, M.H.; vom Baur, E.; Struhl, K.; Allis, C.D. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Mol. Cell 2000, 6, 1309–1320. [Google Scholar] [CrossRef]
- Qiu, H.; Chereji, R.V.; Hu, C.; Cole, H.A.; Rawal, Y.; Clark, D.J.; Hinnebusch, A.G. Genome-wide cooperation by HAT Gcn5, remodeler SWI/SNF, and chaperone Ydj1 in promoter nucleosome eviction and transcriptional activation. Genome Res. 2016, 26, 211–225. [Google Scholar] [CrossRef]
- Qi, S.; He, L.; Zhang, Q.; Dong, Q.; Wang, Y.; Yang, Q.; Tian, C.; He, Q.; Wang, Y. Cross-pathway control gene CPC1/GCN4 coordinates with histone acetyltransferase GCN5 to regulate catalase-3 expression under oxidative stress in Neurospora crassa. Free Radic. Biol. Med. 2018, 117, 218–227. [Google Scholar] [CrossRef]
- Zhou, Y.; Shen, S.; Du, C.; Wang, Y.; Liu, Y.; He, Q. A role for the mitotic proteins Bub3 and BuGZ in transcriptional regulation of catalase-3 expression. PLoS Genet. 2022, 18, e1010254. [Google Scholar] [CrossRef]
- Lian, L.; Wang, L.; Song, S.; Zhu, J.; Liu, R.; Shi, L.; Ren, A.; Zhao, M. GCN4 Regulates Secondary Metabolism through Activation of Antioxidant Gene Expression under Nitrogen Limitation Conditions in Ganoderma lucidum. Appl. Environ. Microbiol. 2021, 87, e0015621. [Google Scholar] [CrossRef] [PubMed]
- Timpner, C.; Braus-Stromeyer, S.A.; Tran, V.T.; Braus, G.H. The Cpc1 regulator of the cross-pathway control of amino acid biosynthesis is required for pathogenicity of the vascular pathogen Verticillium longisporum. Mol. Plant-Microbe Interact. MPMI 2013, 26, 1312–1324. [Google Scholar] [CrossRef] [PubMed]
- Elliott, C.E.; Fox, E.M.; Jarvis, R.S.; Howlett, B.J. The cross-pathway control system regulates production of the secondary metabolite toxin, sirodesmin PL, in the ascomycete, Leptosphaeria maculans. BMC Microbiol. 2011, 11, 169. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Larson, T.G.; Chen, C.H.; Pawlyk, D.M.; Clark, J.A.; Nuss, D.L. Cloning and characterization of a general amino acid control transcriptional activator from the chestnut blight fungus Cryphonectria parasitica. Fungal Genet. Biol. FG B 1998, 23, 81–94. [Google Scholar] [CrossRef]
- Hussain, S.; Tai, B.; Hussain, A.; Jahan, I.; Yang, B.; Xing, F. Genome-Wide Identification and Expression Analysis of the Basic Leucine Zipper (bZIP) Transcription Factor Gene Family in Fusarium graminearum. Genes 2022, 13, 607. [Google Scholar] [CrossRef]
- Zhao, K.; Liu, L.; Huang, S. Genome-Wide Identification and Functional Analysis of the bZIP Transcription Factor Family in Rice Bakanae Disease Pathogen, Fusarium fujikuroi. Int. J. Mol. Sci. 2022, 23, 6658. [Google Scholar] [CrossRef]
- Gai, Y.; Li, L.; Liu, B.; Ma, H.; Chen, Y.; Zheng, F.; Sun, X.; Wang, M.; Jiao, C.; Li, H. Distinct and essential roles of bZIP transcription factors in the stress response and pathogenesis in Alternaria alternata. Microbiol. Res. 2022, 256, 126915. [Google Scholar] [CrossRef]
- Zhang, C.C.; Zhou, C.Z.; Burnap, R.L.; Peng, L. Carbon/Nitrogen Metabolic Balance: Lessons from Cyanobacteria. Trends Plant Sci. 2018, 23, 1116–1130. [Google Scholar] [CrossRef]
- Yadav, K.K.; Rajasekharan, R. The transcription factor GCN4 regulates PHM8 and alters triacylglycerol metabolism in Saccharomyces cerevisiae. Curr. Genet. 2016, 62, 841–851. [Google Scholar] [CrossRef]
- Price-Lloyd, N.; Elvin, M.; Heintzen, C. Synchronizing the Neurospora crassa circadian clock with the rhythmic environment. Biochem. Soc. Trans. 2005, 33 Pt 5, 949–952. [Google Scholar] [CrossRef]
- Karki, S.; Castillo, K.; Ding, Z.; Kerr, O.; Lamb, T.M.; Wu, C.; Sachs, M.S.; Bell-Pedersen, D. Circadian clock control of eIF2α phosphorylation is necessary for rhythmic translation initiation. Proc. Natl. Acad. Sci. USA 2020, 117, 10935–10945. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.L.; Yang, Y.; Hu, Y.; Wu, J.; Han, C.; Lu, Q.; Gan, X.; Qi, S.; Guo, J.; He, Q.; et al. The nutrient-sensing GCN2 signaling pathway is essential for circadian clock function by regulating histone acetylation under amino acid starvation. eLife 2023, 12, e85241. [Google Scholar] [CrossRef] [PubMed]
- Loewith, R.; Hall, M.N. Target of rapamycin (TOR) in nutrient signaling and growth control. Genetics 2011, 189, 1177–1201. [Google Scholar] [CrossRef] [PubMed]
- Hara, K.; Yonezawa, K.; Weng, Q.P.; Kozlowski, M.T.; Belham, C.; Avruch, J. Amino acid sufficiency and mTOR regulate p70 S6 kinase and eIF-4E BP1 through a common effector mechanism. J. Biol. Chem. 1998, 273, 14484–14494. [Google Scholar] [CrossRef]
- Wang, X.; Campbell, L.E.; Miller, C.M.; Proud, C.G. Amino acid availability regulates p70 S6 kinase and multiple translation factors. Biochem. J. 1998, 334 Pt 1, 261–267. [Google Scholar] [CrossRef]
- Cherkasova, V.A.; Hinnebusch, A.G. Translational control by TOR and TAP42 through dephosphorylation of eIF2alpha kinase GCN2. Genes Dev. 2003, 17, 859–872. [Google Scholar] [CrossRef]
- Godard, P.; Urrestarazu, A.; Vissers, S.; Kontos, K.; Bontempi, G.; van Helden, J.; André, B. Effect of 21 different nitrogen sources on global gene expression in the Saccharomyces cerevisiae. Mol. Cell. Biol. 2007, 27, 3065–3086. [Google Scholar] [CrossRef]
- Mitchell, A.P.; Magasanik, B. Three regulatory systems control production of glutamine synthetase in Saccharomyces cerevisiae. Mol. Cell. Biol. 1984, 4, 2767–2773. [Google Scholar]
- Riego, L.; Avendaño, A.; DeLuna, A.; Rodríguez, E.; González, A. GDH1 expression is regulated by GLN3, GCN4, and HAP4 under respiratory growth. Biochem. Biophys. Res. Commun. 2002, 293, 79–85. [Google Scholar] [CrossRef]
- Ishida, C.; Aranda, C.; Valenzuela, L.; Riego, L.; Deluna, A.; Recillas-Targa, F.; Filetici, P.; López-Revilla, R.; González, A. The UGA3-GLT1 intergenic region constitutes a promoter whose bidirectional nature is determined by chromatin organization in Saccharomyces cerevisiae. Mol. Microbiol. 2006, 59, 1790–1806. [Google Scholar] [CrossRef]
- Staschke, K.A.; Dey, S.; Zaborske, J.M.; Palam, L.R.; McClintick, J.N.; Pan, T.; Edenberg, H.J.; Wek, R.C. Integration of general amino acid control and target of rapamycin (TOR) regulatory pathways in nitrogen assimilation in Saccharomyces cerevisiae. J. Biol. Chem. 2010, 285, 16893–16911. [Google Scholar] [CrossRef]
- Tate, J.J.; Buford, D.; Rai, R.; Cooper, T.G. General Amino Acid Control and 14-3-3 Proteins Bmh1/2 Are Required for Nitrogen Catabolite Repression-Sensitive Regulation of Gln3 and Gat1 Localization. Genetics 2017, 205, 633–655. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Yang, Y.; Chen, B.; Zhao, M.; Zhu, J. The GCN4 Transcription Factor: A Review of Its Functional Progress in Fungi. Horticulturae 2024, 10, 1113. https://doi.org/10.3390/horticulturae10101113
Li Y, Yang Y, Chen B, Zhao M, Zhu J. The GCN4 Transcription Factor: A Review of Its Functional Progress in Fungi. Horticulturae. 2024; 10(10):1113. https://doi.org/10.3390/horticulturae10101113
Chicago/Turabian StyleLi, Yanqiu, Yuzhen Yang, Bin Chen, Mingwen Zhao, and Jing Zhu. 2024. "The GCN4 Transcription Factor: A Review of Its Functional Progress in Fungi" Horticulturae 10, no. 10: 1113. https://doi.org/10.3390/horticulturae10101113
APA StyleLi, Y., Yang, Y., Chen, B., Zhao, M., & Zhu, J. (2024). The GCN4 Transcription Factor: A Review of Its Functional Progress in Fungi. Horticulturae, 10(10), 1113. https://doi.org/10.3390/horticulturae10101113




