Rust Disease Changes the Abundance and Composition of Bacterial Community in Iris lactea Rhizosphere
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Sites and Samples of Iris Lactea
2.2. Analysis of Amplicon Sequencing Data
2.3. Statistical Analysis
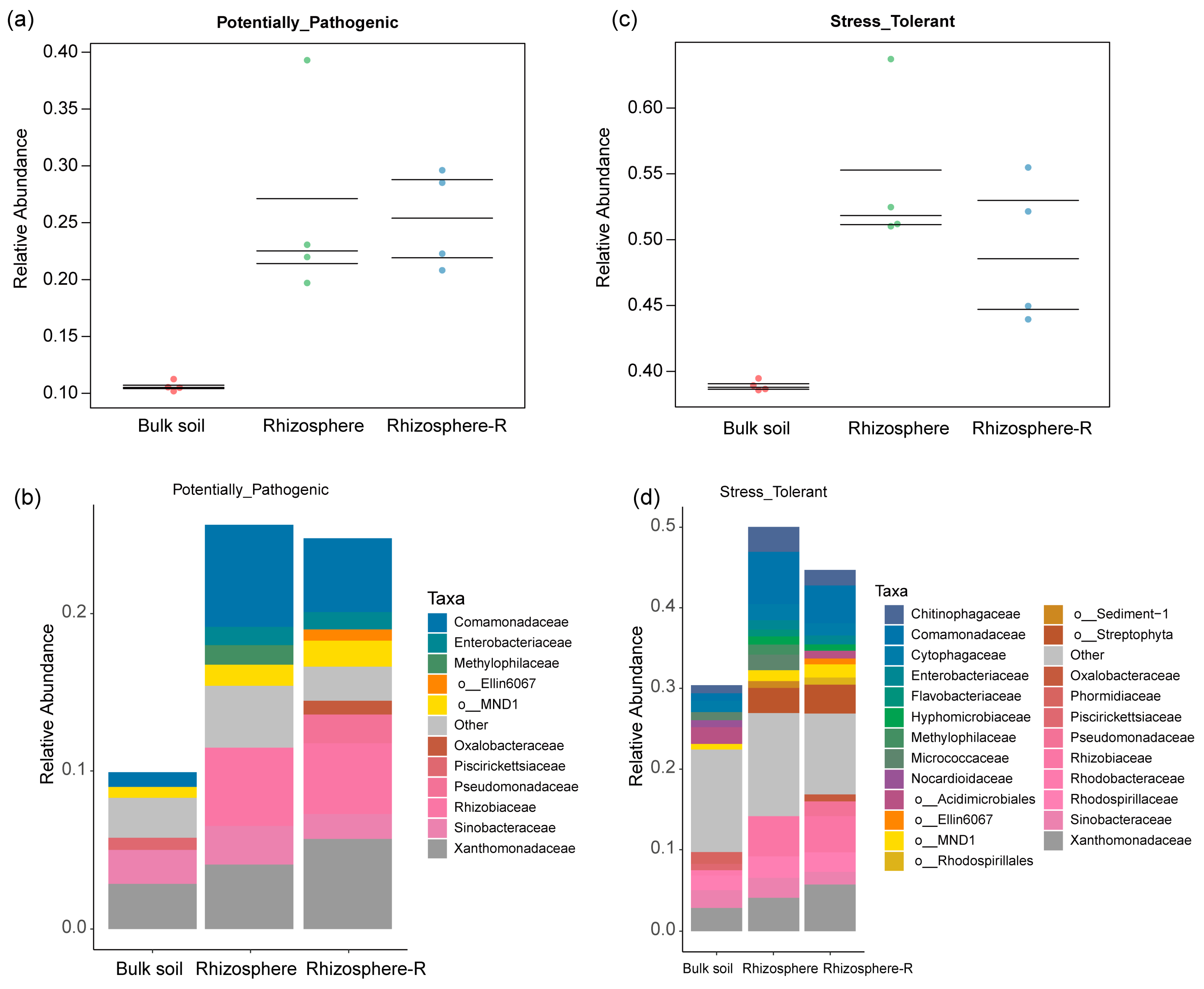
3. Results
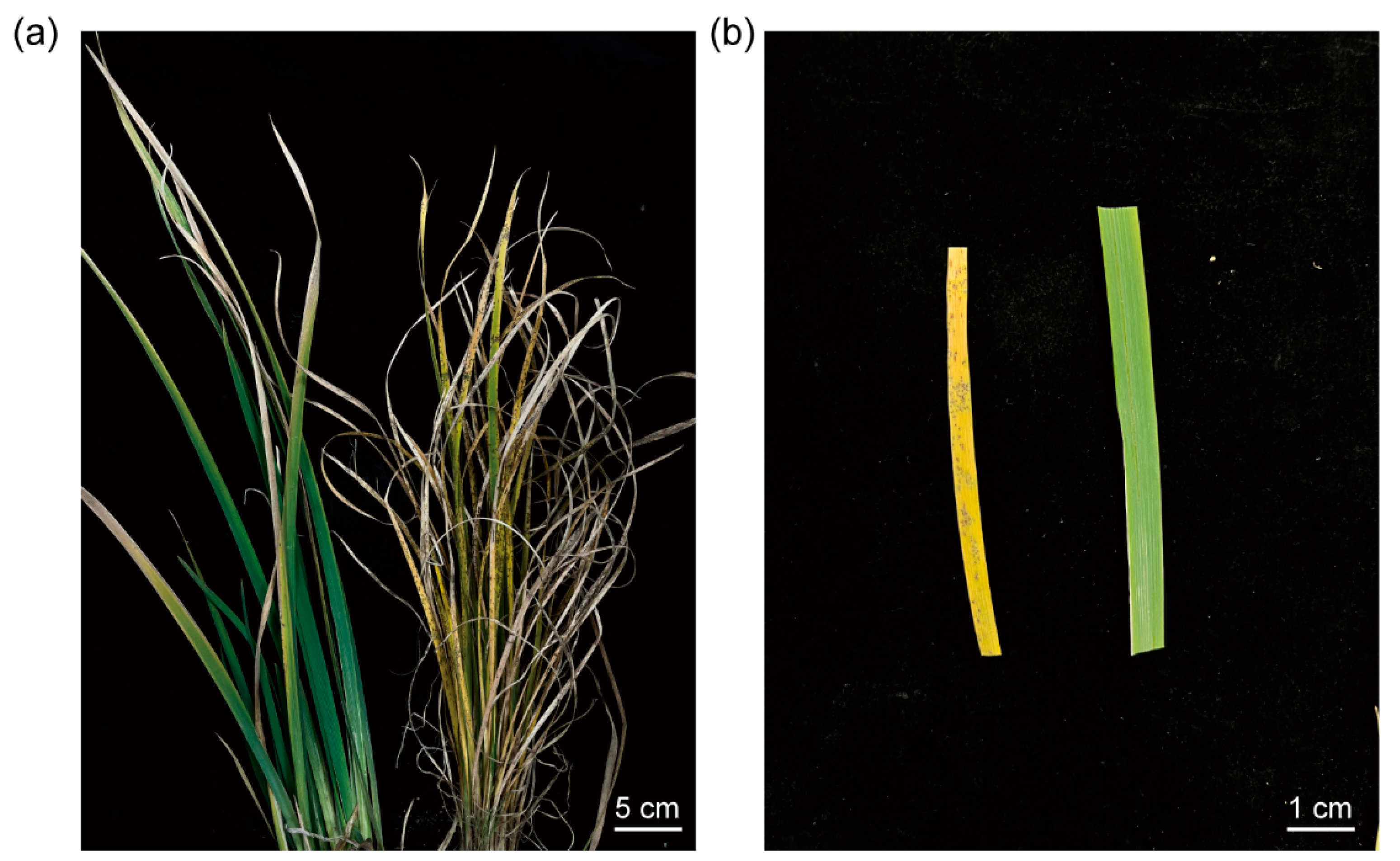
3.1. The Phenotype of Rust Disease and Raw Data Quality Control of Bacterial Community
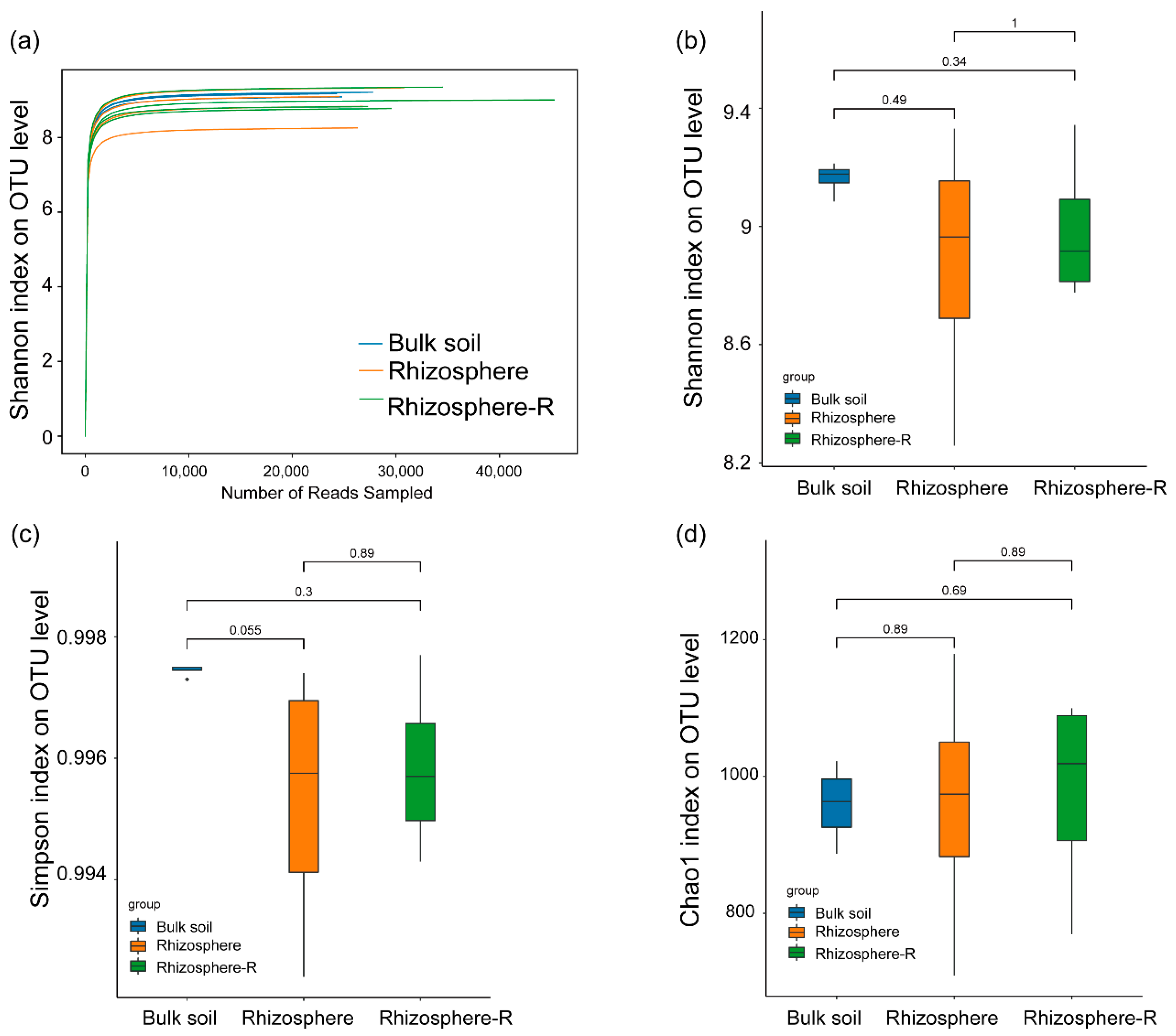
3.2. Microbial α-Diversity and Rarefaction Curve
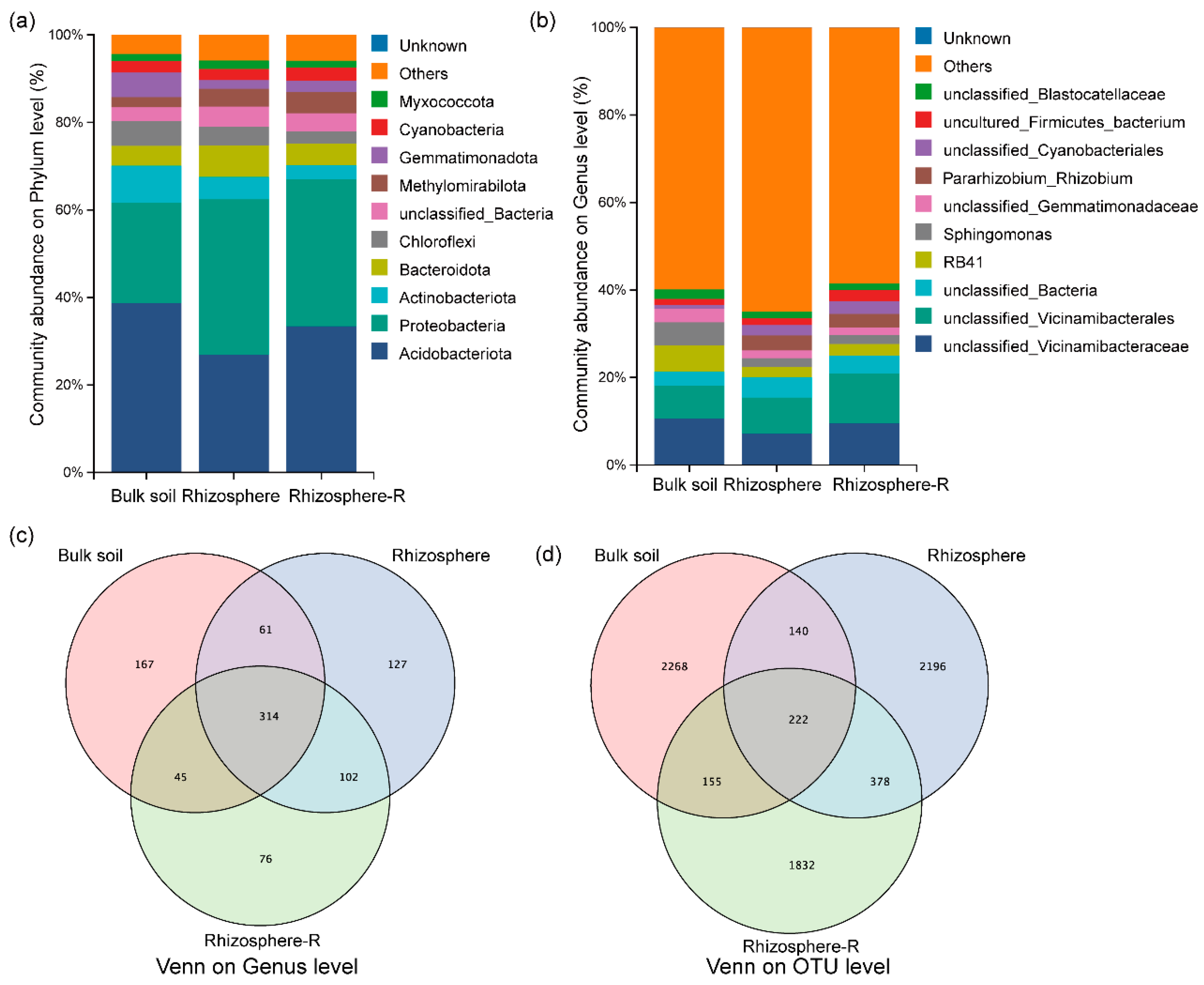
3.3. Composition of Bacterial Communities in the Rhizosphere
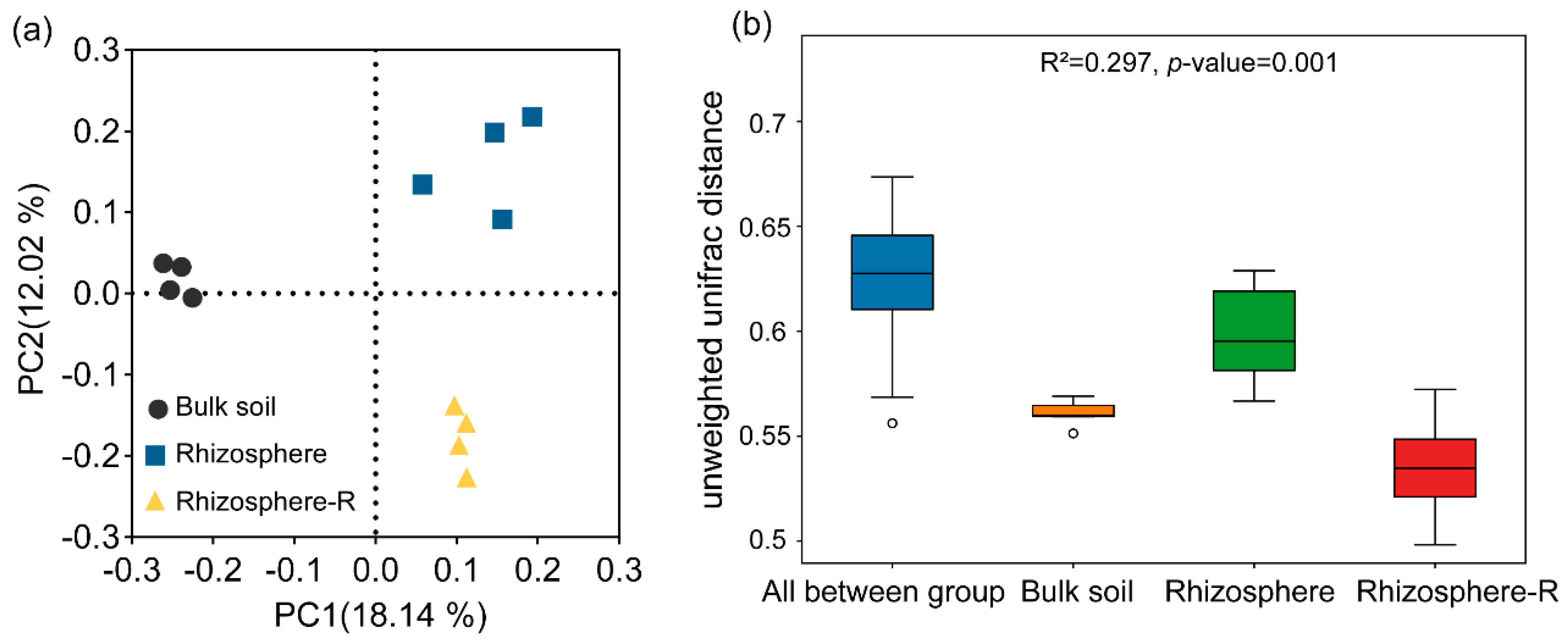
3.4. Rust Disease Alters the β-Diversity of Rhizosphere Soil
3.5. BugBase Phenotype Prediction of Rhizosphere Bacterial Community
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Berendsen, R.L.; Pieterse, C.M.J.; Bakker, P.A.H.M. The Rhizosphere Microbiome and Plant Health. Trends Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef]
- Curl, E.A.; Truelove, B. The Rhizosphere; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2012; ISBN 978-3-642-70722-3. [Google Scholar]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The Rhizosphere Microbiome: Significance of Plant Beneficial, Plant Pathogenic, and Hu man Pathogenic Microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef] [PubMed]
- Lugtenberg, B.; Kamilova, F. Plant-Growth-Promoting Rhizobacteria. Annu. Rev. Microbiol. 2009, 63, 541–556. [Google Scholar] [CrossRef] [PubMed]
- Berendsen, R.L.; Vismans, G.; Yu, K.; Song, Y.; de Jonge, R.; Burgman, W.P.; Burmølle, M.; Herschend, J.; Bakker, P.A.H.M.; Pieterse, C.M.J. Disease-Induced Assemblage of a Plant-Beneficial Bacterial Consortium. ISME J. 2018, 12, 1496–1507. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.; Zhao, J.; Wen, T.; Zhao, M.; Li, R.; Goossens, P.; Huang, Q.; Bai, Y.; Vivanco, J.M.; Kowalchuk, G.A.; et al. Root Exudates Drive the Soil-Borne Legacy of Aboveground Pathogen Infection. Microbiome 2018, 6, 156. [Google Scholar] [CrossRef]
- de Vries, F.T.; Wallenstein, M.D. Below-Ground Connections Underlying above-Ground Food Production: A Framework for Optimising Ecological Connections in the Rhizosphere. J. Ecol. 2017, 105, 913–920. [Google Scholar] [CrossRef]
- Fitzpatrick, C.R.; Copeland, J.; Wang, P.W.; Guttman, D.S.; Kotanen, P.M.; Johnson, M.T.J. Assembly and Ecological Function of the Root Microbiome across Angiosperm Plant Species. Proc. Natl. Acad. Sci. USA 2018, 115, E1157–E1165. [Google Scholar] [CrossRef]
- van der Heijden, M.G.A.; de Bruin, S.; Luckerhoff, L.; van Logtestijn, R.S.P.; Schlaeppi, K. A Widespread Plant-Fungal-Bacterial Sym biosis Promotes Plant Biodiversity, Plant Nutrition and Seedling Recruitment. ISME J. 2016, 10, 389–399. [Google Scholar] [CrossRef]
- Durán, P.; Thiergart, T.; Garrido-Oter, R.; Agler, M.; Kemen, E.; Schulze-Lefert, P.; Hacquard, S. Microbial Interkingdom Interactions in Roots Promote Arabidopsis Survival. Cell 2018, 175, 973–983.e14. [Google Scholar] [CrossRef]
- Helfer, S. Rust Fungi and Global Change. New Phytol. 2014, 201, 770–780. [Google Scholar] [CrossRef]
- Jin, Y.; Feng, G.; Luo, J.; Yan, H.; Sun, M.; Jing, T.; Yang, Y.; Jia, J.; Zhu, X.; Wang, X.; et al. Combined Genome-Wide Association Study and Transcriptome Analysis Reveal Candidate Genes for Resistance to Rust (Puccinia graminis) in Dactylis glomerata. Plant Dis. 2024, 108, 2197–2205. [Google Scholar] [CrossRef] [PubMed]
- Kong, H.G.; Song, G.C.; Sim, H.-J.; Ryu, C.-M. Achieving Similar Root Microbiota Composition in Neighbouring Plants through Air borne Signalling. ISME J. 2021, 15, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Yao, F.; Yang, Y.; Zhou, Y.; Hou, K.; Chen, Y.; Feng, D.; Wu, W. Analysis of the Rhizosphere Bacterial Diversity of Angel ica Dahurica Var. Formosana from Different Experimental Sites and Varieties (Strains). PeerJ 2023, 11, e15997. [Google Scholar] [CrossRef] [PubMed]
- Bardgett, R.D.; van der Putten, W.H. Belowground Biodiversity and Ecosystem Functioning. Nature 2014, 515, 505–511. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Mannaa, M.; Kim, N.; Jeon, H.W.; Jung, H.; Lee, H.-H.; Kim, J.; Park, J.; Park, A.R.; Kim, J.-C.; et al. Response of Pine Rhizo sphere Microbiota to Foliar Treatment with Resistance-Inducing Bacteria against Pine Wilt Disease. Microorganisms 2021, 9, 688. [Google Scholar] [CrossRef]
- De Tender, C.; Haegeman, A.; Vandecasteele, B.; Clement, L.; Cremelie, P.; Dawyndt, P.; Maes, M.; Debode, J. Dynamics in the Straw berry Rhizosphere Microbiome in Response to Biochar and Botrytis Cinerea Leaf Infection. Front. Microbiol. 2016, 7, 2062. [Google Scholar] [CrossRef]
- Sharma, I.; Kashyap, S.; Agarwala, N. Biotic Stress-Induced Changes in Root Exudation Confer Plant Stress Tolerance by Altering Rhi zospheric Microbial Community. Front. Plant Sci. 2023, 14, 1132824. [Google Scholar] [CrossRef]
- Fierer, N.; Bradford, M.A.; Jackson, R.B. Toward an Ecological Classification of Soil Bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef]
- Baud, A.; Kennedy, S.P. Targeted Metagenomic Databases Provide Improved Analysis of Microbiota Samples. Microorganisms 2024, 12, 135. [Google Scholar] [CrossRef]
- Wang, C.; Kuzyakov, Y. Mechanisms and Implications of Bacterial–Fungal Competition for Soil Resources. ISME J. 2024, 18, wrae073. [Google Scholar] [CrossRef]
- Walters, K.E.; Martiny, J.B.H. Alpha-, Beta-, and Gamma-Diversity of Bacteria Varies across Habitats. PLoS ONE 2020, 15, e0233872. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.J.; Ellingsen, K.E.; McArdle, B.H. Multivariate Dispersion as a Measure of Beta Diversity. Ecol. Lett. 2006, 9, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Niu, B.; Paulson, J.N.; Zheng, X.; Kolter, R. Simplified and Representative Bacterial Community of Maize Roots. Proc. Natl. Acad. Sci. USA 2017, 114, E2450–E2459. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Ge, J.; Li, D.; Ding, J.; Xiao, X.; Liang, Y. Microbial Coexistence in the Rhizosphere and the Promotion of Plant Stress Resistance: A Review. Environ. Res. 2023, 222, 115298. [Google Scholar] [CrossRef]
- Liu, H.; Brettell, L.E.; Qiu, Z.; Singh, B.K. Microbiome-Mediated Stress Resistance in Plants. Trends Plant Sci. 2020, 25, 733–743. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; Ver Loren van Themaat, E.; Schulze-Lefert, P. Structure and Functions of the Bacterial Micro biota of Plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef]
- Liu, H.; Macdonald, C.A.; Cook, J.; Anderson, I.C.; Singh, B.K. An Ecological Loop: Host Microbiomes across Multitrophic Interactions. Trends Ecol. Evol. 2019, 34, 1118–1130. [Google Scholar] [CrossRef]
- La, S.; Li, J.; Ma, S.; Liu, X.; Gao, L.; Tian, Y. Protective Role of Native Root-Associated Bacterial Consortium against Root-Knot Nematode Infection in Susceptible Plants. Nat. Commun. 2024, 15, 6723. [Google Scholar] [CrossRef] [PubMed]
- Carrión, V.J.; Perez-Jaramillo, J.; Cordovez, V.; Tracanna, V.; de Hollander, M.; Ruiz-Buck, D.; Mendes, L.W.; van Ijcken, W.F.J.; Gomez-Exposito, R.; Elsayed, S.S.; et al. Pathogen-Induced Activation of Disease-Suppressive Functions in the Endophytic Root Microbiome. Science 2019, 366, 606–612. [Google Scholar] [CrossRef] [PubMed]
- Yin, C.; Casa Vargas, J.M.; Schlatter, D.C.; Hagerty, C.H.; Hulbert, S.H.; Paulitz, T.C. Rhizosphere Community Selection Reveals Bacteria Associated with Reduced Root Disease. Microbiome 2021, 9, 86. [Google Scholar] [CrossRef] [PubMed]
- Agler, M.T.; Ruhe, J.; Kroll, S.; Morhenn, C.; Kim, S.-T.; Weigel, D.; Kemen, E.M. Microbial Hub Taxa Link Host and Abiotic Factors to Plant Microbiome Variation. PLoS Biol. 2016, 14, e1002352. [Google Scholar] [CrossRef] [PubMed]
- Mendes, R.; Kruijt, M.; de Bruijn, I.; Dekkers, E.; van der Voort, M.; Schneider, J.H.M.; Piceno, Y.M.; DeSantis, T.Z.; Andersen, G.L.; Bakker, P.A.H.M.; et al. Deciphering the Rhizosphere Microbiome for Disease-Suppressive Bacteria. Science 2011, 332, 1097–1100. [Google Scholar] [CrossRef]
- Xiong, C.; Zhu, Y.-G.; Wang, J.-T.; Singh, B.; Han, L.-L.; Shen, J.-P.; Li, P.-P.; Wang, G.-B.; Wu, C.-F.; Ge, A.-H.; et al. Host Selection Shapes Crop Microbiome Assembly and Network Complexity. New Phytol. 2021, 229, 1091–1104. [Google Scholar] [CrossRef]
- Leff, J.W.; Bardgett, R.D.; Wilkinson, A.; Jackson, B.G.; Pritchard, W.J.; De Long, J.R.; Oakley, S.; Mason, K.E.; Ostle, N.J.; Johnson, D.; et al. Predicting the Structure of Soil Communities from Plant Community Taxonomy, Phylogeny, and Traits. ISME J. 2018, 12, 1794–1805. [Google Scholar] [CrossRef]
- Stefani, E.; Obradović, A.; Gašić, K.; Altin, I.; Nagy, I.K.; Kovács, T. Bacteriophage-Mediated Control of Phytopathogenic Xanthomonads: A Promising Green Solution for the Future. Microorganisms 2021, 9, 1056. [Google Scholar] [CrossRef]
- Ali, S.; Tyagi, A.; Bae, H. Plant Microbiome: An Ocean of Possibilities for Improving Disease Resistance in Plants. Microorganisms 2023, 11, 392. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Zhang, X. Rust Disease Changes the Abundance and Composition of Bacterial Community in Iris lactea Rhizosphere. Horticulturae 2024, 10, 1065. https://doi.org/10.3390/horticulturae10101065
Zhang H, Zhang X. Rust Disease Changes the Abundance and Composition of Bacterial Community in Iris lactea Rhizosphere. Horticulturae. 2024; 10(10):1065. https://doi.org/10.3390/horticulturae10101065
Chicago/Turabian StyleZhang, Haiyan, and Xu Zhang. 2024. "Rust Disease Changes the Abundance and Composition of Bacterial Community in Iris lactea Rhizosphere" Horticulturae 10, no. 10: 1065. https://doi.org/10.3390/horticulturae10101065
APA StyleZhang, H., & Zhang, X. (2024). Rust Disease Changes the Abundance and Composition of Bacterial Community in Iris lactea Rhizosphere. Horticulturae, 10(10), 1065. https://doi.org/10.3390/horticulturae10101065



