Chip Digital PCR (cdPCR) to Identify and Quantify Botrytis cinerea Infection in Tomatoes
Abstract
1. Introduction
- In comparison with qPCR-based protocols already available, cdPCR does not require standard reference and is less sensitive to PCR inhibitors;
- There are several commercially accessible dPCR systems available at the moment, and they all use different techniques to obtain absolute quantification. A comparatively new technique for dPCR is called Quant Studio® 3D digital PCR. It involves loading a PCR sample onto a microchip, where it is spread among 20,000 reaction wells, enabling the execution of 20,000 distinct PCR reactions. The Quant Studio® technology allows for absolute quantification without requiring reference to a standard control by using Poisson statistical analysis of fluorescent signals from positive and negative wells. Previous studies adopted a droplet digital PCR (ddPCR) platform for B. cinerea quantification. The availability of a new protocol based on Quant Studio® 3D digital PCR can be beneficial for laboratories that use such a platform and for comparisons among methods;
- Previous studies were focused on the B. cinerea quantification in fruits such as strawberries and cherries. The cdPCR assay developed in this study is directed at diagnostics for tomatoes. Both tomato samples, naturally contaminated or spiked with fungus, were analyzed.
2. Materials and Methods
2.1. Fungal Samples
2.2. Plant Samples
2.3. DNA Extraction and Quantification
2.4. Primers and Probes
2.5. Chip Digital PCR for B. cinerea Diagnostic in Tomatoes
3. Results
- Test samples, obtained by spiking tomato DNA with B. cinerea DNA dilutions;
- Tomato seedlings obtained from commercial seed stocks;
- Tomato seedlings artificially contaminated with B. cinerea.
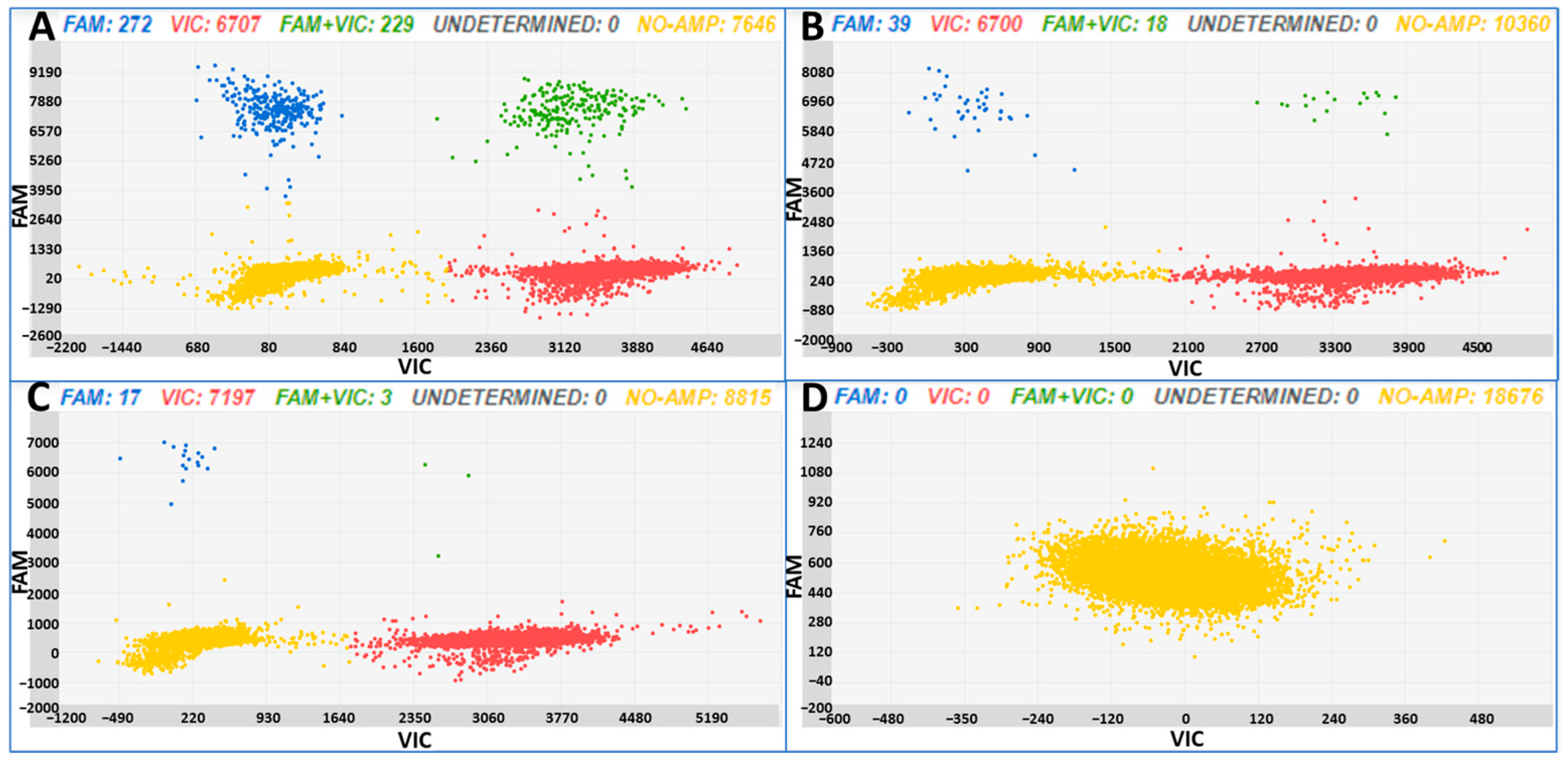
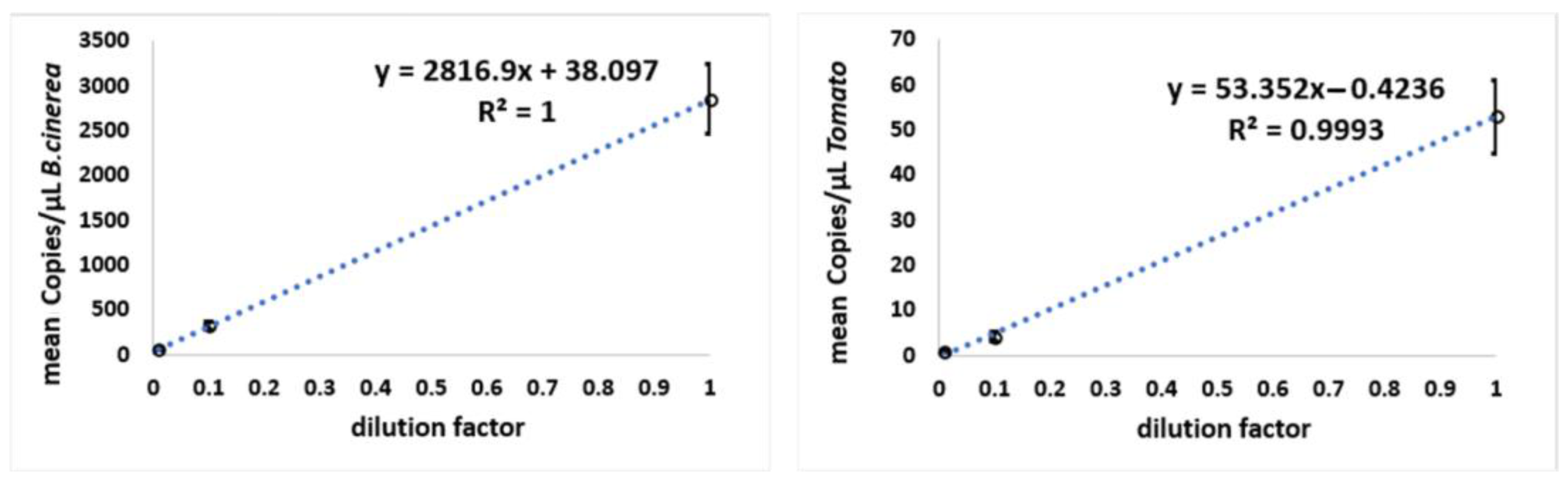
3.1. Test Samples, Obtained Spiking Tomato DNA with B. cinerea DNA Dilutions
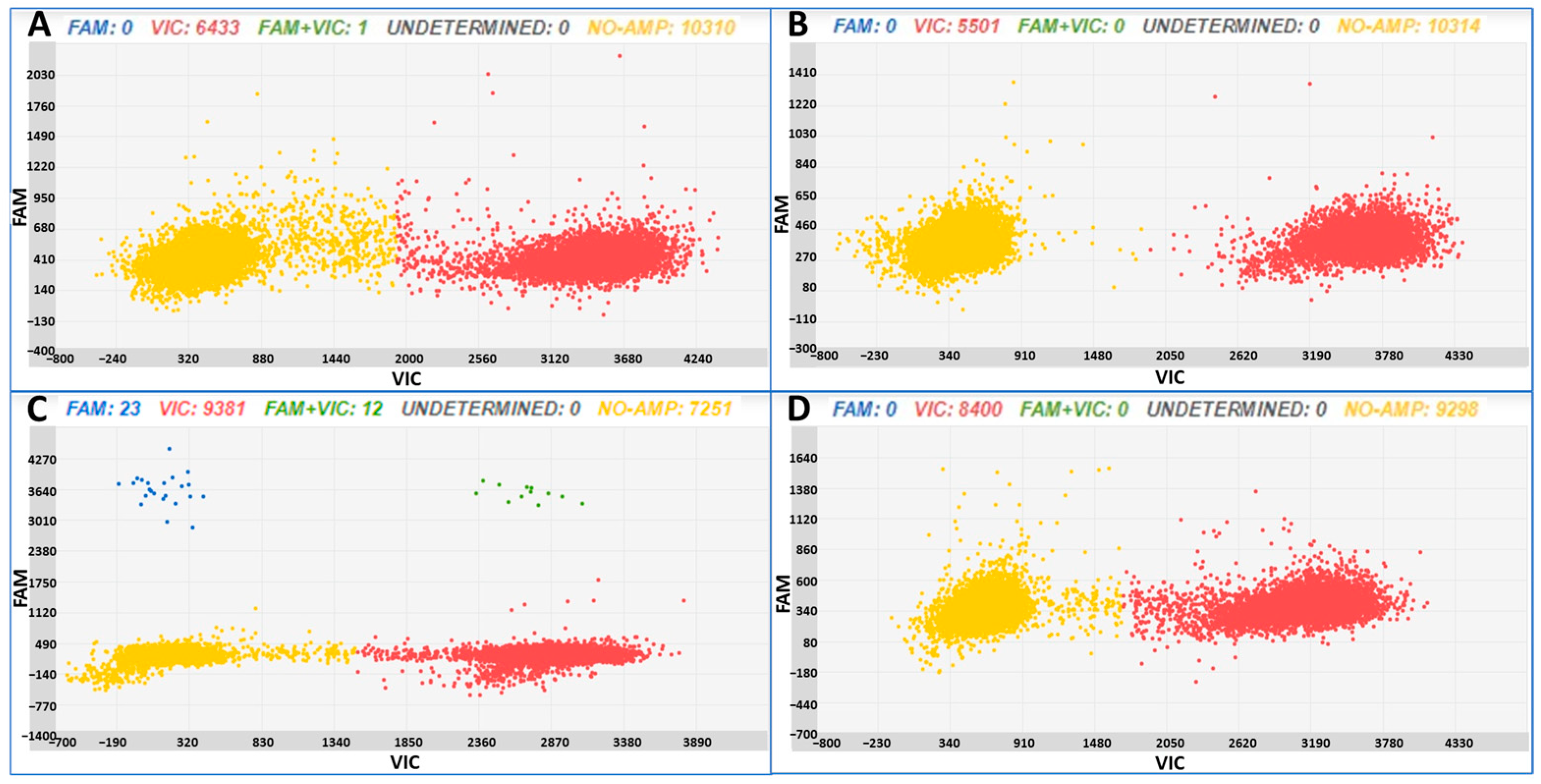
3.2. Tomato Seedling Samples Obtained from Commercial Seed Stocks
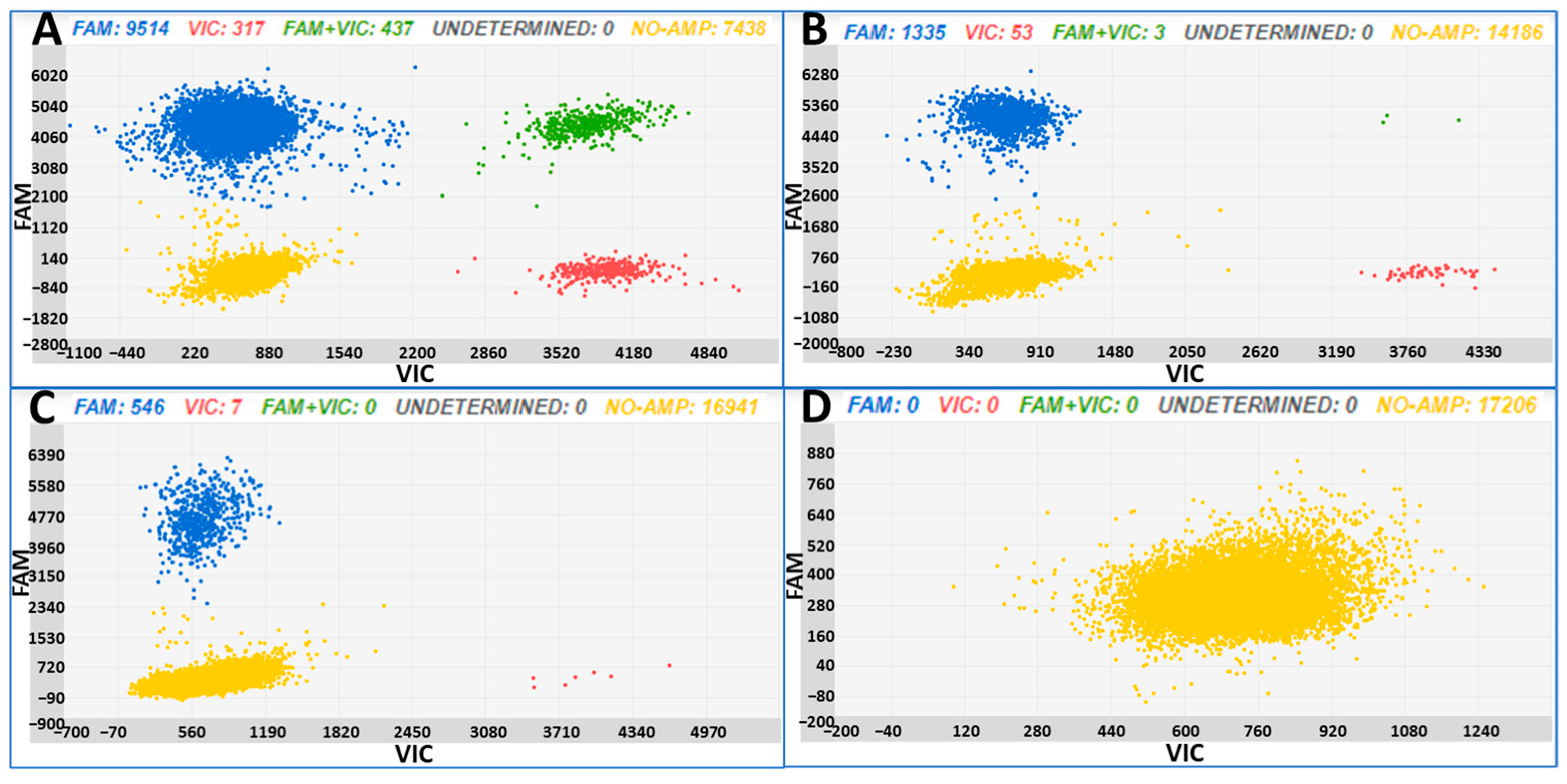
3.3. Tomato Seedlings Artificially Contaminated with B. cinerea
4. Discussion
“it is crucial that the diagnostic assays are thoroughly validated regarding specificity and sensitivity, not only with pure cultures or pure DNA samples, but also with plant samples spiked with the target pathogen”Venbrux et al. [36]
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Williamson, B.; Tudzynski, B.; Tudzynski, P.; Van Kan, J.A. Botrytis cinerea: The cause of grey mould disease. Mol. Plant Pathol. 2007, 8, 561–580. [Google Scholar] [PubMed]
- Romanazzi, G.; Smilanick, J.L.; Feliziani, E.; Droby, S. Integrated management of postharvest gray mold on fruit crops. Postharvest Biol. Technol. 2016, 113, 69–76. [Google Scholar] [CrossRef]
- Weiberg, A.; Wang, M.; Lin, F.M.; Zhao, H.; Zhang, Z.; Kaloshian, I.; Huang, H.D.; Jin, H. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 2013, 342, 118–123. [Google Scholar] [CrossRef]
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Hernández, A.; Rodríguez, A.; Córdoba, M.G.; Martín, A.; Ruiz-Moyano, S. Fungal control in foods through biopreservation. Curr. Opin. Food Sci. 2022, 47, 100904. [Google Scholar] [CrossRef]
- Suarez, M.B.; Walsh, K.; Boonham, N.; O’Neill, T.; Pearson, S.; Barker, I. Development of real-time PCR (TaqMan®) assays for the detection and quantification of Botrytis cinerea in planta. Plant Physiol. Biochem. 2005, 43, 890–899. [Google Scholar] [CrossRef]
- Leyronas, C.; Duffaud, M.; Nicot, P.C. Compared efficiency of the isolation methods for Botrytis cinerea. Mycology 2012, 3, 221–225. [Google Scholar]
- Luchi, N.; Ioos, R.; Santini, A. Fast and reliable molecular methods to detect fungal pathogens in woody plants. Appl. Microbiol. Biotechnol. 2020, 104, 2453–2468. [Google Scholar]
- Morcia, C.; Tumino, G.; Gasparo, G.; Ceresoli, C.; Fattorini, C.; Ghizzoni, R.; Carnevali, P.; Terzi, V. Moving from qPCR to chip digital PCR assays for tracking of some Fusarium species causing Fusarium Head Blight in cereals. Microorganisms 2020, 8, 1307. [Google Scholar] [CrossRef]
- Hindson, C.M.; Chevillet, J.R.; Briggs, H.A.; Gallichotte, E.N.; Ruf, I.K.; Hindson, B.J.; Vessella, R.L.; Tewari, M. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods 2013, 10, 1003–1005. [Google Scholar] [CrossRef]
- Strain, M.C.; Lada, S.M.; Luong, T.; Rought, S.E.; Gianella, S.; Terry, V.H.; Spina, C.A.; Woelk, C.H.; Richman, D.D. Highly precise measurement of HIV DNA by droplet digital PCR. PLoS ONE 2013, 8, e55943. [Google Scholar] [CrossRef] [PubMed]
- Morcia, C.; Ghizzoni, R.; Delogu, C.; Andreani, L.; Carnevali, P.; Terzi, V. Digital PCR: What relevance to plant studies? Biology 2020, 9, 433. [Google Scholar] [CrossRef]
- Diguta, C.F.; Rousseaux, S.; Weidmann, S.; Bretin, N.; Vincent, B.; Guilloux-Benatier, M.; Alexandre, H. Development of a qPCR assay for specific quantification of Botrytis cinerea on grapes. FEMS Microbiol. Lett. 2010, 313, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Si Ammour, M.; Fedele, G.; Morcia, C.; Terzi, V.; Rossi, V. Quantification of Botrytis cinerea in grapevine bunch trash by real-time PCR. Phytopathology 2019, 109, 1312–1319. [Google Scholar] [CrossRef] [PubMed]
- Moretti, C.; Quaglia, M.; Cerri, M.; Nicosia, D.E.; Buonaurio, R. A real-time PCR assay for detection and quantification of Botrytis cinerea in Pelargonium x hortorum plants, and its use for evaluation of plant resistance. Eur. J. Plant Pathol. 2015, 143, 159–171. [Google Scholar] [CrossRef]
- Carmichael, P.C.; Siyoum, N.; Jongman, M.; Korsten, L. Prevalence of Botrytis cinerea at different phenological stages of table grapes grown in the northern region of South Africa. Sci. Hortic. 2018, 239, 57–63. [Google Scholar] [CrossRef]
- Larrabee, M.M.; Voegel, T.M.; Nelson, L.M. A duplex droplet digital PCR assay for quantification of Alternaria spp. and Botrytis cinerea on sweet cherry at different growth stages. Can. J. Plant Pathol. 2021, 43, 734–748. [Google Scholar] [CrossRef]
- Panno, S.; Davino, S.; Caruso, A.G.; Bertacca, S.; Crnogorac, A.; Mandić, A.; Noris, E.; Matić, S. A review of the most common and economically important diseases that undermine the cultivation of tomato crop in the mediterranean basin. Agronomy 2021, 11, 2188. [Google Scholar] [CrossRef]
- Available online: http://dati.istat.it/Index.aspx?QueryId=33703 (accessed on 29 September 2023).
- Al-Samarrai, T.H.; Schmid, J. A simple method for extraction of fungal genomic DNA. Lett. Appl. Microbiol. 2000, 30, 53–56. [Google Scholar] [CrossRef]
- Morcia, C.; Piazza, I.; Ghizzoni, R.; Terzi, V.; Carrara, I.; Bolli, G.; Chiusa, G. Molecular diagnostics in tomato: Chip digital PCR assays targeted to identify and quantify Clavibacter michiganensis subsp. michiganensis and Ralstonia solanacearum in planta. Horticulturae 2023, 9, 553. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Collier, R.; Dasgupta, K.; Xing, Y.P.; Hernandez, B.T.; Shao, M.; Rohozinski, D.; Kovak, E.; Lin, J.; de Oliveira, M.L.P.; Stover, E.; et al. Accurate measurement of transgene copy number in crop plants using droplet digital PCR. Plant J. 2017, 90, 1014–1025. [Google Scholar] [CrossRef] [PubMed]
- Hougs, L.; Gatto, F.; Goerlich, O.; Grohmann, L.; Lieske, K.; Mazzara, M.; Narendja, F.; Ovesna, J.; Papazova, N.; Scholtens, I.M.J.; et al. Verification of analytical methods. In Testing and Analysis of GMO-Containing Foods and Feed; Mahgoub, S.E.O., Nollet, L.M.L., Eds.; CRC Press: Boca Raton, FL, USA, 2017; pp. 245–266. ISBN 978-1-315-17859-2. [Google Scholar]
- Wakeham, A.; Langton, A.; Adams, S.; Kennedy, R. Interface of the environment and occurrence of Botrytis cinerea in pre-symptomatic tomato crops. Crop Prot. 2016, 90, 27–33. [Google Scholar]
- Yahaya, S.M.; Mardiyya, A.Y.; Sakina, S.B.; Hayatu, L.W. Disease cycle and infection strategies of systemic plant pathogen Botrytis cinerea. Nov. Res. Microbiol. J. 2019, 3, 204–214. [Google Scholar]
- El Oirdi, M.; El Rahman, T.A.; Rigano, L.; El Hadrami, A.; Rodriguez, M.C.; Daayf, F.; Vojnov, A.; Bouarab, K. Botrytis cinerea manipulates the antagonistic effects between immune pathways to promote disease development in tomato. Plant Cell 2011, 23, 2405–2421. [Google Scholar] [CrossRef]
- Cheung, N.; Tian, L.; Liu, X.; Li, X. The destructive fungal pathogen Botrytis cinerea—Insights from genes studied with mutant analysis. Pathogens 2020, 9, 923. [Google Scholar] [CrossRef] [PubMed]
- Cotoras, M.; Silva, E. Differences in the initial events of infection of Botrytis cinerea strains isolated from tomato and grape. Mycologia 2005, 97, 485–492. [Google Scholar] [CrossRef]
- Bilkiss, M.; Shiddiky, M.J.A.; Ford, R. Advanced diagnostic approaches for necrotrophic fungal pathogens of temperate legumes with a focus on Botrytis spp. Front. Microbiol. 2019, 10, 1889. [Google Scholar]
- Chilvers, M.I.; du Toit, L.J.; Akamatsu, H.; Peever, T.L. A real-time, quantitative PCR seed assay for Botrytis spp. that cause neck rot of onion. Plant Dis. 2007, 91, 599–608. [Google Scholar] [CrossRef]
- Carisse, O.; Tremblay, D.M.; Lévesque, C.A.; Gindro, K.; Ward, P.; Houde, A. Development of a TaqMan real-time PCR assay for quantification of airborne conidia of Botrytis squamosa and management of Botrytis leaf blight of onion. Phytopathology 2009, 99, 1273–1280. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, M.D.; Li, G.Q.; Yang, L.; Yu, L.; Jiang, D.H.; Huang, H.C.; Zhuang, W.Y. Botrytis fabiopsis, a new species causing chocolate spot of broad bean in central China. Mycologia 2010, 102, 1114–1126. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Zhang, J.; Yang, L.; Wu, M.; Chen, W.; Li, G. Development of PCR-based assays for detecting and differentiating three species of Botrytis infecting broad bean. Plant Dis. 2015, 99, 691–698. [Google Scholar] [CrossRef]
- Malarczyk, D.G.; Panek, J.; Frąc, M. Triplex real-time PCR approach for the detection of crucial fungal berry pathogens—Botrytis spp., Colletotrichum spp. and Verticillium spp. Int. J. Mol. Sci. 2020, 21, 8469. [Google Scholar] [CrossRef] [PubMed]
- Venbrux, M.; Crauwels, S.; Rediers, H. Current and emerging trends in techniques for plant pathogen detection. Front. Plant Sci. 2023, 14, 1120968. [Google Scholar] [PubMed]
- Dreischhoff, S.; Das, I.S.; Häffner, F.; Wolf, A.M.; Polle, A.; Kasper, K.H. Fast and easy bioassay for the necrotizing fungus Botrytis cinerea on poplar leaves. Plant Methods 2023, 19, 32. [Google Scholar] [CrossRef] [PubMed]
- West, J.; Kimber, R. Innovations in air sampling to detect plant pathogens. Ann. Appl. Biol. 2015, 166, 4–17. [Google Scholar]
- Quesada, T.; Hughes, J.; Smith, K.; Shin, K.; James, P.; Smith, J. A low-cost spore trap allows collection and Real-Time PCR quantification of airborne Fusarium circinatum spores. Forests 2018, 9, 586. [Google Scholar] [CrossRef]
- Klosterman, S.J.; Anchieta, A.; McRoberts, N.; Koike, S.T.; Subbarao, K.V.; Voglmayr, H.; Choi, Y.J.; Thines, M.; Martin, F.N. Coupling spore traps and quantitative PCR assays for detection of the Downy Mildew pathogens of spinach (Peronospora effusa) and beet (P. schachtii). Phytopathology 2014, 104, 1349–1359. [Google Scholar] [CrossRef]
| Assay ID | Primers and Probes ID | Primers and Probes Sequences | Biological Target | Target Gene | Amplicon Size | References |
|---|---|---|---|---|---|---|
| Tom-dig | Tom-F | gcaatatcaagagccccgtc | Solanum lycopersicum | Prosystemin GenBank: M84800.1.1 | 91 bp | [21,23] |
| Tom-R | ggagcgcttagcacacat | |||||
| Tom-pr | VIC-tgcaacatccttctttcttctcgtg-MGB | |||||
| BC3-dig | BC3-F | gctgtaatttcaatgtgcagaatcc | Botrytis cinerea | Ribosomal IGS spacer GenBank: AM233400.1 | 94 bp | [6] |
| BC3-R | ggagcaacaattaatcgcatttc | |||||
| BC3-Pr | FAM-tcaccttgcaatgagtgg-MGB |
| Sample | B. cinerea ng | Tomato ng | Copies/µL B. cinerea ± SD | CI Copies/µL B. cinerea | Copies/µL Tomato ± SD | CI Copies/µL Tomato |
|---|---|---|---|---|---|---|
| A | 0.001 | 20 | 46.23 ± 3.02 | 42.39–50.42 | 836.24 ± 18.14 | 816.41–856.54 |
| B | 0.0001 | 20 | 4.73 ± 0.97 | 3.68–6.08 | 662.27 ± 13.13 | 646.42–678.52 |
| C | 0.00001 | 20 | 2.89 ± 0.78 | 2.08–4.03 | 792.90 ± 14.8 | 774.47–811.77 |
| D | 0 | 0 | 0 | 0 | 0 | 0 |
| Sample | Varieties | Tomato ng | Copies/µL B. cinerea ± SD | CI Copie/µL B. cinerea | Copies/µL Tomato ± SD | CI Copie/µL Tomato |
|---|---|---|---|---|---|---|
| A | Mariner | 20 | 0.27 ± 0.26 | 0.08–0.73 | 644.08 ± 14.6 | 628.35–660.21 |
| B | Sailor | 20 | 0 | – | 568.13 ± 12.9 | 553.17–583.50 |
| C | Rossoro | 20 | 3.49 ± 0.87 | 2.60–4.70 | 1102.50 ± 12.2 | 1097.70–1125.70 |
| D | Wilson | 20 | 0 | – | 856 ± 17.1 | 837.70–875.11 |
| Sample | Total DNA (ng) | Copies/µL B. cinerea ± SD | CI Copie/µL B. cinerea | Copies/µL Tomato ± SD | CI Copie/µL Tomato |
|---|---|---|---|---|---|
| A | 2 | 1097.70 ± 17.98 | 1075.7–1120.2 | 56.65 ± 3.71 | 53.63–61.92 |
| B | 0.2 | 119.07 ± 4.79 | 112.86–125.63 | 4.95 ± 1.07 | 3.81–6.40 |
| C | 0.02 | 42.08 ± 2.68 | 38.69–45.76 | 0.53 ± 0.39 | 0.25–1.11 |
| D | 0 | 0 | – | 0 | – |
| Dilution Factor | Copies/µL B. cinerea ± SD | CI Copies/µL B. cinerea | Copies/µL Tomato ± SD | CI Copies/µL Tomato |
|---|---|---|---|---|
| 10 | 2854 ± 384.1 | 2592–3423 | 53 ± 8.12 | 43–61 |
| 327 ± 40.81 | 281–380 | 4 ± 0.98 | 3–5 | |
| 59 ± 10.01 | 47–71 | 0.8 ± 0.24 | 0.51–1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morcia, C.; Carrara, I.; Ghizzoni, R.; Terzi, V.; Bolli, G.; Chiusa, G. Chip Digital PCR (cdPCR) to Identify and Quantify Botrytis cinerea Infection in Tomatoes. Horticulturae 2024, 10, 91. https://doi.org/10.3390/horticulturae10010091
Morcia C, Carrara I, Ghizzoni R, Terzi V, Bolli G, Chiusa G. Chip Digital PCR (cdPCR) to Identify and Quantify Botrytis cinerea Infection in Tomatoes. Horticulturae. 2024; 10(1):91. https://doi.org/10.3390/horticulturae10010091
Chicago/Turabian StyleMorcia, Caterina, Ilaria Carrara, Roberta Ghizzoni, Valeria Terzi, Giovanni Bolli, and Giorgio Chiusa. 2024. "Chip Digital PCR (cdPCR) to Identify and Quantify Botrytis cinerea Infection in Tomatoes" Horticulturae 10, no. 1: 91. https://doi.org/10.3390/horticulturae10010091
APA StyleMorcia, C., Carrara, I., Ghizzoni, R., Terzi, V., Bolli, G., & Chiusa, G. (2024). Chip Digital PCR (cdPCR) to Identify and Quantify Botrytis cinerea Infection in Tomatoes. Horticulturae, 10(1), 91. https://doi.org/10.3390/horticulturae10010091







