Identification of the Loci Associated with Resistance to Banana Xanthomonas Wilt (Xanthomonas vasicola pv. musacearum) Using DArTSeq Markers and Continuous Mapping
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Phenotyping
2.3. Analysis of the Phenotypic Data
2.4. Genotyping
2.5. Continuous Mapping
2.6. Identification of Putative Genes Linked with the Resistance Loci
3. Results
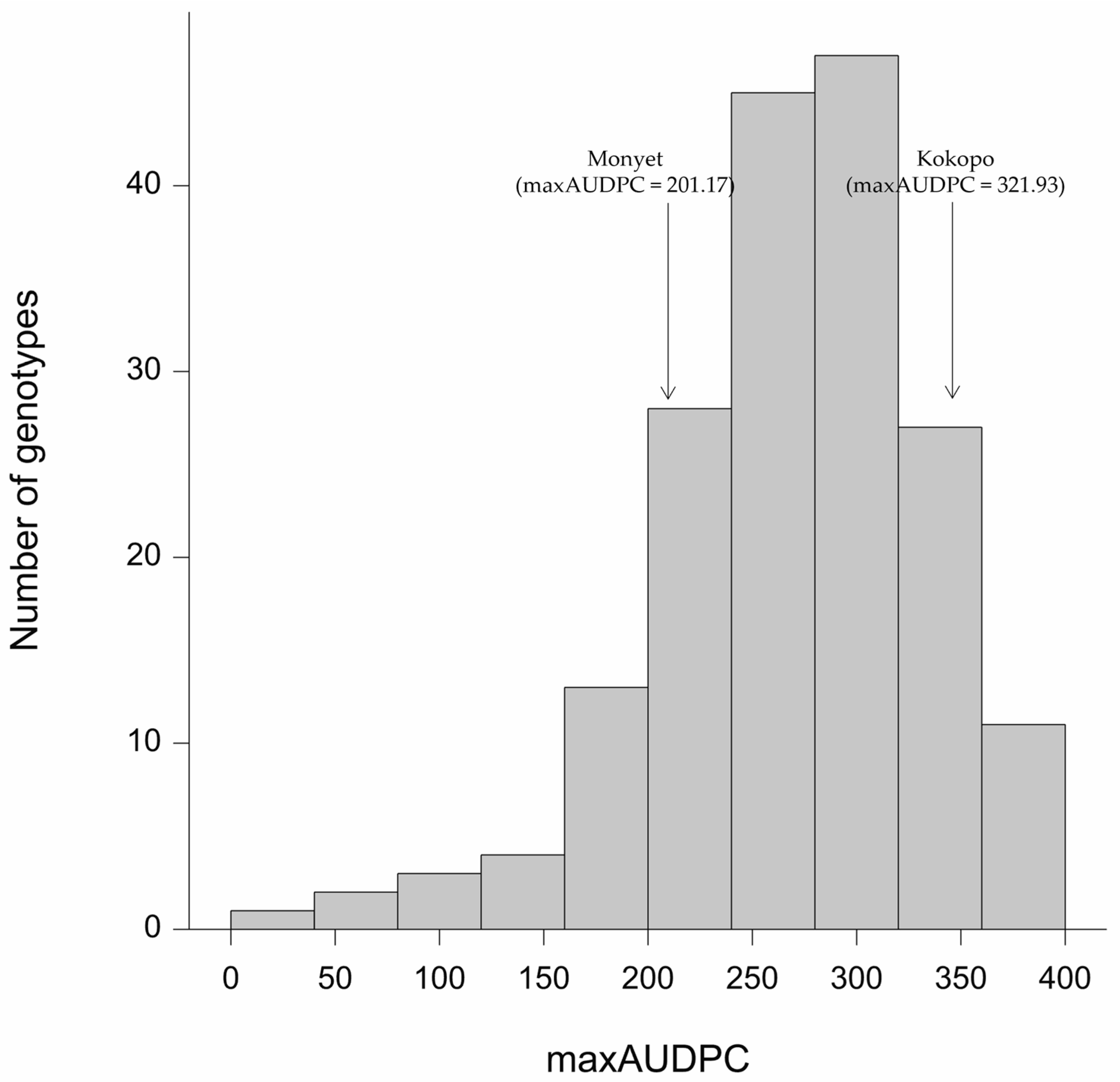
3.1. Phenotypic Variation and Heritability
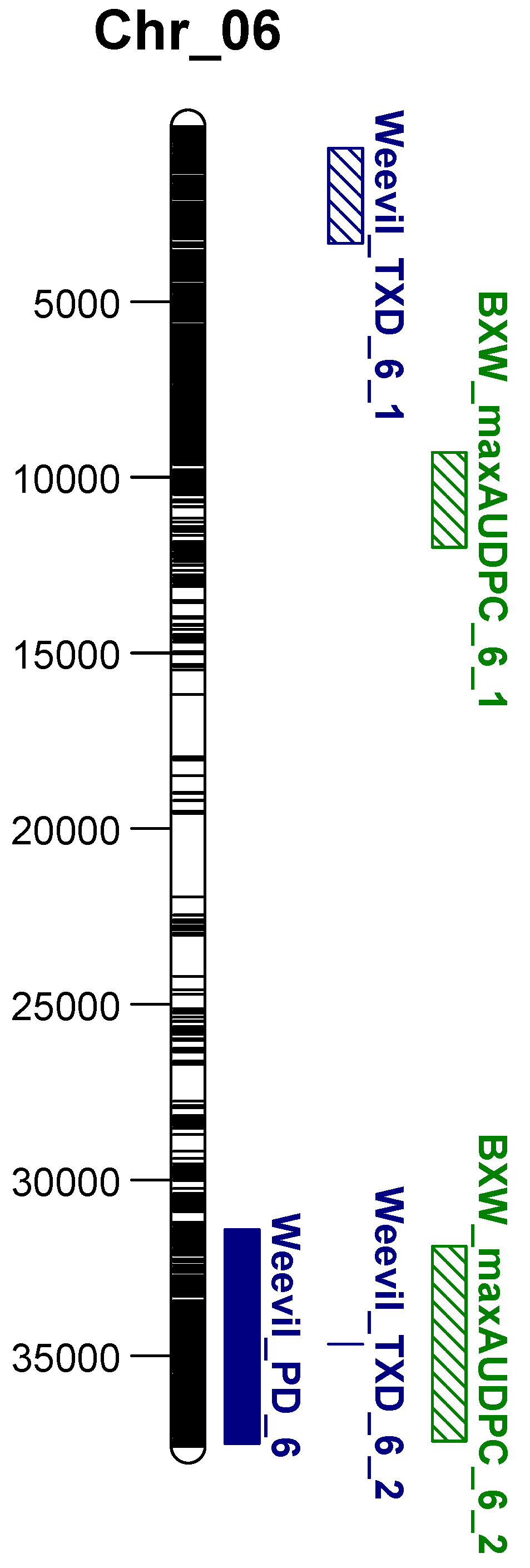
3.2. Continuous Mapping
3.3. Comparison between Weevil and BXW Resistance
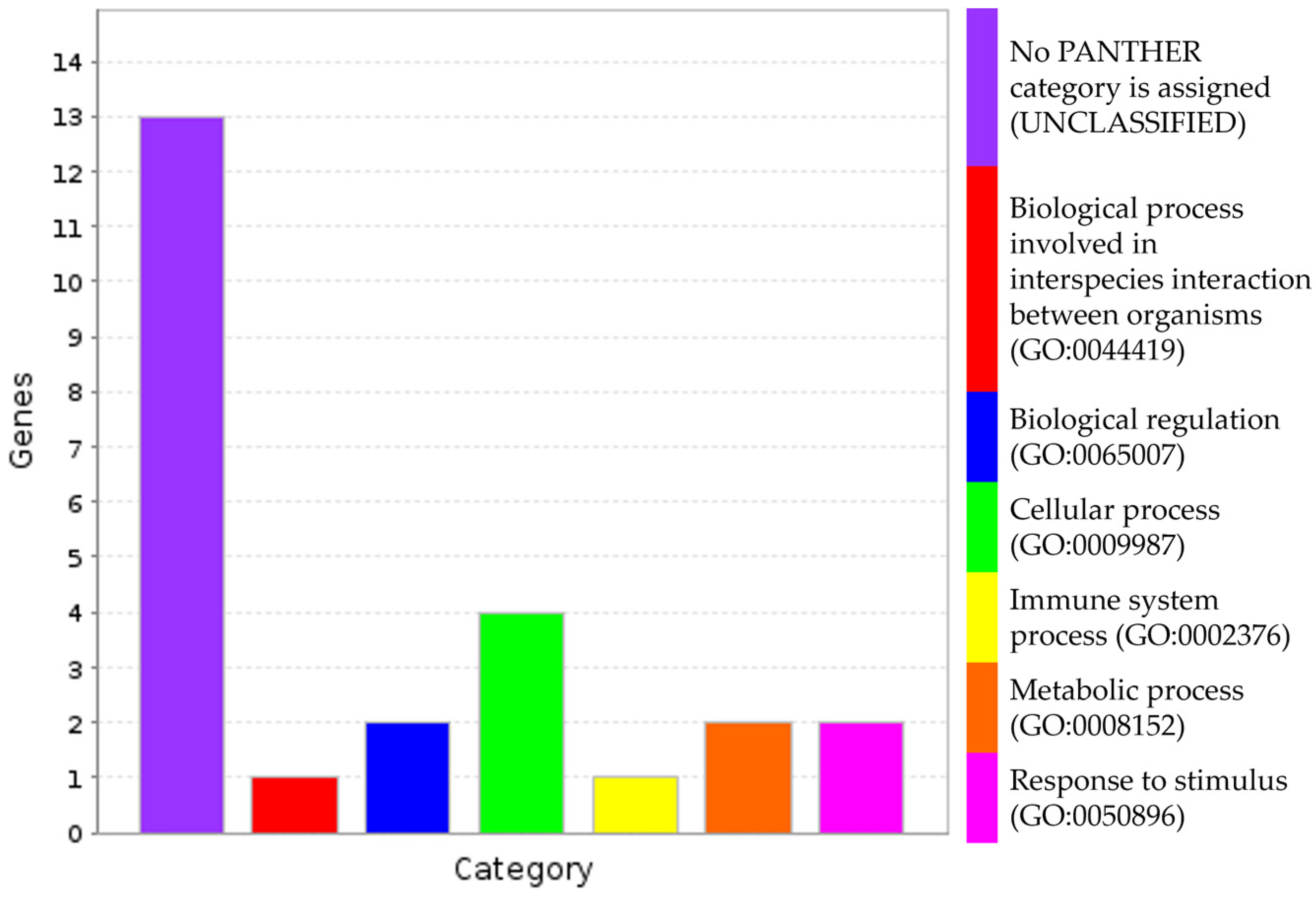
3.4. Putative Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Geberewold, A.Z. Review on impact of banana bacterial wilt (Xhantomonas campestris pv. Musacerum) in East and Central Africa. Cogent Food Agric. 2019, 5, 1586075. [Google Scholar] [CrossRef]
- Yirgou, D.; Bradbury, J. Bacterial wilt of enset (Ensete ventricosum) incited by Xanthomonas musacearum sp. n. Phytopathology 1968, 58, 111–112. [Google Scholar]
- Tushemereirwe, W.; Kangire, A.; Ssekiwoko, F.; Offord, L.; Crozier, J.; Boa, E.; Rutherford, M.; Smith, J. First report of Xanthomonas campestris pv. musacearum on banana in Uganda. Plant Pathol. 2004, 53, 802. [Google Scholar] [CrossRef]
- Reeder, R.; Muhinyuza, J.; Opolot, O.; Aritua, V.; Crozier, J.; Smith, J. Presence of banana bacterial wilt (Xanthomonas campestris pv. musacearum) in Rwanda. Plant Pathol. 2007, 56, 1038. [Google Scholar] [CrossRef]
- Carter, B.A.; Reeder, R.; Mgenzi, S.; Mwangi, M.; Aritua, V.; Smith, J.; Beed, F.D.; Kinyua, Z.; Nakato, V.; Miller, S. Identification of Xanthomonas vasicola (formerly X. campestris pv. musacearum), causative organism of banana xanthomonas wilt, in Tanzania, Kenya and Burundi. Plant Pathol. 2010, 59, 403. [Google Scholar] [CrossRef]
- Ndungo, V.; Eden-Green, S.; Blomme, G.; Crozier, J.; Smith, J. Presence of banana Xanthomonas wilt (Xanthomonas campestris pv. musacearum) in the Democratic Republic of Congo (DRC). Plant Pathol. 2006, 55, 294. [Google Scholar] [CrossRef]
- Nkuba, J.; Tinzaara, W.; Night, G.; Niko, N.; Jogo, W.; Ndyetabula, I.; Mukandala, L.; Ndayihazamaso, P.; Niyongere, C.; Gaidashova, S.; et al. Adverse impact of Banana Xanthomonas Wilt on farmers’ livelihoods in Eastern and Central Africa. Afr. J. Plant Sci. 2015, 9, 279–286. [Google Scholar] [CrossRef]
- Karamura, E.; Kayobyo, G.; Tushemereirwe, W.; Benin, S.; Blomme, G.; Green, S.E.; Markham, R. Assessing the impacts of banana bacterial wilt disease on banana (Musa spp.) productivity and livelihoods of Ugandan farm households. In Proceedings of the IV International Symposium on Banana: International Conference on Banana and Plantain in Africa: Harnessing International 879, Mombasa, Kenya, 5 October 2008; pp. 749–755. [Google Scholar]
- Tinzaara, W.; Karamura, E.; Kubiriba, J.; Ochola, D.; Ocimati, W.; Blomme, G.; Ssekiwoko, F. The banana Xanthomonas wilt epidemic in east and central Africa: Current research and development efforts. In Proceedings of the XXIX International Horticultural Congress on Horticulture: Sustaining Lives, Livelihoods and Landscapes (IHC2014): IX 1114, Brisbane, Australia, 17 August 2014; pp. 267–274. [Google Scholar]
- Tripathi, L.; Tripathi, J. Relative susceptibility of banana cultivars to Xanthomonas campestris pv. musacearum. Afr. J. Biotechnol. 2009, 8, 5343–5350. [Google Scholar]
- Tripathi, L.; Mwangi, M.; Abele, S.; Aritua, V.; Tushemereirwe, W.K.; Bandyopadhyay, R. Xanthomonas wilt: A threat to banana production in East and Central Africa. Plant Dis. 2009, 93, 440–451. [Google Scholar] [CrossRef]
- Tripathi, L.; Tripathi, J.N.; Tushemereirwe, W. Strategies for resistance to bacterial wilt disease of bananas through genetic engineering. Afr. J. Biotechnol. 2004, 3, 688–692. [Google Scholar]
- Biruma, M.; Pillay, M.; Tripathi, L.; Blomme, G.; Abele, S.; Mwangi, M.; Bandyopadhyay, R.; Muchunguzi, P.; Kassim, S.; Nyine, M. Banana Xanthomonas wilt: A review of the disease, management strategies and future research directions. Afr. J. Biotechnol. 2007, 6, 953–962. [Google Scholar]
- Blomme, G.; Jacobsen, K.; Ocimati, W.; Beed, F.; Ntamwira, J.; Sivirihauma, C.; Ssekiwoko, F.; Nakato, V.; Kubiriba, J.; Tripathi, L. Fine-tuning banana Xanthomonas wilt control options over the past decade in East and Central Africa. Eur. J. Plant Pathol. 2014, 139, 271–287. [Google Scholar] [CrossRef]
- Kubiriba, J.; Karamura, E.B.; Jogo, W.; Tushemereirwe, W.K.; Tinzaara, W. Community mobilization: A key to effective control of banana xanthomonas wilt. J. Dev. Agric. Econ. 2012, 4, 125–131. [Google Scholar] [CrossRef]
- Mwangi, M.; Nakato, V. Key factors responsible for the xanthomonas wilt epidemic on banana in East and Central Africa. In Proceedings of the III International Symposium on Banana: ISHS-ProMusa Symposium on Recent Advances in Banana Crop Protection for Sustainable 828, White River, South Africa, 10–14 September 2007; pp. 395–404. [Google Scholar]
- Tinzaara, W.; Karamura, E.B.; Blomme, G.; Jogo, W.; Ocimati, W.; Rietveld, A.; Kubiriba, J.; Opio, F. Why sustainable management of Xanthomonas wilt of banana in east and central Africa has been elusive. In Proceedings of the VII International Symposium on Banana: ISHS-ProMusa Symposium on Bananas and Plantains: Towards Sustainable Global Production 986, Salvador, Brazil, 10–14 October 2011; pp. 157–164. [Google Scholar]
- Nakato, G.V.; Christelová, P.; Were, E.; Nyine, M.; Coutinho, T.A.; Doležel, J.; Uwimana, B.; Swennen, R.; Mahuku, G. Sources of resistance in Musa to Xanthomonas campestris pv. musacearum, the causal agent of banana xanthomonas wilt. Plant Pathol. 2019, 68, 49–59. [Google Scholar] [CrossRef]
- Gayral, P.; Noa-Carrazana, J.-C.; Lescot, M.; Lheureux, F.; Lockhart, B.E.L.; Matsumoto, T.; Piffanelli, P.; Iskra-Caruana, M.-L. A single banana streak virus integration event in the banana genome as the origin of infectious endogenous pararetrovirus. J. Virol. 2008, 82, 6697–6710. [Google Scholar] [CrossRef] [PubMed]
- Batte, M.; Swennen, R.; Uwimana, B.; Akech, V.; Brown, A.; Tumuhimbise, R.; Hovmalm, H.P.; Geleta, M.; Ortiz, R. Crossbreeding East African Highland Bananas: Lessons learnt relevant to the botany of the crop after 21 years of genetic enhancement. Front. Plant Sci. 2019, 10, 81. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, L.; Tripathi, J.N.; Kiggundu, A.; Korie, S.; Shotkoski, F.; Tushemereirwe, W.K. Field trial of Xanthomonas wilt disease-resistant bananas in East Africa. Nat. Biotechnol. 2014, 32, 868–870. [Google Scholar] [CrossRef]
- Tripathi, L.; Atkinson, H.; Roderick, H.; Kubiriba, J.; Tripathi, J.N. Genetically engineered bananas resistant to Xanthomonas wilt disease and nematodes. Food Energy Secur. 2017, 6, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, J.N.; Lorenzen, J.; Bahar, O.; Ronald, P.; Tripathi, L. Transgenic expression of the rice Xa21 pattern-recognition receptor in banana (Musa sp.) confers resistance to Xanthomonas campestris pv. musacearum. Plant Biotechnol. J. 2014, 12, 663–673. [Google Scholar] [CrossRef]
- Ainembabazi, J.H.; Tripathi, L.; Rusike, J.; Abdoulaye, T.; Manyong, V. Ex-ante economic impact assessment of genetically modified banana resistant to Xanthomonas wilt in the Great Lakes region of Africa. PLoS ONE 2015, 10, e0138998. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, J.N.; Ntui, V.O.; Shah, T.; Tripathi, L. CRISPR/Cas9-mediated editing of DMR6 orthologue in banana (Musa spp.) confers enhanced resistance to bacterial disease. Plant Biotechnol. J. 2021, 19, 1291–1293. [Google Scholar] [CrossRef] [PubMed]
- Avalos, A.; Fang, M.; Pan, H.; Lluch, A.R.; Lipka, A.E.; Zhao, S.D.; Giray, T.; Robinson, G.E.; Hudson, M.E. Genomic regions influencing aggressive behavior in honey bees are defined by colony allele frequencies. Proc. Natl. Acad. Sci. USA 2020, 117, 17135–17141. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, B.H.; Jensen, J.; Asp, T.; Janss, L.L. Association studies using family pools of outcrossing crops based on allele-frequency estimates from DNA sequencing. Theor. Appl. Genet. 2014, 127, 1331–1341. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Uwimana, B.; Mwanje, G.; Batte, M.; Akech, V.; Shah, T.; Vuylsteke, M.; Swennen, R. Continuous mapping identifies loci associated with weevil resistance [Cosmopolites sordidus (Germar)] in a triploid banana population. Front. Plant Sci. 2021, 12, 2520. [Google Scholar] [CrossRef] [PubMed]
- Arinaitwe, I.K.; Teo, C.H.; Kayat, F.; Tumuhimbise, R.; Uwimana, B.; Kubiriba, J.; Swennen, R.; Harikrishna, J.A.; Othman, R.Y. Evaluation of banana germplasm and genetic analysis of an F1 population for resistance to Fusarium oxysporum f. sp. cubense race 1. Euphytica 2019, 215, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Adriko, J.; Aritua, V.; Mortensen, C.N.; Tushemereirwe, W.K.; Kubiriba, J.; Lund, O.S. Multiplex PCR for specific and robust detection of Xanthomonas campestris pv. musacearum in pure culture and infected plant material. Plant Pathol. 2012, 61, 489–497. [Google Scholar] [CrossRef]
- Wasukira, A.; Tayebwa, J.; Thwaites, R.; Paszkiewicz, K.; Aritua, V.; Kubiriba, J.; Smith, J.; Grant, M.; Studholme, D.J. Genome-wide sequencing reveals two major sub-lineages in the genetically monomorphic pathogen Xanthomonas campestris pathovar musacearum. Genes 2012, 3, 361–377. [Google Scholar] [CrossRef]
- Nakato, G.V.; Fuentes Rojas, J.L.; Verniere, C.; Blondin, L.; Coutinho, T.; Mahuku, G.; Wicker, E. A new multi locus variable number of tandem repeat analysis scheme for epidemiological surveillance of Xanthomonas vasicola pv. musacearum, the plant pathogen causing bacterial wilt on banana and enset. PLoS ONE 2019, 14, e0215090. [Google Scholar] [CrossRef] [PubMed]
- Ceballos, H.; Deutsch, J.A.; Gutiérrez, H. Recurrent selection for resistance to Exserohilum turcicum in eight subtropical maize populations. Crop Sci. 1991, 31, 964–971. [Google Scholar] [CrossRef]
- Forbes, G.; Trillos, O.; Turkensteen, L.; Hidalgo, O. Field inoculation of potatoes with Phytophthora infestans and its effect on the efficiency of selection for quantitative resistance in the plants. Fitopatología 1993, 28, 117–120. [Google Scholar]
- VSN International. Genstat for Windows, 22nd ed.; VSN International: Hemel Hempstead, UK, 2022. [Google Scholar]
- Cullis, B.R.; Smith, A.B.; Coombes, N.E. On the design of early generation variety trials with correlated data. J. Agric. Biol. Environ. Stat. 2006, 11, 381–393. [Google Scholar] [CrossRef]
- Das, B.; Jena, R.; Samal, K. Optimization of DNA isolation and PCR protocol for RAPD analysis of banana/plantain (Musa spp.). Int. J. Agric. Sci. 2009, 1, 21. [Google Scholar]
- Martin, G.; Baurens, F.-C.; Droc, G.; Rouard, M.; Cenci, A.; Kilian, A.; Hastie, A.; Doležel, J.; Aury, J.-M.; Alberti, A.; et al. Improvement of the banana “Musa acuminata” reference sequence using NGS data and semi-automated bioinformatics methods. BMC Genomics 2016, 17, 243. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.D.; Ebert, D.; Muruganujan, A.; Mushayahama, T.; Albou, L.-P.; Mi, H. PANTHER: Making genome-scale phylogenetics accessible to all. Protein Sci. 2022, 31, 8–22. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]
- de Vicente, M.; Tanksley, S. QTL analysis of transgressive segregation in an interspecific tomato cross. Genetics 1993, 134, 585–596. [Google Scholar] [CrossRef]
- Mackay, I.J.; Cockram, J.; Howell, P.; Powell, W. Understanding the classics: The unifying concepts of transgressive segregation, inbreeding depression and heterosis and their central relevance for crop breeding. Plant Biotechnol. J. 2021, 19, 26–34. [Google Scholar] [CrossRef]
- Comai, L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 2005, 6, 836–846. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, D.; Willis, J.H. The genetics of inbreeding depression. Nat. Rev. Genet. 2009, 10, 783–796. [Google Scholar] [CrossRef]
- Sverrisdóttir, E.; Byrne, S.; Sundmark, E.H.R.; Johnsen, H.Ø.; Kirk, H.G.; Asp, T.; Janss, L.; Nielsen, K.L. Genomic prediction of starch content and chipping quality in tetraploid potato using genotyping-by-sequencing. Theor. Appl. Genet. 2017, 130, 2091–2108. [Google Scholar] [CrossRef] [PubMed]
- Jighly, A. When do autopolyploids need poly-sequencing data? Mol. Ecol. 2022, 31, 1021–1027. [Google Scholar] [CrossRef]
- Ahmad, F.; Martawi, N.M.; Poerba, Y.S.; de Jong, H.; Schouten, H.; Kema, G.H.J. Genetic mapping of Fusarium wilt resistance in a wild banana Musa acuminata ssp. malaccensis accession. Theor. Appl. Genet. 2020, 133, 3409–3418. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Sun, J.; Martin, G.; Gray, L.-A.; Hřibová, E.; Christelová, P.; Yahiaoui, N.; Rounsley, S.; Lyons, R.; Batley, J.; et al. Identification of a major QTL controlling resistance to the subtropical race 4 of Fusarium oxysporum f. sp. cubense in Musa acuminata ssp. malaccensis. Pathogens 2023, 12, 289. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Sun, J.; Viljoen, A.; Mostert, D.; Xie, Y.; Mangila, L.; Bothma, S.; Lyons, R.; Hřibová, E.; Christelová, P.; et al. Genetic mapping, candidate gene identification and marker validation for host plant resistance to the race 4 of Fusarium oxysporum f. sp. cubense using Musa acuminata ssp. malaccensis. Pathogens 2023, 12, 820. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-M.; Ku, H.-M.; Schafleitner, R.; Bains, T.S.; George Kuo, C.; Liu, C.-A.; Nair, R.M. The major quantitative trait locus for mungbean yellow mosaic Indian virus resistance is tightly linked in repulsion phase to the major bruchid resistance locus in a cross between mungbean [Vigna radiata (L.) Wilczek] and its wild relative Vigna radiata ssp. sublobata. Euphytica 2013, 192, 205–216. [Google Scholar] [CrossRef]
- Guo, L.; Wang, K.; Chen, J.; Huang, D.; Fan, Y.; Zhuang, J. Dissection of two quantitative trait loci for grain weight linked in repulsion on the long arm of chromosome 1 of rice (Oryza sativa L.). Crop J. 2013, 1, 70–76. [Google Scholar] [CrossRef]
- Bernstein, M.R.; Zdraljevic, S.; Andersen, E.C.; Rockman, M.V. Tightly linked antagonistic-effect loci underlie polygenic phenotypic variation in C. elegans. Evol. Lett. 2019, 3, 462–473. [Google Scholar] [CrossRef]
- Zhang, X.; Dai, Y.-S.; Wang, Y.-X.; Su, Z.-Z.; Yu, L.-J.; Zhang, Z.-F.; Xiao, S.; Chen, Q.-F. Overexpression of the arabidopsis MACPF Protein AtMACP2 promotes pathogen resistance by activating SA signaling. Int. J. Mol. Sci. 2022, 23, 8784. [Google Scholar] [CrossRef]
- Morita-Yamamuro, C.; Tsutsui, T.; Sato, M.; Yoshioka, H.; Tamaoki, M.; Ogawa, D.; Matsuura, H.; Yoshihara, T.; Ikeda, A.; Uyeda, I.; et al. The Arabidopsis gene CAD1 controls programmed cell death in the plant immune system and encodes a protein containing a MACPF domain. Plant Cell Physiol. 2005, 46, 902–912. [Google Scholar] [CrossRef] [PubMed]
- Soderlund, C.; Descour, A.; Kudrna, D.; Bomhoff, M.; Boyd, L.; Currie, J.; Angelova, A.; Collura, K.; Wissotski, M.; Ashley, E.; et al. Sequencing, mapping, and analysis of 27,455 maize full-length cDNAs. PLoS Genet. 2009, 5, e1000740. [Google Scholar] [CrossRef]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Gookin, T.E.; Kim, J.; Assmann, S.M. Whole proteome identification of plant candidate G-protein coupled receptors in Arabidopsis, rice, and poplar: Computational prediction and in-vivo protein coupling. Genome Biol. 2008, 9, R120. [Google Scholar] [CrossRef] [PubMed]
- Zimin, A.V.; Puiu, D.; Hall, R.; Kingan, S.; Clavijo, B.J.; Salzberg, S.L. The first near-complete assembly of the hexaploid bread wheat genome, Triticum aestivum. GigaScience 2017, 6, gix097. [Google Scholar] [CrossRef] [PubMed]
- Perrier, X.; Bakry, F.; Carreel, F.; Jenny, C.; Horry, J.-P.; Lebot, V.; Hippolyte, I. Combining biological approaches to shed light on the evolution of edible bananas. Ethnobot. Res. Appl. 2009, 7, 199–216. [Google Scholar] [CrossRef]
- Němečková, A.; Christelová, P.; Čížková, J.; Nyine, M.; Van den houwe, I.; Svačina, R.; Uwimana, B.; Swennen, R.; Doležel, J.; Hřibová, E. Molecular and cytogenetic study of East African highland banana. Front. Plant Sci. 2018, 9, 1371. [Google Scholar] [CrossRef]
- Christelová, P.; De Langhe, E.; Hřibová, E.; Čížková, J.; Sardos, J.; Hušáková, M.; Van den houwe, I.; Sutanto, A.; Kepler, A.K.; Swennen, R.; et al. Molecular and cytological characterization of the global Musa germplasm collection provides insights into the treasure of banana diversity. Biodivers. Conserv. 2016, 26, 801–824. [Google Scholar] [CrossRef]
- Li, X.; Yu, S.; Cheng, Z.; Chang, X.; Yun, Y.; Jiang, M.; Chen, X.; Wen, X.; Li, H.; Zhu, W.; et al. Origin and evolution of the triploid cultivated banana genome. Nat. Genet. 2024, 56, 136–142. [Google Scholar] [CrossRef]


| sn | SNP ID | Chrom i | Position i | −LOG(p) | Genotype of the Parents | Effect ii | R iii | A iii | expl_var. iv | |
|---|---|---|---|---|---|---|---|---|---|---|
| Monyet | Kokopo | |||||||||
| 1 | chr02_23268545 | 2 | 3,268,545 | 3.04 | RRAA | RR | −29.88 | G | A | 7.22 |
| 2 | chr02_23894612 | 2 | 3,894,612 | 3.14 | RRAA | RR | −30.89 | A | G | 7.53 |
| 3 | chr03_25228251 | 3 | 5,228,251 | 3.15 | AAAA | RA | −37.80 | A | G | 7.54 |
| 4 | chr06_9287277 | 6 | 9,287,277 | 3.16 | AAAA | RA | −40.01 | A | G | 7.58 |
| 5 | chr06_10081954 | 6 | 10,081,954 | 3.66 | RRRR | RA | 36.90 | T | A | 9.18 |
| 6 | chr06_10081996 | 6 | 10,081,996 | 3.72 | RRRR | RA | 37.22 | G | T | 9.36 |
| 7 | chr06_11466145 | 6 | 11,466,145 | 3.25 | RRRR | RA | 34.98 | A | G | 8.20 |
| 8 | chr06_11997622 | 6 | 11,997,622 | 3.12 | RRRR | RA | 40.21 | G | T | 7.52 |
| 9 | chr06_31895299 | 6 | 31,895,299 | 3.53 | RRAA | RR | 30.47 | G | T | 8.66 |
| 10 | chr06_35307807 | 6 | 35,307,807 | 4.49 | RRRR | RA | 41.98 | T | C | 11.55 |
| 11 | chr06_35399010 | 6 | 35,399,010 | 3.08 | AAAA | RA | −33.41 | A | T | 7.35 |
| 12 | chr06_35734979 | 6 | 35,734,979 | 3.96 | AAAA | RA | −35.05 | G | A | 10.00 |
| 13 | chr06_35735014 | 6 | 35,735,014 | 3.74 | RRRR | RA | 33.67 | A | G | 9.36 |
| 14 | chr06_35773242 | 6 | 35,773,242 | 3.11 | RRRR | RA | 38.85 | G | C | 7.53 |
| 15 | chr06_36095415 | 6 | 36,095,415 | 3.13 | RRRR | RA | 43.69 | T | C | 7.47 |
| 16 | chr06_36497284 | 6 | 36,497,284 | 3.24 | AAAA | RA | −38.84 | G | T | 8.18 |
| 17 | chr06_36497290 | 6 | 36,497,290 | 3.22 | AAAA | RA | −38.89 | G | T | 8.17 |
| 18 | chr06_36497314 | 6 | 36,497,314 | 3.24 | RRRR | RA | 41.56 | T | C | 8.15 |
| 19 | chr06_37429651 | 6 | 37,429,651 | 3.22 | AAAA | RA | −38.61 | A | G | 7.86 |
| 20 | chr07_2838116 | 7 | 2,838,116 | 3.38 | RRRA | RR | 42.82 | C | T | 8.21 |
| 21 | chr07_2838135 | 7 | 2,838,135 | 3.47 | RRRA | RR | 43.45 | G | C | 8.49 |
| 22 | chr07_2838162 | 7 | 2,838,162 | 3.45 | RRRA | RR | 43.26 | T | A | 8.43 |
| 23 | chr07_5962374 | 7 | 5,962,374 | 3.02 | AAAA | RA | −39.48 | A | G | 7.42 |
| 24 | chr07_6863391 | 7 | 6,863,391 | 3.11 | RRRR | RA | 45.63 | C | A | 7.49 |
| 25 | chr07_6863416 | 7 | 6,863,416 | 3.25 | RRRR | RA | 46.44 | A | C | 7.90 |
| sn | Gene_ID | Chrom | Gene Position (bp) | Strand | Linked Marker | Distance (bp) | Annotation |
|---|---|---|---|---|---|---|---|
| 1 | Ma02_g15610 | chr02 | 23,270,730–23,272,339 | + | chr02_23268545 | 2185 | Remorin_C domain-containing protein |
| 2 | Ma02_g16600 | chr02 | 23,890,072–23,894,146 | + | chr02_23894612 | 466 | Lipoyl synthase, mitochondrial |
| 3 | Ma03_g19990 | chr03 | 25,228,695–25,230,557 | + | chr03_25228251 | 444 | Abscisic stress-ripening protein 5 |
| 4 | Ma06_g13550 | chr06 | 9,286,287–9,294,510 | − | chr06_9287277 | 0 | MACPF domain-containing protein At4g24290-like |
| 5 | Ma06_g14760 | chr06 | 10,081,045–10,082,123 | + | chr06_10081954 and chr06_10081996 | 0 | Conserved hypothetical protein |
| 6 | Ma06_g16910 | chr06 | 11,467,120–11,467,747 | − | chr06_11466145 | 975 | Zinc finger protein ZAT12-like |
| 7 | Ma06_g17690 | chr06 | 11,985,303–12,004,743 | + | chr06_11997622 | 0 | HUA2-like protein 2 |
| 8 | Ma06_g30680 | chr06 | 31,884,480–31,898,896 | + | chr06_31895299 | 0 | Non-lysosomal glucosylceramidase-like |
| 9 | Ma06_g35490 | chr06 | 35,304,891–35,315,265 | + | chr06_35307807 | 0 | Amino-acid N-acetyltransferase |
| 10 | Ma06_g35620 | chr06 | 35,399,155–35,400,631 | + | chr06_35399010 | 145 | Transcription factor MYB44 |
| 11 | Ma06_g36200 | chr06 | 35,734,408–35,738,511 | − | chr06_35734979 and chr06_35735014 | 0 | RRM domain-containing protein |
| 12 | Ma06_g36300 | chr06 | 35,774,897–357,76,327 | + | chr06_35773242 | 1655 | HMA domain-containing protein |
| 13 | Ma06_g36840 | chr06 | 36,091,779–36,096,147 | + | chr06_36095415 | 0 | Transmembrane protein adipocyte-associated 1-like |
| 14 | Ma06_g37450 | chr06 | 36,492,067–36,495,406 | − | chr06_36497284, chr06_36497290, and chr06_36497314 | 1878 | Clp R domain-containing protein |
| 15 | Ma06_g38780 | chr06 | 37,415,987–37,427,552 | − | chr06_37429651 | 2099 | Oxysterol-binding protein-related protein 2A |
| 16 | Ma07_g03700 | chr07 | 2,836,251–2,840,247 | + | chr07_2838116, chr07_2838135, and chr07_2838162 | 0 | M20_dimer domain-containing protein |
| 17 | Ma07_g08000 | chr07 | 5,961,864–5,962,783 | + | chr07_5962374 | 0 | CASP-like protein |
| 18 | Ma07_g09140 | chr07 | 6,857,345–6,870,535 | − | chr07_6863391 and chr07_6863416 | 0 | Na_H_Exchanger domain-containing protein |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Uwimana, B.; Nakato, G.V.; Kanaabi, R.; Nasuuna, C.; Mwanje, G.; Mahuku, G.S.; Akech, V.; Vuylsteke, M.; Swennen, R.; Shah, T. Identification of the Loci Associated with Resistance to Banana Xanthomonas Wilt (Xanthomonas vasicola pv. musacearum) Using DArTSeq Markers and Continuous Mapping. Horticulturae 2024, 10, 87. https://doi.org/10.3390/horticulturae10010087
Uwimana B, Nakato GV, Kanaabi R, Nasuuna C, Mwanje G, Mahuku GS, Akech V, Vuylsteke M, Swennen R, Shah T. Identification of the Loci Associated with Resistance to Banana Xanthomonas Wilt (Xanthomonas vasicola pv. musacearum) Using DArTSeq Markers and Continuous Mapping. Horticulturae. 2024; 10(1):87. https://doi.org/10.3390/horticulturae10010087
Chicago/Turabian StyleUwimana, Brigitte, Gloria Valentine Nakato, Reagan Kanaabi, Catherine Nasuuna, Gerald Mwanje, George Simba Mahuku, Violet Akech, Marnik Vuylsteke, Rony Swennen, and Trushar Shah. 2024. "Identification of the Loci Associated with Resistance to Banana Xanthomonas Wilt (Xanthomonas vasicola pv. musacearum) Using DArTSeq Markers and Continuous Mapping" Horticulturae 10, no. 1: 87. https://doi.org/10.3390/horticulturae10010087
APA StyleUwimana, B., Nakato, G. V., Kanaabi, R., Nasuuna, C., Mwanje, G., Mahuku, G. S., Akech, V., Vuylsteke, M., Swennen, R., & Shah, T. (2024). Identification of the Loci Associated with Resistance to Banana Xanthomonas Wilt (Xanthomonas vasicola pv. musacearum) Using DArTSeq Markers and Continuous Mapping. Horticulturae, 10(1), 87. https://doi.org/10.3390/horticulturae10010087






