Selenocysteine Formation by Enterococcus faecium ABMC-05 Follows a Mechanism That Is Not Dependent on Genes selA and selD but on Gene cysK
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Growing Conditions
2.3. Bacterium Identification
2.4. MALDI-TOF MS
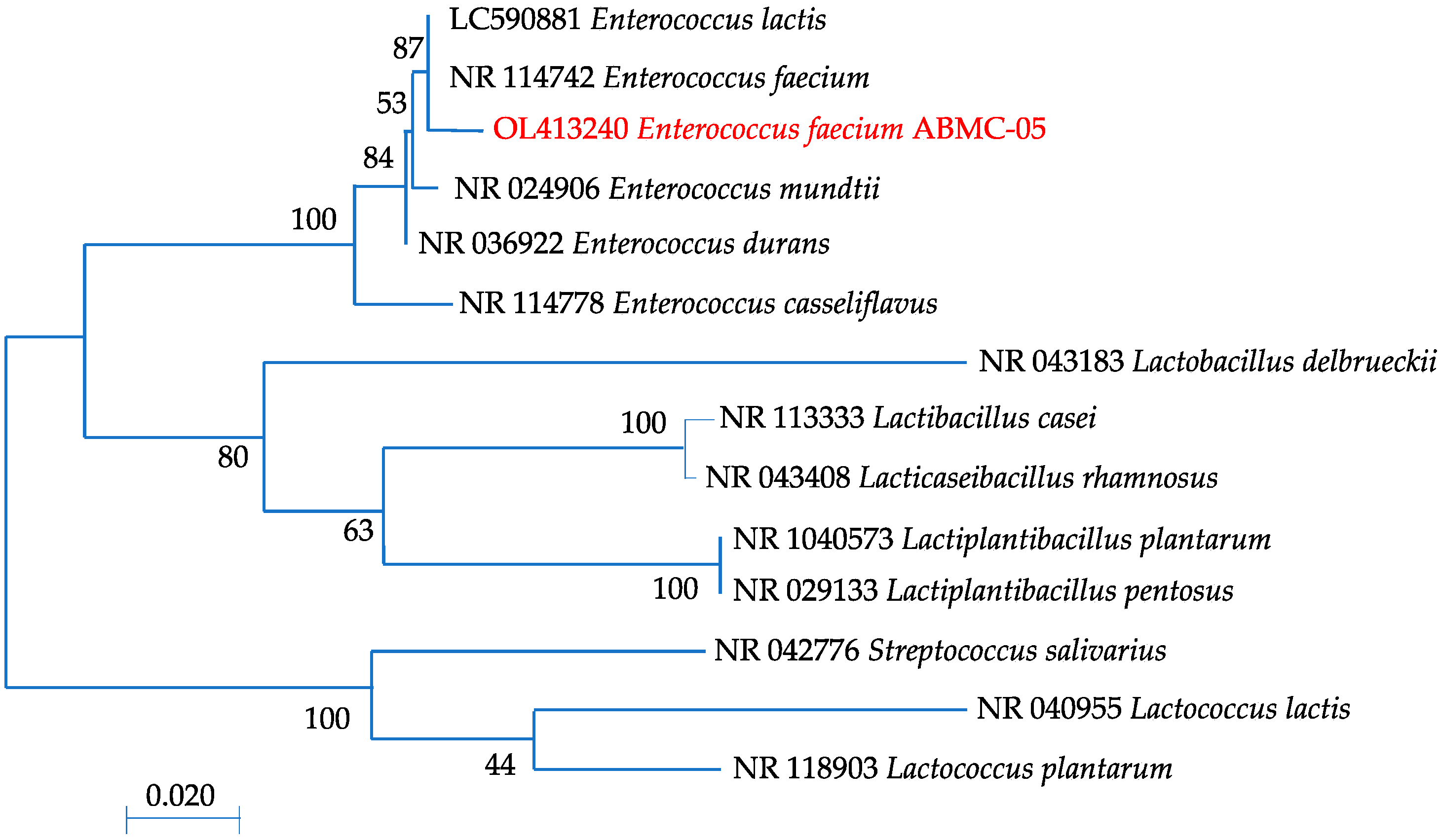
2.5. 16S rDNA Identification and Phylogenetic Analysis
2.6. Minimum Inhibitory Concentration of Na2SeO3
2.7. Selenization of E. faecium ABMC-05
2.8. Quantification of Total Selenium Content by Inductively Coupled Optical Emission Spectrometry (ICP-OES)
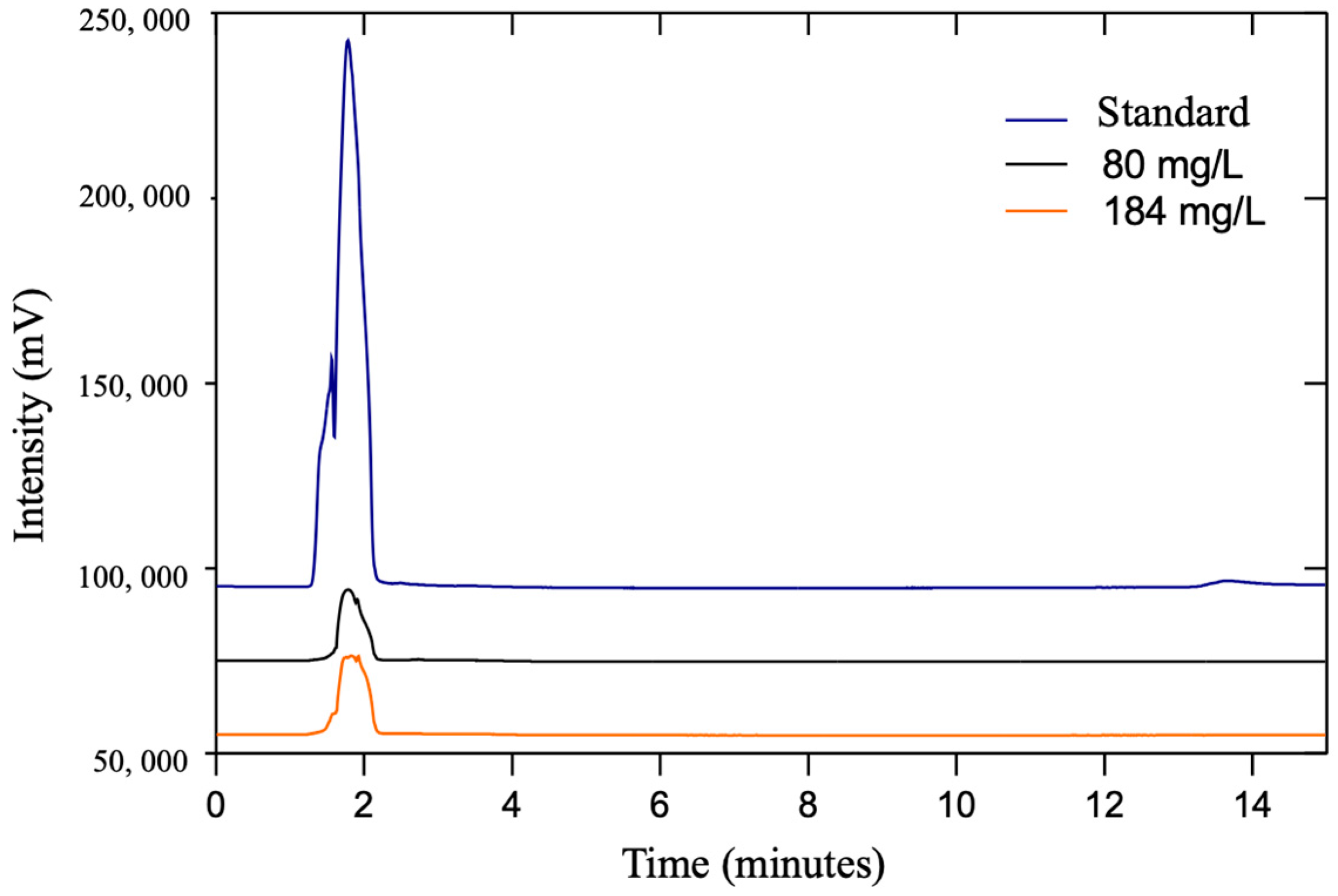
2.9. Selenocysteine Determination
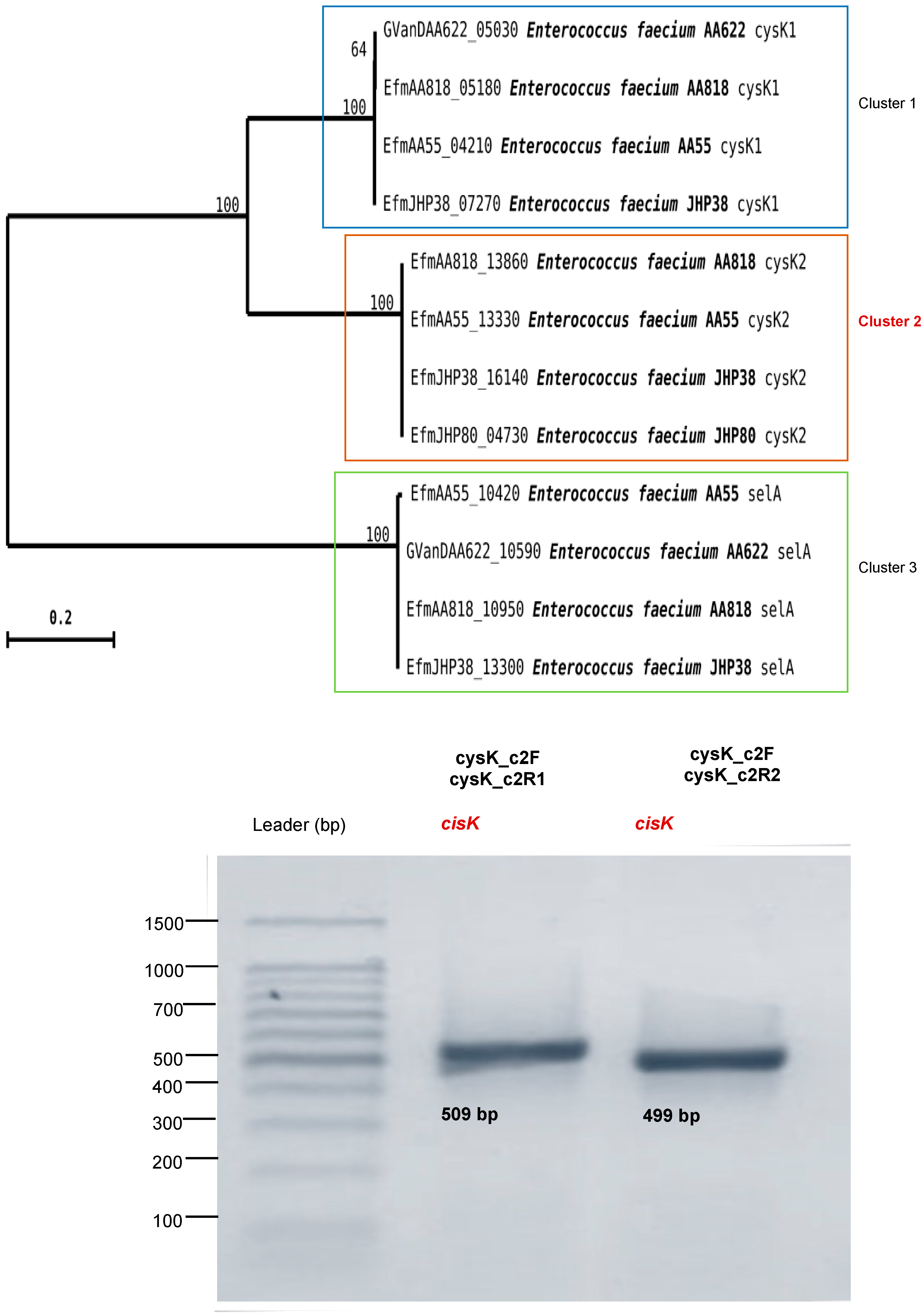
2.10. Gene Amplification: selA, selD, and cysK
2.11. Statistical Analysis
3. Results and Discussion
3.1. Identification
3.2. Minimum Concentration of the Inhibition and Selenization of E. faecium ABMC-05
3.3. Determination of Selenium Accumulation in Bacteria by ICP
3.4. Selenocysteine Determination
3.5. Determination of the Presence of the Genes selD, selA, and cysK in E. faecium ABMC-05
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Misra, S.; Boylan, M.; Selvam, A.; Spallholz, J.E.; Björnstedt, M. Redox-active selenium compounds from toxicity and cell death to cancer treatment. Nutrients 2015, 7, 3536–3556. [Google Scholar] [CrossRef] [PubMed]
- Vinceti, M.; Filippini, T.; Wise, L.A. Environmental selenium and human health: An update. Curr. Environ. Health Rep. 2018, 5, 464–485. [Google Scholar] [CrossRef] [PubMed]
- Pescuma, M.; Gomez-Gomez, B.; Perez-Corona, T.; Font, G.; Madrid, Y.; Mozzi, F. Food prospects of selenium enriched-Lactobacillus acidophilus CRL 636 and Lactobacillus reuteri CRL 1101. J. Funct. Foods 2017, 35, 466–473. [Google Scholar] [CrossRef]
- Maseko, T.; Callahan, D.L.; Dunshea, F.R.; Doronila, A.; Kolev, S.D.; Ng, K. Chemical characterisation and speciation of organic selenium in cultivated selenium-enriched Agaricus bisporus. Food Chem. 2013, 141, 3681–3687. [Google Scholar] [CrossRef] [PubMed]
- Galano, E.; Mangiapane, E.; Bianga, J.; Palmese, A.; Pessione, E.; Szpunar, J.; Lobinski, R.; Amoresano, A. Privileged incorporation of selenium as selenocysteine in Lactobacillus reuteri proteins demonstrated by selenium-specific imaging and proteomics. Mol. Cell Proteom. 2013, 12, 2196–2204. [Google Scholar] [CrossRef]
- Yang, J.; Wang, J.; Yang, K.; Liu, M.; Qi, Y.; Zhang, T.; Fan, M.; Wei, X. Antibacterial activity of selenium-enriched lactic acid bacteria against common food-borne pathogens in vitro. J. Dairy Sci. 2017, 101, 1930–1942. [Google Scholar] [CrossRef]
- Turner, R.T.; Weiner, J.H.; Taylor, D.E. Selenium metabolism in Escherichia coli. Biometals 1998, 11, 223–227. [Google Scholar] [CrossRef]
- Rigger, L.; Schmidt, R.L.; Holman, K.M.; Simonović, M.; Micura, R. The synthesis of methylated, phosphorylated, and phosphonated 3′-aminoacyl-tRNASec mimics. Chem. Eur. J. 2013, 19, 15872–15878. [Google Scholar] [CrossRef]
- Castañeda-Ovando, A.; Segovia-Cruz, J.A.; Flores-Aguilar, J.F.; Rodríguez-Serrano, G.M.; Salazar-Pereda, V.; Ramírez-Godínez, J.; Contreras-López, E.; Jaimez-Ordaz, J.; González-Olivares, L.G. Serine-enriched minimal medium enhances conversion of selenium into selenocysteine by Streptococcus thermophilus. J. Dairy Sci. 2019, 102, 6781–6789. [Google Scholar] [CrossRef]
- Mariotti, M.; Ridge, P.G.; Zhang, Y.; Lobanov, A.V.; Pringle, T.H.; Guigo, R.; Hatfield, D.L.; Gladyshev, V.N. Composition and evolution of the vertebrate and mammalian selenoproteomes. PLoS ONE 2012, 7, e33066. [Google Scholar] [CrossRef]
- Santesmasses, D.; Mariotti, M.; Guigó, R. Computational identification of the selenocysteine tRNA (tRNASec) in genomes. PLoS Comput. Biol. 2017, 13, e1005383. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Turanov, A.A.; Hatfield, D.L.; Gladyshev, V.N. In silico identification of genes involved in selenium metabolism: Evidence for a third selenium utilization trait. BMC Genom. 2008, 9, 11–13. [Google Scholar] [CrossRef] [PubMed]
- Estrada, A.M.; Olivares, L.G.G.; López, E.C.; Serrano, G.R. SelA and SelD genes involved in selenium absorption metabolism in lactic acid bacteria isolated from Mexican cheeses. Int. Dairy J. 2020, 103, 104629. [Google Scholar] [CrossRef]
- Palomo, M.; Gutiérrez, A.M.; Pérez-Conde, M.C.; Cámara, C.; Madrid, Y. Se metallomics during lactic fermentation of Se-enriched yogurt. Food Chem. 2014, 164, 371–379. [Google Scholar] [CrossRef]
- Deng, Y.; Man, C.; Fan, Y.; Wang, Z.; Li, L.; Ren, H.; Cheng, W.; Jiang, Y. Preparation of elemental selenium-enriched fermented milk by newly isolated Lactobacillus brevis from kefir grains. Int. Dairy J. 2015, 44, 31–36. [Google Scholar] [CrossRef]
- Calomme, M.R.; Van den Branden, K.; Vanden Berghe, D.A. Selenium and Lactobacillus species. J. Appl. Microbiol. 1995, 79, 331–340. [Google Scholar] [CrossRef]
- Alzate, A.; Cañas, B.; Pérez-Munguía, S.; Hernández-Mendoza, H.; Pérez-Conde, C.; Gutiérrez, A.M.; Cámara, C. Evaluation of the inorganic selenium biotransformation in selenium-enriched yogurt by HPLC-ICP-MS. J. Agric. Food Chem. 2007, 55, 9776–9783. [Google Scholar] [CrossRef]
- Krittaphol, W.; Wescombe, P.A.; Thomson, C.D.; McDowell, A.; Tagg, J.R.; Fawcett, J.P. Metabolism of L-selenomethionine and selenite by probiotic bacteria: In vitro and in vivo studies. Biol. Trace Elem. Res. 2011, 144, 358–1369. [Google Scholar] [CrossRef]
- Yin, X.; Zhou, Y.; Yang, H.; Liao, Y.; Ma, T.; Wang, F. Enhanced selenocysteine biosynthesis for seleno-methylselenocysteine production in Bacillus subtilis. Appl. Microbiol. Biotechnol. 2023, 107, 2843–2854. [Google Scholar] [CrossRef]
- Escobar-Ramírez, M.C.; Jaimez-Ordaz, J.; Escorza-Iglesias, V.A.; Rodríguez-Serrano, G.M.; Contreras-López, E.; Ramírez-Godínez, J.; Castañeda-Ovando, A.; Morales-Estrada, A.I.; Felix-Reyes, N.; González-Olivares, L.G. Lactobacillus pentosus ABHEAU-05: An in vitro digestion resistant lactic acid bacterium isolated from a traditional fermented Mexican beverage. Rev. Argent Microbiol. 2020, 52, 305–314. [Google Scholar] [CrossRef]
- Escobar-Ramírez, M.C.; Rodríguez-Serrano, G.M.; Salazar-Pereda, V.; Castañeda-Ovando, A.; Pérez-Escalante, E.; Jaimez-Ordaz, J.; González-Olivares, L.G. Biogenic production of selenocysteine by Enterococcus faecium ABMC-05: An indigenous lactic acid bacterium from fermented Mexican beverage. Food Sci. Technol. 2022, 43, e63622. [Google Scholar] [CrossRef]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Strahsburger, E.; Retamales, P.; Estrada, J.; Seeger, M. Microdot method: Used with chromogenic agar is a useful procedure for sanitary monitoring in aquaculture. Lat. Am. J. Aquat. Res. 2016, 44, 742–749. [Google Scholar] [CrossRef]
- Turło, J.; Gutkowska, B.; Malinowska, E. Relationship between the selenium, selenomethionine, and selenocysteine content of submerged cultivated mycelium of Lentinula edodes (Berk.). Acta Chromatogr. 2007, 18, 36–48. [Google Scholar]
- Vázquez-Ortíz, F.A.; Caire, G.; Higuera-Ciapara, I.; Hernández, G. High-performance liquid chromatographic determination of free amino acids in shrimp. J. Liq. Chromatogr. 1995, 18, 2059–2068. [Google Scholar] [CrossRef]
- Jones, B.N. Amino acid analysis by o-phthaldialdehyde precolumn derivatization and reverse-phase HPLC. In Methods Protein Microcharacterization: Biological Methods, 1st ed.; Shively, J.E., Ed.; Humana Press: Totowa, NJ, USA, 1986; pp. 121–151. [Google Scholar] [CrossRef]
- Castillo-Portela, G.; Villar-Delgado, J.; Montano-Martínez, R.; Martínez, C.; Pérez-Alfocea, F.; Albacete, A.; Sánchez-Bravo, J.; Acosta-Echeverria, M. Cuantificación por HPLC del contenido de aminoácidos presentes en el FOTOMAS-E. ICIDCA Sobre Los Deriv. Caña Azúcar 2011, 45, 64–67. [Google Scholar]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Stackebrandt, E.; Goebel, B.M. Taxonomic Note: A Place for DNA-DNA Reassociation and 16s rRNA Sequence Analysis in the Present Species Definition in Bacteriology. Int. J. Syst. Evol. Microbiol. 1994, 44, 846–849. [Google Scholar] [CrossRef]
- Gutiérrez-Sarmiento, W.; Peña-Ocaña, B.A.; Lam-Gutiérrez, A.; Guzmán-Albores, J.M.; Jasso-Chávez, R.; Ruíz-Valdiviezo, V.M. Microbial community structure, physicochemical characteristics and predictive functionalities of the Mexican tepache fermented beverage. Microbiol. Res. 2022, 260, 127045. [Google Scholar] [CrossRef]
- Moreno-Terrazas, R.D. Determinación de las características microbiológicas, bioquímicas y sensoriales para la estandarización del proceso de elaboración de tepache. Doctoral Dissertation, Universidad Autónoma Metropolitana (Unidad Xochimilco), México City, Mexico, 27 January 2005. [Google Scholar]
- Cervantes-Contreras, M.; Pedroza, A.M. Caracterización microbiológica del pulque y cuantificación de su contenido de etanol mediante espectroscopia Raman. Superf. Y Vacio. 2008, 21, 1–5. [Google Scholar]
- de la Fuente-Salcido, N.M.; Castañeda-Ramírez, J.C.; García-Almendárez, B.E.; Bideshi, D.K.; Salcedo-Hernández, R.; Barboza-Corona, J.E. Isolation and characterization of bacteriocinogenic lactic bacteria from M-Tuba and Tepache, two traditional fermented beverages in México. Food Sci. Nutr. 2015, 3, 434–442. [Google Scholar] [CrossRef] [PubMed]
- Pusztahelyi, T.; Kovács, S.; Pócsi, I.; Prokisch, J. Selenite-stress selected mutant strains of probiotic bacteria for Se source production. J. Trace Elem. Med. Biol. 2015, 30, 96–101. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Wang, C. Factors affecting selenium-enrichment efficiency, metabolic mechanisms and physiological functions of selenium-enriched lactic acid bacteria. J. Future Foods 2022, 2, 285–293. [Google Scholar] [CrossRef]
- Martínez, F.G.; Cuencas-Barrientos, M.E.; Mozzi, F.; Pescuma, M. Survival of selenium-enriched lactic acid bacteria in a fermented drink under storage and simulated gastro-intestinal digestion. Food Res. Int. 2019, 123, 115–124. [Google Scholar] [CrossRef]
- Krausova, G.; Kana, A.; Hyrslova, I.; Mrvikova, I.; Kavkova, M. Development of selenized lactic acid bacteria and their selenium bioaccummulation capacity. Fermentation. 2020, 6, 91. [Google Scholar] [CrossRef]
- Kim, E.K.; Cha, C.J.; Cho, Y.J.; Cho, Y.B.; Roe, J.H. Synthesis of gama-glutamylcysteine as a major low-molecular-weight thiol in lactic acid bacteria Leuconostoc spp. Biochem. Biophys. Res. Commun. 2008, 369, 1047–1051. [Google Scholar] [CrossRef]
- Masip, L.; Veeravalli, K.; Georgiou, G. The many faces of glutathione in bacteria. Antioxid. Redox Signal. 2006, 8, 753–762. [Google Scholar] [CrossRef]
- Xia, S.K.; Chen, L.; Liang, J.Q. Enriched selenium and its effects on growth and biochemical composition in Lactobacillus bulgaricus. J. Agric. Food Chem. 2007, 55, 2413–2417. [Google Scholar] [CrossRef]
- Lampis, S.; Zonaro, E.; Bertolini, C.; Cecconi, D.; Monti, F.; Micaroni, M.; Turner, R.J.; Butler, C.S.; Vallini, G. Selenite biotransformation and detoxification by Stenotrophomonas maltophilia SeITE02: Novel clues on the route to bacterial biogenesis of selenium nanoparticles. J. Hazar. Mater. 2017, 324, 3–14. [Google Scholar] [CrossRef]
- Martínez, F.G.; Moreno-Martin, G.; Pescuma, M.; Madrid-Albarrán, Y.; Mozzi, F. Biotransformation of selenium by lactic acid bacteria: Formation of seleno-nanoparticles and seleno-amino acids. Front. Bioeng. Biotechnol. 2020, 8, 506. [Google Scholar] [CrossRef] [PubMed]
- Pieniz, S.; Andreazza, R.; Mann, M.B.; Camargo, F.; Brandelli, A. Bioaccumulation and distribution of selenium in Enterococcus durans. J. Trace Elem. Med. Biol. 2017, 40, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Stabnikova, O.; Khonkiv, M.; Kovshar, I.; Stabnikov, V. Biosynthesis of selenium nanoparticles by lactic acid bacteria and areas of their possible applications. World J. Microbiol. Biotechnol. 2023, 39, 230. [Google Scholar] [CrossRef]
- Wadhwani, S.A.; Shedbalkar, U.U.; Singh, R.; Chopade, B.A. Biogenic selenium nanoparticles: Current status and future prospects. Appl. Microbiol. Biotechnol. 2016, 100, 2555–2566. [Google Scholar] [CrossRef] [PubMed]
- Palomo-Siguero, M.; Gutiérrez, A.M.; Pérez-Conde, C.; Madrid, Y. Effect of selenite and selenium nanoparticles on lactic bacteria: A multi-analytical study. Microchem. J. 2016, 126, 488–495. [Google Scholar] [CrossRef]
- Lampis, S.; Zonaro, E.; Bertolini, C.; Bernardi, P.; Butler, C.S.; Vallini, G. Delayed formation of zero-valent selenium nanoparticles by Bacillus mycoides SeiTE01 as a consequence of selenite reduction under aerobic conditions. Microb. Cell Factories 2014, 13, 35–49. [Google Scholar] [CrossRef]
- Fernández-Llamosas, H.; Castro, L.; Blázquez, M.L.; Díaz, E.; Carmona, M. Speeding up bioproduction of selenium nanoparticles by using Vibrio natriegens as microbial factory. Sci. Rep. 2017, 7, 16046. [Google Scholar] [CrossRef]
- Kousha, M.; Yeganeh, S.; Amirkolaie, A.K. Effect of sodium selenite on the bacteria growth, selenium accumulation, and selenium biotransformation in Pediococcus acidilactici. Food Sci. Biotechnol. 2017, 26, 1013–1018. [Google Scholar] [CrossRef]
- Sarret, G.; Avoscan, L.; Carrière, M.; Collins, R.; Geoffroy, N.; Carrot, F.; Covès, J.; Gouget, B. Chemical forms of selenium in the metal-resistant bacterium Ralstonia metallidurans CH34 exposed to selenite and selenate. Appl. Environ. Microbiol. 2005, 71, 2331–2337. [Google Scholar] [CrossRef]
- Kessi, J.; Hanselmann, K.W. Similarities between the abiotic reduction of selenite with glutathione and the dissimilatory reaction mediated by Rhodospirillum rubrum and Escherichia coli. J. Biol. Chem. 2004, 279, 50662–50669. [Google Scholar] [CrossRef]
- Kessi, J.; Ramuz, M.; Wehrli, E.; Spycher, M.; Bachofen, R. Reduction of selenite and detoxification of elemental selenium by the phototrophic bacterium Rhodospirillum rubrum. Appl. Environ. Microbiol. 1999, 65, 4734–4740. [Google Scholar] [CrossRef] [PubMed]
- Pophaly, S.D.; Singh, R.; Pophaly, S.D.; Kaushik, J.K.; Tomar, S.K. Current status and emerging role of glutathione in food grade lactic acid bacteria. Microb. Cell Factories 2012, 11, 114. [Google Scholar] [CrossRef] [PubMed]
- Scortecci, J.F.; Serrão, V.H.B.; Fernandes, A.F.; Basso, L.G.M.; Gutierrez, R.F.; Araujo, A.P.U.; Neto, M.O.; Thiemann, O.H. Initial steps in selenocysteine biosynthesis: The interaction between selenocysteine lyase and selenophosphate synthetase. Int. J. Biol. Macromol. 2020, 156, 18–26. [Google Scholar] [CrossRef]
- Manzine, L.R.; Cassago, A.; da Silva, M.T.A.; Thiemann, O.H. An efficient protocol for the production of tRNA-free recombinant Selenocysteine Synthase (SELA) from Escherichia coli and its biophysical characterization. Protein Expr. Purif. 2013, 88, 80–84. [Google Scholar] [CrossRef]
- Müller, S.; Heider, J.; Böck, A. The path of unspecific incorporation of selenium in Escherichia coli. Arch. Microbiol. 1997, 168, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Young, P.A.; Kaiser, I.I. Aminoacylation of Escherichia coli cysteine tRNA by selenocysteine. Arch. Biochem. Biophys. 1975, 171, 483–489. [Google Scholar] [CrossRef] [PubMed]


| Log CFU/mL | ||
|---|---|---|
| Na2SeO3 (mg/L) | 36 h | 48 h |
| 0 | 8.48 a ± 0.13 | 8.28 a ± 0.13 |
| 100 | 5.29 bc ± 0.08 | 5.63 c ± 0.10 |
| 200 | 5.95 b ± 0.13 | 6.13 b ± 0.13 |
| 300 | 5.15 c ± 0.09 | 5.78 bc ± 0.13 |
| 400 | 6.10 b ± 0.24 | 5.65 c ± 0.33 |
| 500 | 5.40 bc ± 0.52 | 4.59 d ± 0.06 |
| R2 | 0.8774 | 0.9363 |
| 80 mg/L | 102 mg/L | 184 mg/L | ||||
|---|---|---|---|---|---|---|
| Time (h) | Selenium in Cells (mg/L) | Bioaccumulation (µg Se/Log CFU) | Selenium in Cells (mg/L) | Bioaccumulation (µg Se/Log CFU) | Selenium in Cells (mg/L) | Bioaccumulation (µg de Se/Log CFU) |
| 24 | 10.87 ± 0.13 a | 1.85 ± 0.70 ab | 4.26 ± 0.51 a | 0.72 ± 0.38 b | 13.34 ± 2.59 a | 2.19 ± 0.44 a |
| 48 | 15.99 ± 1.04 b | 2.77 ± 0.63 a | 7.33 ± 1.78 b | 1.64 ± 0.80 a | 15.25 ± 1.32 a | 2.76 ± 0.92 a |
| 72 | 15.89 ± 0.55 b | 2.71 ± 0.52 a | 14.72 ± 0.56 c | 2.59 ± 0.10 a | 20.26 ± 1.89 b | 3.35 ± 0.63 a |
| 96 | 15.98 ± 0.10 b | 2.64 ± 0.68 b | 13.39 ± 0.76 c | 2.16 ± 0.24 b | 26.08 ± 3.53 b | 4.28 ± 0.86 a |
| 120 | 21.44 ± 0.13 c | 3.40 ± 0.88 b | 13.27 ± 0.41 c | 2.31 ± 0.37 c | 28.88 ± 2.99 a | 4.82 ± 0.87 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Escobar-Ramírez, M.C.; Rodríguez-Serrano, G.M.; Zúñiga-León, E.; García-Montes, M.A.; Pérez-Escalante, E.; González-Olivares, L.G. Selenocysteine Formation by Enterococcus faecium ABMC-05 Follows a Mechanism That Is Not Dependent on Genes selA and selD but on Gene cysK. Fermentation 2023, 9, 684. https://doi.org/10.3390/fermentation9070684
Escobar-Ramírez MC, Rodríguez-Serrano GM, Zúñiga-León E, García-Montes MA, Pérez-Escalante E, González-Olivares LG. Selenocysteine Formation by Enterococcus faecium ABMC-05 Follows a Mechanism That Is Not Dependent on Genes selA and selD but on Gene cysK. Fermentation. 2023; 9(7):684. https://doi.org/10.3390/fermentation9070684
Chicago/Turabian StyleEscobar-Ramírez, Meyli Claudia, Gabriela Mariana Rodríguez-Serrano, Eduardo Zúñiga-León, Mario Adolfo García-Montes, Emmanuel Pérez-Escalante, and Luis Guillermo González-Olivares. 2023. "Selenocysteine Formation by Enterococcus faecium ABMC-05 Follows a Mechanism That Is Not Dependent on Genes selA and selD but on Gene cysK" Fermentation 9, no. 7: 684. https://doi.org/10.3390/fermentation9070684
APA StyleEscobar-Ramírez, M. C., Rodríguez-Serrano, G. M., Zúñiga-León, E., García-Montes, M. A., Pérez-Escalante, E., & González-Olivares, L. G. (2023). Selenocysteine Formation by Enterococcus faecium ABMC-05 Follows a Mechanism That Is Not Dependent on Genes selA and selD but on Gene cysK. Fermentation, 9(7), 684. https://doi.org/10.3390/fermentation9070684







