Biosynthesis of Nicotinamide Mononucleotide: Current Metabolic Engineering Strategies, Challenges, and Prospects
Abstract
1. Introduction
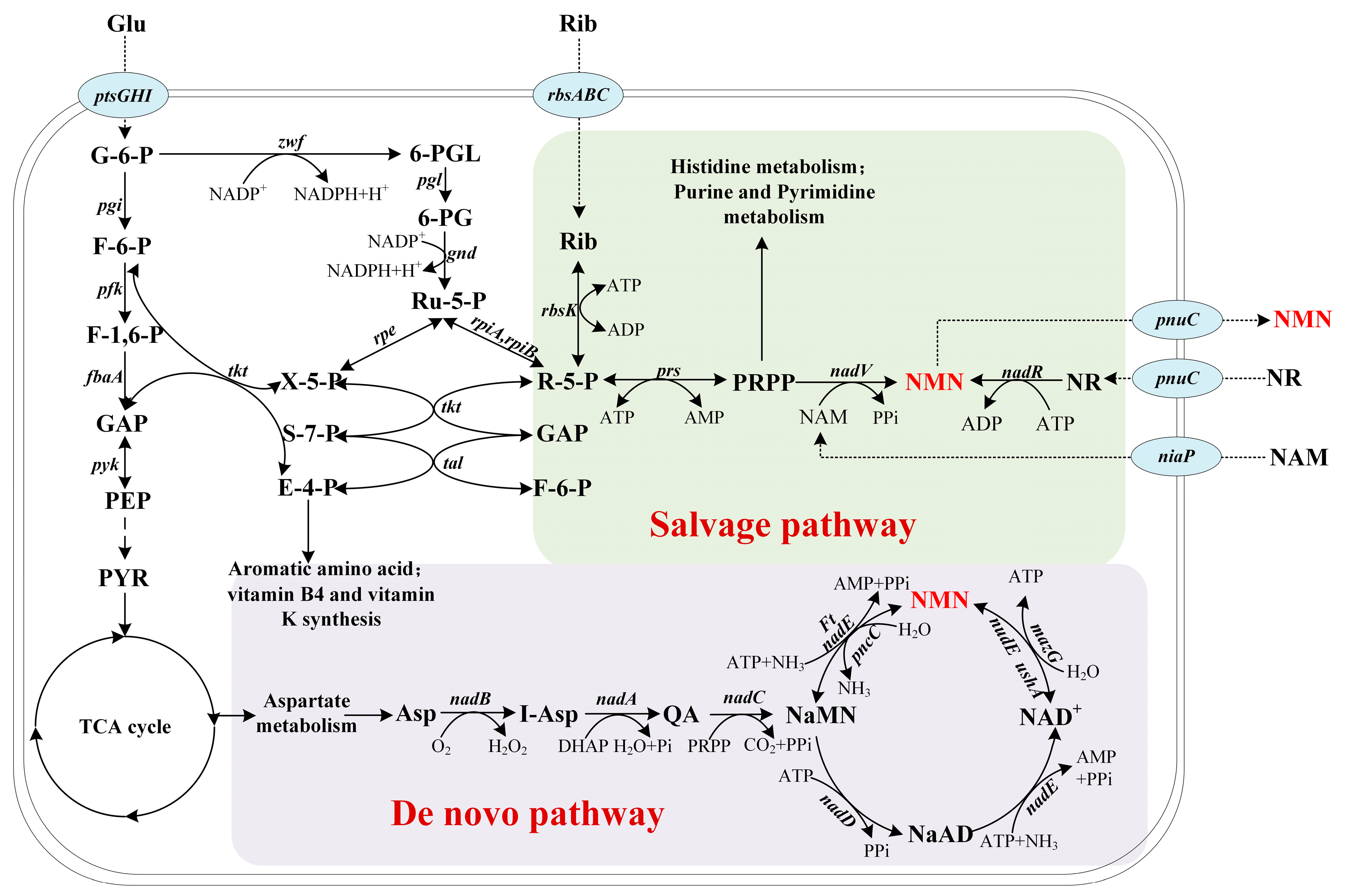
2. Metabolism of Nicotinamide Mononucleotide
2.1. De Novo Pathway
2.2. Salvage Pathway
3. Metabolic Engineering Strategies for Nicotinamide Mononucleotide Production
3.1. Selection and Directed Evolution of the Key Enzyme NAMPT
3.2. Improving the Supply of Essential Precursors
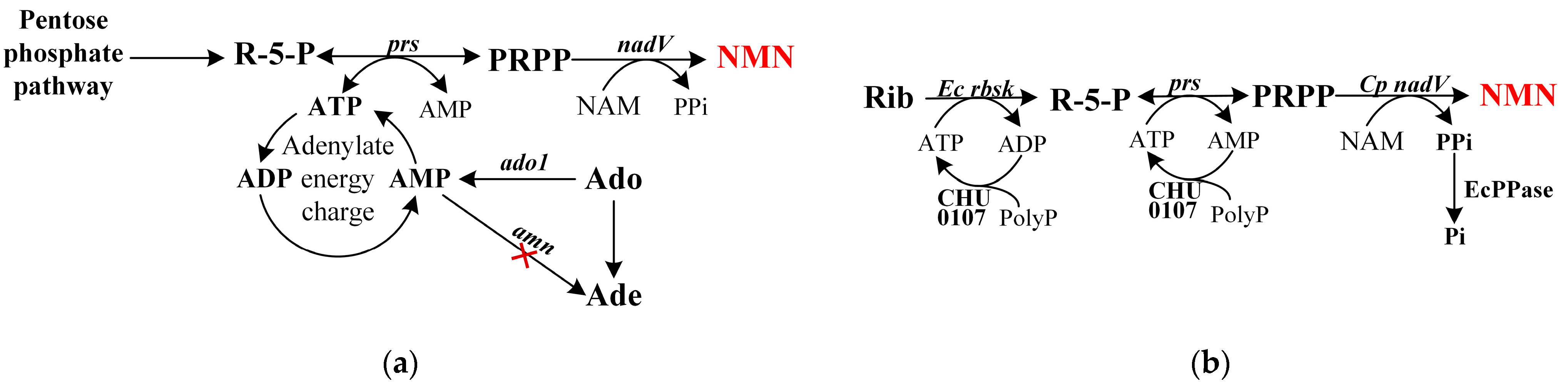
3.3. Constructing ATP Regeneration Systems
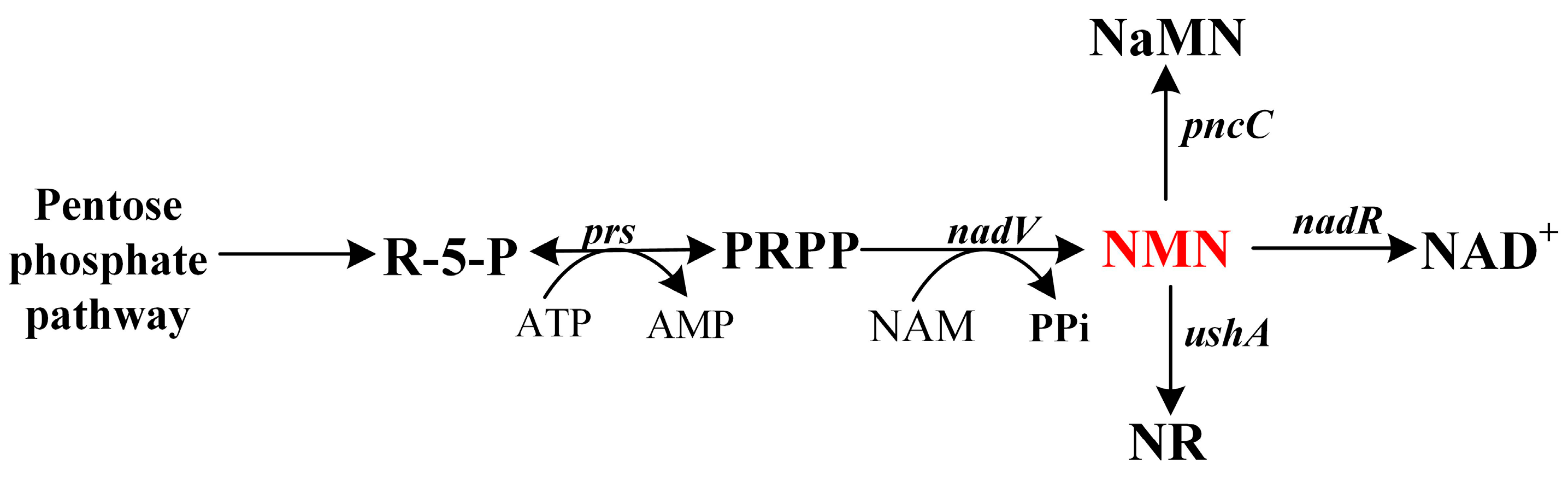
3.4. Blocking the Formation of Byproducts
3.5. Expressing Membrane Transporters to Export NMN form the Cell
4. Discussion
4.1. Utilizing in Silico Analysis and High-throughput Screening to Evolve Key Enzymes
4.2. Enhancing Synergetic Carbon Utilization to Improve Precursor Supply
4.3. Constructing Artificial Microbial Consortia to Reduce the Metabolic Burden
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dölle, C.; Skoge, R.H.; Vanlinden, M.R.; Ziegler, M. NAD biosynthesis in humans--enzymes, metabolites and therapeutic aspects. Curr. Top. Med. Chem. 2013, 13, 2907–2917. [Google Scholar] [CrossRef] [PubMed]
- Okabe, K.; Yaku, K.; Tobe, K.; Nakagawa, T. Implications of altered NAD metabolism in metabolic disorders. J. Biomed. Sci. 2019, 26, 34. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.M. Supplementation with NAD(+) and Its Precursors to Prevent Cognitive Decline across Disease Contexts. Nutrients 2022, 14, 3231. [Google Scholar] [CrossRef] [PubMed]
- Tang, E.; Shen, X.; Wang, J.; Sun, X.; Yuan, Q. Synergetic utilization of glucose and glycerol for efficient myo-inositol biosynthesis. Biotechnol. Bioeng. 2020, 117, 1247–1252. [Google Scholar] [CrossRef]
- Feng, Z.; Qin, Y.; Huo, F.; Jian, Z.; Li, X.; Geng, J.; Li, Y.; Wu, J. NMN recruits GSH to enhance GPX4-mediated ferroptosis defense in UV irradiation induced skin injury. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1868, 166287. [Google Scholar] [CrossRef]
- Shen, Q.; Zhang, S.J.; Xue, Y.Z.; Peng, F.; Cheng, D.Y.; Xue, Y.P.; Zheng, Y.G. Biological synthesis of nicotinamide mononucleotide. Biotechnol. Lett. 2021, 43, 2199–2208. [Google Scholar] [CrossRef]
- Preiss, J.; Handler, P. Enzymatic Synthesis of Nicotinamide Mononucleotide. J. Biol. Chem. 1957, 225, 759–770. [Google Scholar] [CrossRef]
- Li, Q.; Meng, D.; You, C. An artificial multi-enzyme cascade biocatalysis for biomanufacturing of nicotinamide mononucleotide from starch and nicotinamide in one-pot. Enzym. Microb. Technol. 2022, 162, 110122. [Google Scholar] [CrossRef]
- Sinclair, D.A. Biological Production of NAD Precursors and Analogs. WO Patent 2015/069860, 14 May 2015. [Google Scholar]
- Mills, K.F.; Yoshida, S.; Stein, L.R.; Grozio, A.; Kubota, S.; Sasaki, Y.; Redpath, P.; Migaud, M.E.; Apte, R.S.; Uchida, K.; et al. Long-Term Administration of Nicotinamide Mononucleotide Mitigates Age-Associated Physiological Decline in Mice. Cell Metab. 2016, 24, 795–806. [Google Scholar] [CrossRef]
- Huang, Z.; Li, N.; Yu, S.; Zhang, W.; Zhang, T.; Zhou, J. Systematic Engineering of Escherichia coli for Efficient Production of Nicotinamide Mononucleotide From Nicotinamide. ACS Synth. Biol. 2022, 11, 2979–2988. [Google Scholar] [CrossRef]
- Pollard, C.-L.; Younan, A.; Swegen, A.; Gibb, Z.; Grupen, C.G. Insights into the NAD(+) biosynthesis pathways involved during meiotic maturation and spindle formation in porcine oocytes. J. Reprod. Dev. 2022, 68, 216–224. [Google Scholar] [CrossRef]
- Groth, B.; Venkatakrishnan, P.; Lin, S.J. NAD(+) Metabolism, Metabolic Stress, and Infection. Front. Mol. Biosci. 2021, 8, 686412. [Google Scholar] [CrossRef]
- Sharma, S.; Hsieh, Y.C.; Dietze, J.; Bockwoldt, M.; Stromland, O.; Ziegler, M.; Heiland, I. Early Evolutionary Selection of NAD Biosynthesis Pathway in Bacteria. Metabolites 2022, 12, 569. [Google Scholar] [CrossRef]
- Yoshino, J.; Baur, J.A.; Imai, S.I. NAD(+) Intermediates: The Biology and Therapeutic Potential of NMN and NR. Cell Metab. 2018, 27, 513–528. [Google Scholar] [CrossRef]
- Navas, L.E.; Carnero, A. NAD(+) metabolism, stemness, the immune response, and cancer. Signal. Transduct. Target. Ther. 2021, 6, 2. [Google Scholar] [CrossRef]
- Zhu, F.Y.; Pena, M.; Bennett, G.N. Metabolic engineering of Escherichia coli for quinolinic acid production by assembling L-aspartate oxidase and quinolinate synthase as an enzyme complex. Metab. Eng. 2021, 67, 164–172. [Google Scholar] [CrossRef]
- Ollagnier-de Choudens, S.; Loiseau, L.; Sanakis, Y.; Barras, F.; Fontecave, M. Quinolinate synthetase, an iron-sulfur enzyme in NAD biosynthesis. FEBS Lett. 2005, 579, 3737–3743. [Google Scholar] [CrossRef]
- Yang, L.; Mu, X.; Nie, Y.; Xu, Y. Improving the production of NAD(+) via multi-strategy metabolic engineering in Escherichia coli. Metab. Eng. 2021, 64, 122–133. [Google Scholar] [CrossRef]
- Sorci, L.; Martynowski, D.; Rodionov, D.A.; Eyobo, Y.; Zogaj, X.; Klose, K.E.; Nikolaev, E.V.; Magni, G.; Zhang, H.; Osterman, A.L. Nicotinamide mononucleotide synthetase is the key enzyme for an alternative route of NAD biosynthesis in Francisella tularensis. Proc. Natl. Acad. Sci. USA 2009, 106, 3083–3088. [Google Scholar] [CrossRef]
- Wang, L.; Zhou, Y.J.; Ji, D.; Lin, X.; Liu, Y.; Zhang, Y.; Liu, W.; Zhao, Z.K. Identification of UshA as a major enzyme for NAD degradation in Escherichia coli. Enzym. Microb. Technol. 2014, 58–59, 75–79. [Google Scholar] [CrossRef]
- De Ingeniis, J.; Kazanov, M.D.; Shatalin, K.; Gelfand, M.S.; Osterman, A.L.; Sorci, L. Glutamine versus ammonia utilization in the NAD synthetase family. PLoS ONE 2012, 7, e39115. [Google Scholar] [CrossRef] [PubMed]
- Martin, P.R.; Shea, R.J.; Mulks, M.H. Identification of a plasmid-encoded gene from Haemophilus ducreyi which confers NAD independence. J. Bacteriol. 2001, 183, 1168–1174. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Li, Z.; Miller, E.S. Vibrio Phage KVP40 Encodes a Functional NAD(+) Salvage Pathway. J. Bacteriol. 2017, 199, e00855-16. [Google Scholar] [CrossRef] [PubMed]
- Bieganowski, P.; Brenner, C. Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a Preiss-Handler independent route to NAD+ in fungi and humans. Cell 2004, 117, 495–502. [Google Scholar] [CrossRef] [PubMed]
- Sonntag, T.; Ancel, S.; Karaz, S.; Cichosz, P.; Jacot, G.; Giner, M.P.; Sanchez-Garcia, J.L.; Pannerec, A.; Moco, S.; Sorrentino, V.; et al. Nicotinamide riboside kinases regulate skeletal muscle fiber-type specification and are rate-limiting for metabolic adaptations during regeneration. Front. Cell Dev. Biol. 2022, 10, 1049653. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, R.; Nakamura, S.; Nakazawa, T.; Minoura, K.; Yoshida, T.; Nishi, Y.; Kobayashi, Y.; Ohkubo, T. Structure and reaction mechanism of human nicotinamide phosphoribosyltransferase. J. Biochem. 2010, 147, 95–107. [Google Scholar] [CrossRef]
- Burgos, E.S.; Schramm, V.L. Weak coupling of ATP hydrolysis to the chemical equilibrium of human nicotinamide phosphoribosyltransferase. Biochemistry 2008, 47, 11086–11096. [Google Scholar] [CrossRef]
- Hove-Jensen, B.; Andersen, K.R.; Kilstrup, M.; Martinussen, J.; Switzer, R.L.; Willemoes, M. Phosphoribosyl Diphosphate (PRPP): Biosynthesis, Enzymology, Utilization, and Metabolic Significance. Microbiol. Mol. Biol. Rev. 2017, 81, e00040-16. [Google Scholar] [CrossRef]
- Shi, T.; Wang, Y.; Wang, Z.; Wang, G.; Liu, D.; Fu, J.; Chen, T.; Zhao, X. Deregulation of purine pathway in Bacillus subtilis and its use in riboflavin biosynthesis. Microb. Cell Factories 2014, 13, 101. [Google Scholar] [CrossRef]
- Zakataeva, N.P.; Romanenkov, D.V.; Skripnikova, V.S.; Vitushkina, M.V.; Livshits, V.A.; Kivero, A.D.; Novikova, A.E. Wild-type and feedback-resistant phosphoribosyl pyrophosphate synthetases from Bacillus amyloliquefaciens: Purification, characterization, and application to increase purine nucleoside production. Appl. Microbiol. Biotechnol. 2012, 93, 2023–2033. [Google Scholar] [CrossRef]
- Rappu, P.; Pullinen, T.; Mantsala, P. In vivo effect of mutations at the PRPP binding site of the Bacillus subtilis purine repressor. J. Bacteriol. 2003, 185, 6728–6731. [Google Scholar] [CrossRef]
- Singh, S.K.; Kurnasov, O.V.; Chen, B.; Robinson, H.; Grishin, N.V.; Osterman, A.L.; Zhang, H. Crystal structure of Haemophilus influenzae NadR protein. A bifunctional enzyme endowed with NMN adenyltransferase and ribosylnicotinimide kinase activities. J. Biol. Chem. 2002, 277, 33291–33299. [Google Scholar] [CrossRef]
- Kurnasov, O.V.; Polanuyer, B.M.; Ananta, S.; Sloutsky, R.; Tam, A.; Gerdes, S.Y.; Osterman, A.L. Ribosylnicotinamide kinase domain of NadR protein: Identification and implications in NAD biosynthesis. J. Bacteriol. 2002, 184, 6906–6917. [Google Scholar] [CrossRef]
- Qian, X.-L.; Dai, Y.-S.; Li, C.-X.; Pan, J.; Xu, J.-H.; Mu, B. Enzymatic synthesis of high-titer nicotinamide mononucleotide with a new nicotinamide riboside kinase and an efficient ATP regeneration system. Bioresour. Bioprocess. 2022, 9, 6906–6917. [Google Scholar] [CrossRef]
- He, J.J.; Liu, X.X.; Li, Y.; Wang, Z.; Shi, H.L.; Kan, Y.C.; Yao, L.G.; Tang, C.D. High level expression of nicotinamide nucleoside kinase from Saccharomyces cerevisiae and its purification and immobilization by one-step method. Front. Bioeng. Biotechnol. 2023, 11, 1134152. [Google Scholar] [CrossRef]
- Dolle, C.; Ziegler, M. Application of a coupled enzyme assay to characterize nicotinamide riboside kinases. Anal. Biochem. 2009, 385, 377–379. [Google Scholar] [CrossRef]
- Tempel, W.; Rabeh, W.M.; Bogan, K.L.; Belenky, P.; Wojcik, M.; Seidle, H.F.; Nedyalkova, L.; Yang, T.; Sauve, A.A.; Park, H.W.; et al. Nicotinamide riboside kinase structures reveal new pathways to NAD+. PLoS Biol. 2007, 5, e263. [Google Scholar] [CrossRef]
- Ratajczak, J.; Joffraud, M.; Trammell, S.A.; Ras, R.; Canela, N.; Boutant, M.; Kulkarni, S.S.; Rodrigues, M.; Redpath, P.; Migaud, M.E.; et al. NRK1 controls nicotinamide mononucleotide and nicotinamide riboside metabolism in mammalian cells. Nat. Commun. 2016, 7, 13103. [Google Scholar] [CrossRef]
- Zhu, N.; Olivera, B.M.; Roth, J.R. Activity of the nicotinamide mononucleotide transport system is regulated in Salmonella typhimurium. J. Bacteriol. 1991, 173, 1311–1320. [Google Scholar] [CrossRef]
- Sauer, E.; Merdanovic, M.; Mortimer, A.P.; Bringmann, G.; Reidl, J. PnuC and the utilization of the nicotinamide riboside analog 3-aminopyridine in Haemophilus influenzae. Antimicrob. Agents Chemother. 2004, 48, 4532–4541. [Google Scholar] [CrossRef]
- Grose, J.H.; Bergthorsson, U.; Xu, Y.; Sterneckert, J.; Khodaverdian, B.; Roth, J.R. Assimilation of nicotinamide mononucleotide requires periplasmic AphA phosphatase in Salmonella enterica. J. Bacteriol. 2005, 187, 4521–4530. [Google Scholar] [CrossRef] [PubMed]
- Gerasimova, A.V.; Gelfand, M.S. Evolution of the NadR regulon in Enterobacteriaceae. J. Bioinform. Comput. Biol. 2005, 3, 1007–1019. [Google Scholar] [CrossRef] [PubMed]
- Jaehme, M.; Slotboom, D.J. Structure, function, evolution, and application of bacterial Pnu-type vitamin transporters. Biol. Chem. 2015, 396, 955–966. [Google Scholar] [CrossRef] [PubMed]
- Todisco, S.; Agrimi, G.; Castegna, A.; Palmieri, F. Identification of the mitochondrial NAD+ transporter in Saccharomyces cerevisiae. J. Biol. Chem. 2006, 281, 1524–1531. [Google Scholar] [CrossRef] [PubMed]
- Belenky, P.A.; Moga, T.G.; Brenner, C. Saccharomyces cerevisiae YOR071C encodes the high affinity nicotinamide riboside transporter Nrt1. J. Biol. Chem. 2008, 283, 8075–8079. [Google Scholar] [CrossRef] [PubMed]
- Shoji, S.; Yamaji, T.; Makino, H.; Ishii, J.; Kondo, A. Metabolic design for selective production of nicotinamide mononucleotide from glucose and nicotinamide. Metab. Eng. 2021, 65, 167–177. [Google Scholar] [CrossRef]
- Shen, Y.P.; Liao, Y.L.; Lu, Q.; He, X.; Yan, Z.B.; Liu, J.Z. ATP and NADPH engineering of Escherichia coli to improve the production of 4-hydroxyphenylacetic acid using CRISPRi. Biotechnol. Biofuels 2021, 14, 100. [Google Scholar] [CrossRef]
- Liang, B.; Sun, G.N.; Zhang, X.P.; Nie, Q.J.; Zhao, Y.K.; Yang, J.M. Recent advances, challenges and metabolic engineering strategies in the biosynthesis of 3-hydroxypropionic acid. Biotechnol. Bioeng. 2022, 119, 2639–2668. [Google Scholar] [CrossRef]
- Jiang, M.; Hong, K.; Mao, Y.; Ma, H.; Chen, T.; Wang, Z. Natural 5-Aminolevulinic Acid: Sources, Biosynthesis, Detection and Applications. Front. Bioeng. Biotechnol. 2022, 10, 841443. [Google Scholar] [CrossRef]
- Marinescu, G.C.; Popescu, R.G.; Stoian, G.; Dinischiotu, A. beta-nicotinamide mononucleotide (NMN) production in Escherichia coli. Sci. Rep. 2018, 8, 12278. [Google Scholar] [CrossRef]
- Black, W.B.; Aspacio, D.; Bever, D.; King, E.; Zhang, L.; Li, H. Metabolic engineering of Escherichia coli for optimized biosynthesis of nicotinamide mononucleotide, a noncanonical redox cofactor. Microb. Cell Fact. 2020, 19, 150. [Google Scholar] [CrossRef]
- Liu, Y.; Yasawong, M.; Yu, B. Metabolic engineering of Escherichia coli for biosynthesis of beta-nicotinamide mononucleotide from nicotinamide. Microb. Biotechnol. 2021, 14, 2581–2591. [Google Scholar] [CrossRef]
- Maharjan, A.; Singhvi, M.; Kim, B.S. Biosynthesis of a Therapeutically Important Nicotinamide Mononucleotide through a Phosphoribosyl Pyrophosphate Synthetase 1 and 2 Engineered Strain of Escherichia coli. ACS Synth. Biol. 2021, 10, 3055–3065. [Google Scholar] [CrossRef]
- Sugiyama, K.; Iijima, K.; Yoshino, M.; Dohra, H.; Tokimoto, Y.; Nishikawa, K.; Idogaki, H.; Yoshida, N. Nicotinamide mononucleotide production by fructophilic lactic acid bacteria. Sci. Rep. 2021, 11, 7662. [Google Scholar] [CrossRef]
- Ngivprom, U.; Lasin, P.; Khunnonkwao, P.; Worakaensai, S.; Jantama, K.; Kamkaew, A.; Lai, R.Y. Synthesis of nicotinamide mononucleotide from xylose via coupling engineered Escherichia coli and a biocatalytic cascade. Chembiochem 2022, 23, e202200071. [Google Scholar] [CrossRef]
- He, Z.; Yang, X.; Tian, X.; Li, L.; Liu, M. Yeast Cell Surface Engineering of a Nicotinamide Riboside Kinase for the Production of beta-Nicotinamide Mononucleotide via Whole-Cell Catalysis. ACS Synth. Biol. 2022, 11, 3451–3459. [Google Scholar] [CrossRef]
- Yi, Y.-C.; Shih, I.T.; Yu, T.-H.; Lee, Y.-J.; Ng, I.S. Challenges and opportunities of bioprocessing 5-aminolevulinic acid using genetic and metabolic engineering: A critical review. Bioresour. Bioprocess. 2021, 8, 100. [Google Scholar] [CrossRef]
- Sharma, A.; Gupta, G.; Ahmad, T.; Mansoor, S.; Kaur, B. Enzyme Engineering: Current Trends and Future Perspectives. Food Rev. Int. 2019, 37, 121–154. [Google Scholar] [CrossRef]
- Fu, R.Z.; Zhang, Q. Nicotinamide Phosphoribosyltransferase Mutant and Application Thereof. WO Patent 2018/023206A1, 8 February 2018. [Google Scholar]
- Wang, Y.; Xue, P.; Cao, M.; Yu, T.; Lane, S.T.; Zhao, H. Directed Evolution: Methodologies and Applications. Chem. Rev. 2021, 121, 12384–12444. [Google Scholar] [CrossRef]
- Zhou, M.; Li, Y.; Che, H.; Sun, Y.; Wang, S.; Zhan, Y.; Cai, D.; Chen, S. Metabolic Engineering of Bacillus licheniformis for the Bioproduction of Nicotinamide Riboside from Nicotinamide and Glucose. ACS Sustain. Chem. Eng. 2023, 11, 6201–6210. [Google Scholar] [CrossRef]
- García-Calvo, L.; Rane, D.V.; Everson, N.; Humlebrekk, S.T.; Mathiassen, L.F.; Mæhlum, A.H.M.; Malmo, J.; Bruheim, P. Central carbon metabolite profiling reveals vector-associated differences in the recombinant protein production host Escherichia coli BL21. Front. Chem. Eng. 2023, 5, 1142226. [Google Scholar] [CrossRef]
- Kim, S.K.; Kim, H.; Woo, S.G.; Kim, T.H.; Rha, E.; Kwon, K.K.; Lee, H.; Lee, S.G.; Lee, D.H. CRISPRi-based programmable logic inverter cascade for antibiotic-free selection and maintenance of multiple plasmids. Nucleic Acids Res. 2022, 50, 13155–13171. [Google Scholar] [CrossRef] [PubMed]
- Shimada, T.; Kori, A.; Ishihama, A. Involvement of the ribose operon repressor RbsR in regulation of purine nucleotide synthesis in Escherichia coli. FEMS Microbiol. Lett. 2013, 344, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Strauch, M.A. AbrB modulates expression and catabolite repression of a Bacillus subtilis ribose transport operon. J. Bacteriol. 1995, 177, 6727–6731. [Google Scholar] [CrossRef] [PubMed]
- Zakataeva, N.P.; Romanenkov, D.V.; Yusupova, Y.R.; Skripnikova, V.S.; Asahara, T.; Gronskiy, S.V. Identification, Heterologous Expression, and Functional Characterization of Bacillus subtilis YutF, a HAD Superfamily 5′-Nucleotidase with Broad Substrate Specificity. PLoS ONE 2016, 11, e0167580. [Google Scholar] [CrossRef]
- Jeanguenin, L.; Lara-Nunez, A.; Rodionov, D.A.; Osterman, A.L.; Komarova, N.Y.; Rentsch, D.; Gregory, J.F., 3rd; Hanson, A.D. Comparative genomics and functional analysis of the NiaP family uncover nicotinate transporters from bacteria, plants, and mammals. Funct. Integr. Genom. 2012, 12, 25–34. [Google Scholar] [CrossRef]
- Zhou, W.J.; Tsai, A.; Dattmore, D.A.; Stives, D.P.; Chitrakar, I.; D’Alessandro, A.M.; Patil, S.; Hicks, K.A.; French, J.B. Crystal structure of E. coli PRPP synthetase. Bmc Struct. Biol. 2019, 19, 1. [Google Scholar] [CrossRef]
- Luo, Z.; Zeng, W.; Du, G.; Chen, J.; Zhou, J. Enhanced Pyruvate Production in Candida glabrata by Engineering ATP Futile Cycle System. ACS Synth. Biol. 2019, 8, 787–795. [Google Scholar] [CrossRef]
- Barrado, P.; Rodriguez, M.J.; Jimenez, A.; Fernandez Lobato, M. Expression in Escherichia coli of a recombinant adenosine kinase from Saccharomyces cerevisiae: Purification, kinetics and substrate analyses. Yeast 2003, 20, 1145–1150. [Google Scholar] [CrossRef]
- Leung, H.B.; Kvalnes-Krick, K.L.; Meyer, S.L.; deRiel, J.K.; Schramm, V.L. Structure and regulation of the AMP nucleosidase gene (amn) from Escherichia coli. Biochemistry 1989, 28, 8726–8733. [Google Scholar] [CrossRef]
- Wan, L.; Wang, X.Y.; Hu, Y.H.; Li, Q.; Zhao, Z.K. Gram-scale biocatalytic preparation of the non-natural cofactor nicotinamide cytosine dinucleotide. Tetrahedron Lett. 2022, 88, 153568. [Google Scholar] [CrossRef]
- Hara, N.; Yamada, K.; Shibata, T.; Osago, H.; Tsuchiya, M. Nicotinamide phosphoribosyltransferase/visfatin does not catalyze nicotinamide mononucleotide formation in blood plasma. PLoS ONE 2011, 6, e22781. [Google Scholar] [CrossRef]
- Kong, L.H.; Liu, T.Y.; Yao, Q.S.; Zhang, X.H.; Xu, W.N.; Qin, J.Y. Enhancing the biosynthesis of nicotinamide mononucleotide in Lactococcus lactis by heterologous expression of FtnadE. J. Sci. Food Agric. 2022, 103, 450–456. [Google Scholar] [CrossRef]
- Panchapakesan, S.S.S.; Corey, L.; Malkowski, S.N.; Higgs, G.; Breaker, R.R. A second riboswitch class for the enzyme cofactor NAD(+). Rna 2021, 27, 99–105. [Google Scholar] [CrossRef]
- Ali, M.; Ishqi, H.M.; Husain, Q. Enzyme engineering: Reshaping the biocatalytic functions. Biotechnol. Bioeng. 2020, 117, 1877–1894. [Google Scholar] [CrossRef]
- Currin, A.; Parker, S.; Robinson, C.J.; Takano, E.; Scrutton, N.S.; Breitling, R. The evolving art of creating genetic diversity: From directed evolution to synthetic biology. Biotechnol. Adv. 2021, 50, 107762. [Google Scholar] [CrossRef]
- Magni, G.; Di Stefano, M.; Orsomando, G.; Raffaelli, N.; Ruggieri, S. NAD(P) Biosynthesis Enzymes as Potential Targets for Selective Drug Design. Curr. Med. Chem. 2009, 16, 1372–1390. [Google Scholar] [CrossRef]
- Ren, F.; Ding, X.; Zheng, M.; Korzinkin, M.; Cai, X.; Zhu, W.; Mantsyzov, A.; Aliper, A.; Aladinskiy, V.; Cao, Z.; et al. AlphaFold accelerates artificial intelligence powered drug discovery: Efficient discovery of a novel CDK20 small molecule inhibitor. Chem. Sci. 2023, 14, 1443–1452. [Google Scholar] [CrossRef]
- Madhavan, A.; Arun, K.B.; Binod, P.; Sirohi, R.; Tarafdar, A.; Reshmy, R.; Kumar Awasthi, M.; Sindhu, R. Design of novel enzyme biocatalysts for industrial bioprocess: Harnessing the power of protein engineering, high throughput screening and synthetic biology. Bioresour. Technol. 2021, 325, 124617. [Google Scholar] [CrossRef]
- Tournier, V.; Topham, C.M.; Gilles, A.; David, B.; Folgoas, C.; Moya-Leclair, E.; Kamionka, E.; Desrousseaux, M.L.; Texier, H.; Gavalda, S.; et al. An engineered PET depolymerase to break down and recycle plastic bottles. Nature 2020, 580, 216–219. [Google Scholar] [CrossRef]
- Zhang, R.Y.; Qin, Y.; Lv, X.Q.; Wang, P.; Xu, T.Y.; Zhang, L.; Miao, C.Y. A fluorometric assay for high-throughput screening targeting nicotinamide phosphoribosyltransferase. Anal. Biochem. 2011, 412, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Nentwich, S.S.; Brinkrolf, K.; Gaigalat, L.; Hüser, A.T.; Rey, D.A.; Mohrbach, T.; Marin, K.; Pühler, A.; Tauch, A.; Kalinowski, J. Characterization of the LacI-type transcriptional repressor RbsR controlling ribose transport in Corynebacterium glutamicum ATCC 13032. Microbiology 2009, 155, 150–164. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Wang, J.; Shen, X.; Wang, J.; Chen, Z.; Sun, X.; Yuan, Q.; Yan, Y. Investigating the strategies for microbial production of trehalose from lignocellulosic sugars. Biotechnol. Bioeng. 2018, 115, 785–790. [Google Scholar] [CrossRef] [PubMed]
- Gawand, P.; Hyland, P.; Ekins, A.; Martin, V.J.; Mahadevan, R. Novel approach to engineer strains for simultaneous sugar utilization. Metab. Eng. 2013, 20, 63–72. [Google Scholar] [CrossRef]
- Luo, H.; Li, P.; Ji, B.; Nielsen, J. Modeling the metabolic dynamics at the genome-scale by optimized yield analysis. Metab. Eng. 2023, 75, 119–130. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, X. Modular co-culture engineering, a new approach for metabolic engineering. Metab. Eng. 2016, 37, 114–121. [Google Scholar] [CrossRef]
- Gardner, J.G.; Escalante-Semerena, J.C. In Bacillus subtilis, the sirtuin protein deacetylase, encoded by the srtN gene (formerly yhdZ), and functions encoded by the acuABC genes control the activity of acetyl coenzyme A synthetase. J. Bacteriol. 2009, 191, 1749–1755. [Google Scholar] [CrossRef]
- Dijamentiuk, A.; Mangavel, C.; Elfassy, A.; Michaux, F.; Burgain, J.; Rondags, E.; Delaunay, S.; Ferrigno, S.; Revol-Junelles, A.M.; Borges, F. Invert emulsions alleviate biotic interactions in bacterial mixed culture. Microb. Cell Factories 2023, 22, 16. [Google Scholar] [CrossRef]
- Özkaya, Ö.; Xavier, K.B.; Dionisio, F.; Balbontín, R. Maintenance of Microbial Cooperation Mediated by Public Goods in Single- and Multiple-Trait Scenarios. J. Bacteriol. 2017, 199, e00297-17. [Google Scholar] [CrossRef]
| Chassis | Strategies | Cultivation | Intracellular NMN Production (mg/L) | Extracellular NMN Production (mg/L) | Reference |
|---|---|---|---|---|---|
| S. cerevisiae | Overexpression of nadV; deletion of nma1 1 | Bioreactor 2 | ND 3 (300–350 nmol per g of wet cells) | ND 3 | [9] |
| E. coli | Overexpression of nadV | 500 mL benchtop bioreactor | 7877.59 | 10.89 | [51] |
| E. coli | Overexpression of nadV and prs (with L135I mutation) | 500 mL benchtop bioreactor | 7563.42 | 15.42 | [51] |
| E. coli | Overexpression of nadV and nadE; deletion of pncC | 2 mL deep-well plate | 501.33 | ND 3 | [52] |
| E. coli | Overexpression of nadV, niaP, pnuC, pig, zwf, pgl, gnd, ripA, ripB, and prs | 2 L fermenter | ND 3 | 6.79 × 103 | [47] |
| E. coli | Overexpression of nadV, ygcS, prs (with L135I mutation), zwf, gnd, and ado1; deletion of nadR, pncC, amn, and purR 4 | Flasks 2 | ND 3 | 496.2 | [53] |
| E. coli | Overexpression nadV, prs1, and prs2; optimization of the amounts of ribose, Mg2+, phosphate, nicotinamide, and lactose inducer (using response surface methodology) | 250 mL Erlenmeyer flask | 772.05 | ND 3 | [54] |
| Fructobacillus durionis | Lactic acid bacteria isolated from natural resources cultivated in MRS medium containing 1% D-fructose for 12 h | Flasks 2 | 2.1 | ND 3 | [55] |
| E. coli | Overexpression of nadV, prs (with L135I mutation), and pnuC; deletion of nadR, pncC, ushA, and purR; controlling the supplementation of nicotinamide and dissolved oxygen level | 5 L fermenter | ND 3 | 1.62 × 104 | [11] |
| E. coli | Deletion of tktA 5, tktB 5, and ptsG 5; cascade bioconversion using EcRBSK, EcPRPS, CpNAMPT, CHU0107, and EcPPase | 500 mL baffled flask and 10 mL scale bioreactor | ND 3 | 284.09 | [56] |
| S. cerevisiae EBY100 | Overexpression of nrk2 6 (displayed on the cell surface); optimization of the amounts of NR, ATP, and Mg2+; optimization of pH and temperature | 30 mL screw vial | ND 3 | 1.26 × 104 | [57] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, S.; Zhao, J.; Zheng, Y.; Chen, T.; Wang, Z. Biosynthesis of Nicotinamide Mononucleotide: Current Metabolic Engineering Strategies, Challenges, and Prospects. Fermentation 2023, 9, 594. https://doi.org/10.3390/fermentation9070594
Luo S, Zhao J, Zheng Y, Chen T, Wang Z. Biosynthesis of Nicotinamide Mononucleotide: Current Metabolic Engineering Strategies, Challenges, and Prospects. Fermentation. 2023; 9(7):594. https://doi.org/10.3390/fermentation9070594
Chicago/Turabian StyleLuo, Shiqi, Juntao Zhao, Yangyang Zheng, Tao Chen, and Zhiwen Wang. 2023. "Biosynthesis of Nicotinamide Mononucleotide: Current Metabolic Engineering Strategies, Challenges, and Prospects" Fermentation 9, no. 7: 594. https://doi.org/10.3390/fermentation9070594
APA StyleLuo, S., Zhao, J., Zheng, Y., Chen, T., & Wang, Z. (2023). Biosynthesis of Nicotinamide Mononucleotide: Current Metabolic Engineering Strategies, Challenges, and Prospects. Fermentation, 9(7), 594. https://doi.org/10.3390/fermentation9070594






