Exploration of Natural Product Repository by Combined Genomics and Metabolomics Profiling of Mangrove-Derived Streptomyces murinus THV12 Strain
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of S. murinus THV12 Strain
2.2. Antimicrobial Activity Assay of S. murinus-Derived Secondary Metabolites
2.3. Determination of Minimum Inhibitory Concentration (MIC) and Minimum Bactericidal Concentration (MBC)
2.4. Whole-Genome Sequencing and Analysis of S. murinus THV12 Strain
2.5. One Strain Many Compounds Strategy for S. murinus THV 12 Strain
2.6. LC-HR-MS/MS-Guided Analysis of Secondary Metabolites
2.7. Principal Component Analysis
3. Results
3.1. Antagonistic Activity of S. murinus THV12
3.2. Minimum Inhibitory Concentration and Minimum Bactericidal Concentration of Metabolite Extract
3.3. Genome Features and Annotation
3.4. Insights into the BGC Organization of S. murinus THV 12
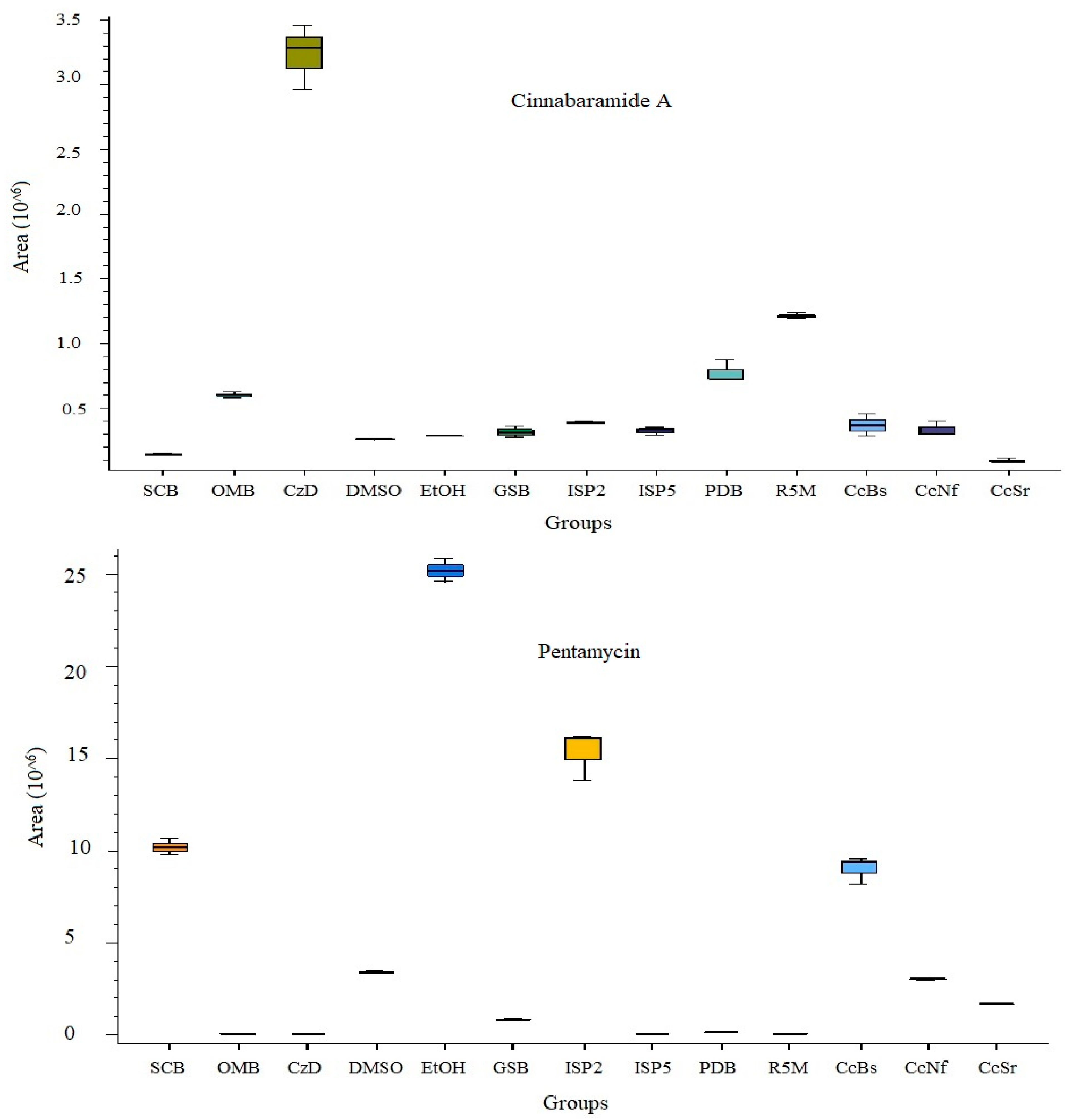
3.5. Secondary Metabolite Profile of S. murinus THV12
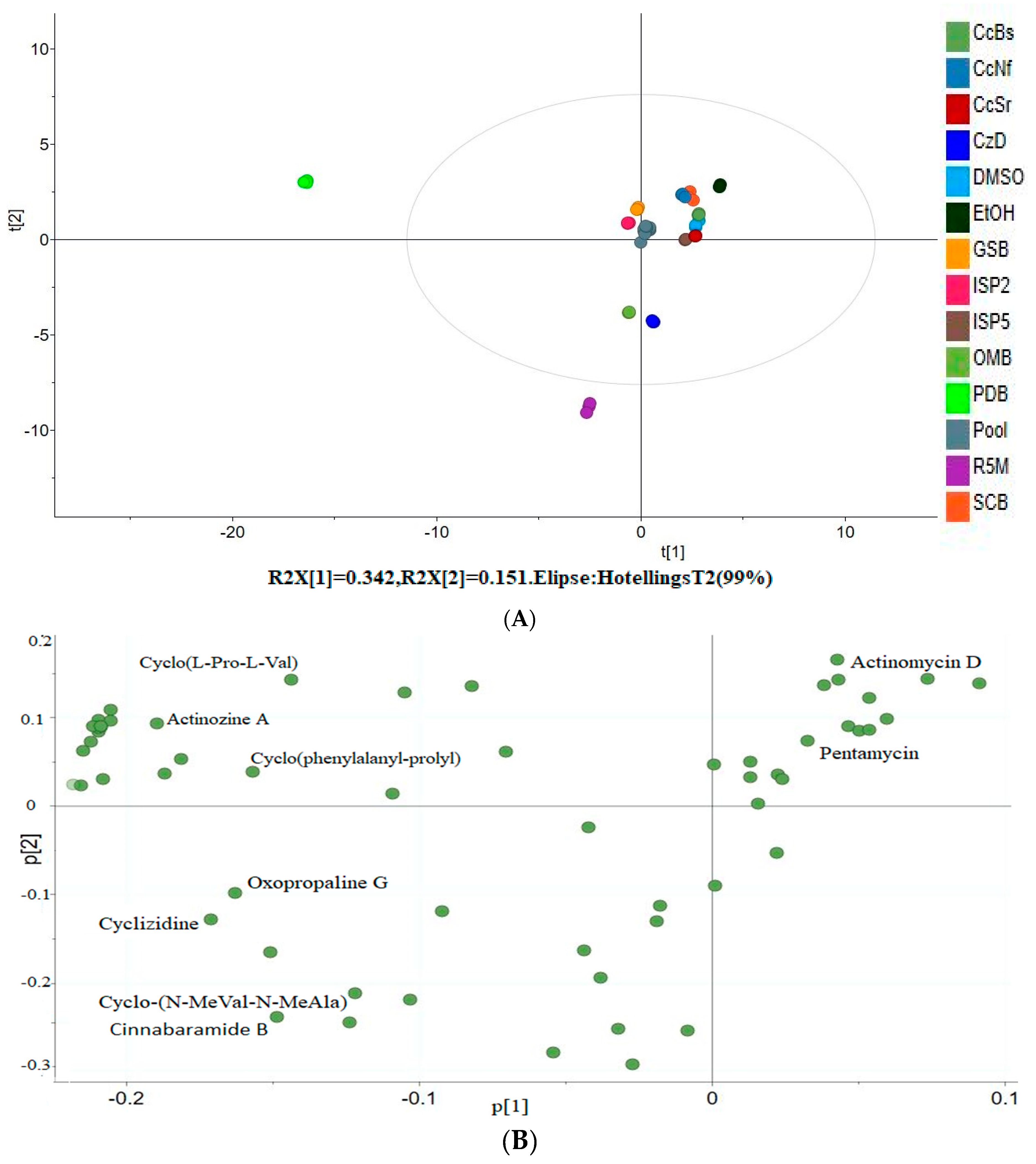
3.6. Principal Component Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Harir, M.; Bendif, H.; Bellahcene, M.; Fortas, Z.; Pogni, R. Streptomyces Secondary Metabolites. In Basic Biology and Applications of Actinobacteria; Enany, S., Ed.; IntechOpen: London, UK, 2018; ISBN 9781789846140. [Google Scholar]
- Manteca, Á.; Yagüe, P. Streptomyces as a Source of Antimicrobials: Novel Approaches to Activate Cryptic Secondary Metabolite Pathways. In Antimicrobials, Antibiotic Resistance, Antibiofilm Strategies and Activity Methods; Kırmusaoğlu, S., Ed.; IntechOpen: London, UK, 2019; ISBN 9781789857894. [Google Scholar]
- Alam, K.; Mazumder, A.; Sikdar, S.; Zhao, Y.-M.; Hao, J.; Song, C.; Wang, Y.; Sarkar, R.; Islam, S.; Zhang, Y.; et al. Streptomyces: The Biofactory of Secondary Metabolites. Front. Microbiol. 2022, 13, 968053. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Ren, Z.; Chunyu, W.-X.; Li, G.-D.; Chen, X.; Zhang, Z.-T.-L.; Sun, H.-B.; Wang, M.; Xie, T.-P.; Wang, M.; et al. Exploration of Diverse Secondary Metabolites from Streptomyces sp. YINM00001, Using Genome Mining and One Strain Many Compounds Approach. Front. Microbiol. 2022, 13, 831174. [Google Scholar] [CrossRef] [PubMed]
- Hemphill, C.F.P.; Sureechatchaiyan, P.; Kassack, M.U.; Orfali, R.S.; Lin, W.; Daletos, G.; Proksch, P. OSMAC Approach Leads to New Fusarielin Metabolites from Fusarium Tricinctum. J. Antibiot. 2017, 70, 726–732. [Google Scholar] [CrossRef] [PubMed]
- Bibb, M.J. Regulation of Secondary Metabolism in Streptomycetes. Curr. Opin. Microbiol. 2005, 8, 208–215. [Google Scholar] [CrossRef]
- Zong, G.; Fu, J.; Zhang, P.; Zhang, W.; Xu, Y.; Cao, G.; Zhang, R. Use of Elicitors to Enhance or Activate the Antibiotic Production in Streptomyces. Crit. Rev. Biotechnol. 2022, 42, 1260–1283. [Google Scholar] [CrossRef]
- Abdelmohsen, U.R.; Grkovic, T.; Balasubramanian, S.; Kamel, M.S.; Quinn, R.J.; Hentschel, U. Elicitation of Secondary Metabolism in Actinomycetes. Biotechnol. Adv. 2015, 33, 798–811. [Google Scholar] [CrossRef]
- Marmann, A.; Aly, A.; Lin, W.; Wang, B.; Proksch, P. Co-Cultivation—A Powerful Emerging Tool for Enhancing the Chemical Diversity of Microorganisms. Mar. Drugs 2014, 12, 1043–1065. [Google Scholar] [CrossRef]
- Moody, S.C. Microbial Co-Culture: Harnessing Intermicrobial Signaling for the Production of Novel Antimicrobials. Future Microbiol. 2014, 9, 575–578. [Google Scholar] [CrossRef]
- Hayakawa, M.; Kajiura, T.; Nonomura, H. New Methods for the Highly Selective Isolation of Streptosporangium and Dactylosporangium from Soil. J. Ferment. Bioeng. 1991, 72, 327–333. [Google Scholar] [CrossRef]
- Chen, X.; Jiang, Y.; Li, Q.; Han, L.; Jiang, C. Molecular Phylogenetic Identification of Actinobacteria. In Actinobacteria—Basics and Biotechnological Applications; Dhanasekaran, D., Jiang, Y., Eds.; IntechOpen: London, UK, 2016; ISBN 9789535122487. [Google Scholar]
- CLSI-M07-A10; Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically. Approved Standard M7-A5. National Committee for Clinical Laboratory Standards: Wayne, PA, USA, 2000.
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. AntiSMASH 6.0: Improving Cluster Detection and Comparison Capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Romano, S.; Jackson, S.; Patry, S.; Dobson, A. Extending the “One Strain Many Compounds” (OSMAC) Principle to Marine Microorganisms. Mar. Drugs 2018, 16, 244. [Google Scholar] [CrossRef]
- Tangerina, M.M.P.; Furtado, L.C.; Leite, V.M.B.; Bauermeister, A.; Velasco-Alzate, K.; Jimenez, P.C.; Garrido, L.M.; Padilla, G.; Lopes, N.P.; Costa-Lotufo, L.V.; et al. Metabolomic Study of Marine Streptomyces Sp.: Secondary Metabolites and the Production of Potential Anticancer Compounds. PLoS ONE 2020, 15, e0244385. [Google Scholar] [CrossRef]
- Pettit, R.K. Small-Molecule Elicitation of Microbial Secondary Metabolites: Elicitation of Microbial Secondary Metabolites. Microb. Biotechnol. 2011, 4, 471–478. [Google Scholar] [CrossRef]
- Chen, G.; Wang, G.-Y.-S.; Li, X.; Waters, B.; Davies, J. Enhanced Production of Microbial Metabolites in the Presence of Dimethyl Sulfoxide. J. Antibiot. 2000, 53, 1145–1153. [Google Scholar] [CrossRef] [PubMed]
- Doull, J.L.; Singh, A.K.; Hoare, M.; Ayer, S.W. Conditions for the Production of Jadomycin B by Streptomyces Venezuelae ISP5230: Effects of Heat Shock, Ethanol Treatment and Phage Infection. J. Ind. Microbiol. 1994, 13, 120–125. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Chen, J.; Wang, Z.; Wang, C. Integrated Metabolomic and Network Analysis to Explore the Potential Mechanism of Three Chemical Elicitors in Rapamycin Overproduction. Microorganisms 2022, 10, 2205. [Google Scholar] [CrossRef] [PubMed]
- Pan, R.; Bai, X.; Chen, J.; Zhang, H.; Wang, H. Exploring Structural Diversity of Microbe Secondary Metabolites Using OSMAC Strategy: A Literature Review. Front. Microbiol. 2019, 10, 294. [Google Scholar] [CrossRef]
- Niu, G.; Chater, K.F.; Tian, Y.; Zhang, J.; Tan, H. Specialised Metabolites Regulating Antibiotic Biosynthesis in Streptomyces Spp. FEMS Microbiol. Rev. 2016, 40, 554–573. [Google Scholar] [CrossRef] [PubMed]
- Onaka, H.; Mori, Y.; Igarashi, Y.; Furumai, T. Mycolic Acid-Containing Bacteria Induce Natural-Product Biosynthesis in Streptomyces Species. Appl. Env. Microbiol. 2011, 77, 400–406. [Google Scholar] [CrossRef]
- Sung, A.; Gromek, S.; Balunas, M. Upregulation and Identification of Antibiotic Activity of a Marine-Derived Streptomyces Sp. via Co-Cultures with Human Pathogens. Mar. Drugs 2017, 15, 250. [Google Scholar] [CrossRef]
- van Heel, A.J.; de Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A User-Friendly Web Server to Thoroughly Mine RiPPs and Bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef] [PubMed]
- Machushynets, N.V.; Elsayed, S.S.; Du, C.; Siegler, M.A.; De La Cruz, M.; Genilloud, O.; Hankemeier, T.; Van Wezel, G.P. Discovery of Actinomycin L, a New Member of the Actinomycin Family of Antibiotics. Sci. Rep. 2022, 12, 2813. [Google Scholar] [CrossRef]
- Felnagle, E.A.; Barkei, J.J.; Park, H.; Podevels, A.M.; McMahon, M.D.; Drott, D.W.; Thomas, M.G. MbtH-Like Proteins as Integral Components of Bacterial Nonribosomal Peptide Synthetases. Biochemistry 2010, 49, 8815–8817. [Google Scholar] [CrossRef] [PubMed]
- Bursy, J.; Kuhlmann, A.U.; Pittelkow, M.; Hartmann, H.; Jebbar, M.; Pierik, A.J.; Bremer, E. Synthesis and Uptake of the Compatible Solutes Ectoine and 5-Hydroxyectoine by Streptomyces Coelicolor A3(2) in Response to Salt and Heat Stresses. Appl. Environ. Microbiol. 2008, 74, 7286–7296. [Google Scholar] [CrossRef] [PubMed]
- Zong, C.; Cheung-Lee, W.L.; Elashal, H.E.; Raj, M.; Link, A.J. Albusnodin: An Acetylated Lasso Peptide from Streptomyces albus. Chem. Commun. 2018, 54, 1339–1342. [Google Scholar] [CrossRef]
- Udwary, D.W.; Zeigler, L.; Asolkar, R.N.; Singan, V.; Lapidus, A.; Fenical, W.; Jensen, P.R.; Moore, B.S. Genome Sequencing Reveals Complex Secondary Metabolome in the Marine Actinomycete Salinispora tropica. Proc. Natl. Acad. Sci. USA 2007, 104, 10376–10381. [Google Scholar] [CrossRef]
- Lackner, G.; Moebius, N.; Partida-Martinez, L.; Hertweck, C. Complete Genome Sequence of Burkholderia Rhizoxinica, an Endosymbiont of Rhizopus microsporus. J. Bacteriol. 2011, 193, 783–784. [Google Scholar] [CrossRef]
- Thomas, D.; Morris, M.; Curtis, J.M.; Boyd, R.K. Fragmentation Mechanisms of Protonated Actinomycins and Their Use in Structural Determination of Unknown Analogues. J. Mass Spectrom. 1995, 30, 1111–1125. [Google Scholar] [CrossRef]
- Moumbock, A.F.A.; Gao, M.; Qaseem, A.; Li, J.; Kirchner, P.A.; Ndingkokhar, B.; Bekono, B.D.; Simoben, C.V.; Babiaka, S.B.; Malange, Y.I.; et al. StreptomeDB 3.0: An Updated Compendium of Streptomycetes Natural Products. Nucleic Acids Res. 2021, 49, D600–D604. [Google Scholar] [CrossRef]
- Borgman, P.; Lopez, R.D.; Lane, A.L. The Expanding Spectrum of Diketopiperazine Natural Product Biosynthetic Pathways Containing Cyclodipeptide Synthases. Org. Biomol. Chem. 2019, 17, 2305–2314. [Google Scholar] [CrossRef]
- Yao, T.; Liu, J.; Liu, Z.; Li, T.; Li, H.; Che, Q.; Zhu, T.; Li, D.; Gu, Q.; Li, W. Genome Mining of Cyclodipeptide Synthases Unravels Unusual TRNA-Dependent Diketopiperazine-Terpene Biosynthetic Machinery. Nat. Commun. 2018, 9, 4091. [Google Scholar] [CrossRef] [PubMed]
- Forner, D.; Berrué, F.; Correa, H.; Duncan, K.; Kerr, R.G. Chemical Dereplication of Marine Actinomycetes by Liquid Chromatography–High Resolution Mass Spectrometry Profiling and Statistical Analysis. Anal. Chim. Acta 2013, 805, 70–79. [Google Scholar] [CrossRef] [PubMed]
- Gill, K.A.; Berrué, F.; Arens, J.C.; Carr, G.; Kerr, R.G. Cystargolides, 20S Proteasome Inhibitors Isolated from Kitasatospora cystarginea. J. Nat. Prod. 2015, 78, 822–826. [Google Scholar] [CrossRef] [PubMed]
- Alanjary, M.; Kronmiller, B.; Adamek, M.; Blin, K.; Weber, T.; Huson, D.; Philmus, B.; Ziemert, N. The Antibiotic Resistant Target Seeker (ARTS), an Exploration Engine for Antibiotic Cluster Prioritization and Novel Drug Target Discovery. Nucleic Acids Res. 2017, 45, W42–W48. [Google Scholar] [CrossRef]
- Song, Z.; Hou, Y.; Yang, Q.; Li, X.; Wu, S. Structures and Biological Activities of Diketopiperazines from Marine Organisms: A Review. Mar. Drugs 2021, 19, 403. [Google Scholar] [CrossRef]
- Stadler, M.; Bitzer, J.; Mayer-Bartschmid, A.; Müller, H.; Benet-Buchholz, J.; Gantner, F.; Tichy, H.-V.; Reinemer, P.; Bacon, K.B. Cinnabaramides A−G: Analogues of Lactacystin and Salinosporamide from a Terrestrial Streptomycete. J. Nat. Prod. 2007, 70, 246–252. [Google Scholar] [CrossRef]
- Lara, A.C.; Corretto, E.; Kotrbová, L.; Lorenc, F.; Petříčková, K.; Grabic, R.; Chroňáková, A. The Genome Analysis of the Human Lung-Associated Streptomyces Sp. TR1341 Revealed the Presence of Beneficial Genes for Opportunistic Colonization of Human Tissues. Microorganisms 2021, 9, 1547. [Google Scholar] [CrossRef]





| Test Microorganisms | MIC Values (µg/mL) | MBC Values (µg/mL) | |
|---|---|---|---|
| Extract | Standard Antibiotic | Extract | |
| MRSA | 4 | 10 | 8 |
| E. cloacae | 8 | 10 | 16 |
| L. monocytogenes | 2 | 10 | 4 |
| E. faecalis | 8 | 20 | 16 |
| B. cereus | 2 | 10 | 4 |
| C. albicans | 4 | 10 | 8 |
| K. pneumoniae | 128 | 20 | 256 |
| A. hydrophila | 8 | 10 | 16 |
| Features | Values |
|---|---|
| Topology | Linear |
| Genome size (bp) | 8,363,247 |
| Largest contig | 735,030 |
| Average G + C content (%) | 71.80 % |
| N50 | 305,657 |
| L50 | 10 |
| Protein-coding genes | 7345 |
| tRNA genes | 85 |
| Cluster No. | Type | Location | Predicted Product | Similarity (%) | Reference Strain | Accession Number |
|---|---|---|---|---|---|---|
| Cluster 1 | Melanin | 660,228–670,839 | melanin | 60 | Streptomyces avermitilis | AB070939 |
| Cluster 2 | NRPS | 161,283–252,556 | kirromycin | 16 | Streptomyces collinus Tu 365 | AM746336 |
| Cluster 3 | Terpene | 346,928–399,702 | hopene | 92 | Streptomyces coelicolor A3(2) | AL645882 |
| Cluster 4 | NRPS | 431,106–460,838 | actinomycin D | 71 | Streptomyces anulatus | HM038106 |
| Cluster 5 | T1PKS | 59,963–101,449 | tetronasin | 3 | Streptomyces longisporoflavus | FJ462704 |
| Cluster 6 | Lanthipepetide | 165,027–175,242 | informatipeptin | 28 | Streptomyces viridochromogenes DSM 40736 | GG657757 |
| Cluster 7 | Ectoine | 309,337–319,747 | ectoine | 100 | Streptomyces chrysomallus | AY524544 |
| Cluster 8 | Lanthipeptide | 17,762–82,339 | bleomycin | 12 | Streptomyces verticillus | AF210249 |
| Cluster 9 | T1PKS-NRPS | 245,312–295,148 | cinnabaramide A | 18 | Streptomyces cinnabarigriseus | FR687018 |
| Cluster 10 | NRPS | 64,888–126,870 | friulimicin A | 21 | Actinoplanes friuliensis | AJ488769 |
| Cluster 11 | T1PKS | 107,606–180,211 | spore pigment | 83 | Streptomyces avermitilis | AB070937 |
| Cluster 12 | Lassopeptide | 46,064–68,617 | albusnodin | 100 | Streptomyces albus | NZ_CP033071 |
| Cluster 13 | Siderophore | 219,062–230,831 | desferrioxamin E | 83 | Streptomyces coelicolor A3(2) | AL645882 |
| Cluster 14 | NRPS | 1–28,514 | A-47934 | 23 | Streptomyces toyocaensis | U82965 |
| Cluster 15 | T1PKS | 129,295–191,261 | meilingmycin | 5 | Streptomyces nanchangensis | FJ952082 |
| Cluster 16 | Terpene | 219,424–239,763 | geosmin | 100 | Streptomyces coelicolor A3(2) | AL645882 |
| Cluster 17 | T1PKS | 150–53,837 | borrelidin | 9 | Streptomyces parvulus | AJ580915 |
| Cluster 18 | NRPS | 58,670–101,440 | diisonitrile antibiotic | 66 | Streptomyces thioluteus | KY427327 |
| Cluster 19 | T1PKS-NRPS | 126,352–208,105 | meridamycin | 21 | Streptomyces sp. NRRL 30748 | DQ351275 |
| Cluster 20 | T1PKS | 43,154–123,144 | kinamycin | 20 | Streptomyces murayamaensis | AH012623 |
| Cluster 21 | Lanthipeptide | 144,862–194,829 | cinnamycin | 19 | Streptomyces cinnamoneus | AJ536588 |
| Cluster 22 | NRPS | 1–42,891 | glycinocin A | 9 | Streptomyces viridochromogenes | HM756254 |
| Cluster 23 | T3PKS | 46,294–87,358 | herboxidiene | 7 | Streptomyces chromofuscus | JN671974 |
| Cluster 24 | Terpene | 49,000–70,085 | julichrome Q3-3 | 25 | Streptomyces afghaniensis 772 | NZ_KE354310 |
| Cluster 25 | PKS | 3934–25,208 | ebelactone | 5 | Kitasatospora aburaviensis | LT608336 |
| Cluster 26 | NRPS | 71,264–112,637 | A-503083 | 7 | Streptomyces sp. SANK 62799 | AB538860 |
| Cluster 27 | T1PKS-NRPS | 84,863–111,768 | pyralomicin | 18 | Nonomuraea spiralis | JX424761 |
| Cluster 28 | T1PKS | 73,773–112,239 | pentamycin | 86 | Streptomyces sp. S816 | QQVZ01000001 |
| Cluster 29 | NRPS | 1–37,425 | showdomycin | 23 | Streptomyces showdoensis | LAQS01000018 |
| Cluster 30 | T1PKS | 1–25,087 | sceliphrolactam | 24 | Streptomyces sp. SD85 | KX230849 |
| Cluster 31 | NRPS | 1–11,557 | atratumycin | 7 | Streptomyces atratus | MK370905 |
| Cluster 32 | NRPS | 1–2297 | RP-1776 | 4 | Streptomyces sp. Acta 2897 | JF430460 |
| Cluster 33 | NRPS | 1–3067 | rhizomide A | 100 | Paraburkholderia rhizoxinica HKI 454 | NC_014718 |
| Cluster 34 | NRPS | 1–3742 | triostin A | 11 | Streptomyces triostinicus | AB366635 |
| Sl No. | Types | Nos. |
|---|---|---|
| 1 | Melanin | 1 |
| 2 | Lanthipeptide class 3 | 1 |
| 3 | NRPS | 14 |
| 4 | T1PKS | 10 |
| 5 | T1PKS-NRPS | 5 |
| 6 | Terpene | 6 |
| 7 | Lanthipeptide class 2 | 1 |
| 8 | Lanthipeptide class 1 | 1 |
| 9 | Ectoine | 1 |
| 10 | Lassopeptide | 1 |
| 11 | Siderophore | 1 |
| 12 | RiPP-like | 1 |
| 13 | T2PKS | 1 |
| 14 | T3PKS | 1 |
| 15 | RiPP-like | 2 |
| Putative Identity | Molecular Formula | Adduct Type | RT [min] | Mass Error (ppm) | Calc. MW | m/z | FISh Coverage | Matched Fragment Ions | Highest Expression in |
|---|---|---|---|---|---|---|---|---|---|
| Actinomycin D | C62 H86 N12 O16 | [M + 2H]+2 | 16.809 | 1.63 | 1254.6305 | 628.3225 | 31.78 | 86.09645, 132.10191, 282.180, 399.26102, 628.32050, 8 57.38312, 956.45129, 957.43744, 1297.09998 | SCB, GSB |
| Actinomycin Z2 | C62 H84 N12 O18 | [M + 2H]+2 | 14.62 | 0.4 | 1284.6032 | 643.3089 | 9.01 | 72.04465, 132.10191, 197.12935, 628.32050, 957.43744 | PDB |
| Cyclizidine | C17 H27 N O4 | [M + H]+1 | 12.993 | 0.27 | 309.19409 | 310.2014 | 50 | 250.15637, 310.20377 | R5M, CzD |
| Cinnabaramide A | C19 H29 N O4 | [M + H]+1 | 15.52 | −0.22 | 335.20958 | 336.2169 | Nil | CzD, R5M | |
| Cinnabaramide B | C19 H29 N O5 | [M + H]+1 | 13.702 | −0.08 | 351.20454 | 352.21182 | Nil | CzD, R5M | |
| Cyclo (phenylalanyl-prolyl) | C14 H16 N2 O2 | [M + H]+1 | 8.059 | −0.72 | 244.121 | 245.1283 | 70.21 | 120.08070, 154.07368, 245.12842 | PDB |
| Cyclo-(N-MeVal-N-MeAla) | C10 H18 N2 O2 | [M + H]+1 | 7.659 | −0.16 | 198.1368 | 199.1441 | 57.58 | 72.08077, 126.12772, 171.14919, 199.14410 | PDB, R5M |
| Pentamycin | C35 H58 O12 | [M + H]+1 | 12.335 | −0.85 | 670.39225 | 671.3996 | 46 | 79.05448, 95.04921, 121.06478, 635.37848 | EtOH, ISP2 |
| Cyclo (4 hydroxy-D-Pro-D-Trp) | C16 H17 N3 O3 | [M + H]+1 | 7.342 | −0.12 | 299.12695 | 300.1342 | 12.5 | 114.05464, 282.12396, 300.13428 | PDB |
| Deferrioxamine E | C27 H48 N6 O9 | [M + H]+1 | 20.793 | 1.75 | 600.34933 | 601.3566 | 22.22 | 601.3562 | OMB, ISP5 |
| Cyclo ((D)-Pro-(D)-Leu) | C11 H18 N2 O2 | [M + H]+1 | 7.798 | −1.77 | 210.13646 | 211.1437 | 76.19 | 86.09640, 98.06000, 211.14415, 183.14903 | PDB |
| Actinozine A | C11 H18 N2 O4 | [M + H]+1 | 6.9 | −0.74 | 242.12648 | 243.1337 | 73.58 | 100.03922, 165.10231, 201.12343, 243.13400 | PDB |
| Diterpene derivative | C24 H30 O7 | [M+NH4]+1 | 11.749 | 0.22 | 430.19925 | 448.2331 | 33.33 | 151.07553, 431.20493 | DMSO |
| Streptazone E | C12 H13 N O | [M + H]+1 | 13.092 | −0.09 | 187.0997 | 188.107 | Nil | CcSr | |
| Actinomycin D | C62 H86 N12 O16 | [M + 2H]+2 | 16.809 | 1.63 | 1254.6305 | 628.3225 | 31.78 | 86.09645, 132.10191, 282.180, 399.26102, 628.32050, 8 57.38312, 956.45129, 957.43744, 1297.09998 | SCB, GSB |
| Actinomycin Z2 | C62 H84 N12 O18 | [M + 2H]+2 | 14.62 | 0.4 | 1284.6032 | 643.3089 | 9.01 | 72.04465, 132.10191, 197.12935, 628.32050, 957.43744 | PDB |
| Cyclizidine | C17 H27 N O4 | [M + H]+1 | 12.993 | 0.27 | 309.19409 | 310.2014 | 50 | 250.15637, 310.20377 | R5M, CzD |
| Cinnabaramide A | C19 H29 N O4 | [M + H]+1 | 15.52 | −0.22 | 335.20958 | 336.2169 | Nil | CzD, R5M | |
| Cinnabaramide B | C19 H29 N O5 | [M + H]+1 | 13.702 | −0.08 | 351.20454 | 352.21182 | Nil | CzD, R5M | |
| Cyclo (phenylalanyl-prolyl) | C14 H16 N2 O2 | [M + H]+1 | 8.059 | −0.72 | 244.121 | 245.1283 | 70.21 | 120.08070, 154.07368, 245.12842 | PDB |
| Cyclo-(N-MeVal-N-MeAla) | C10 H18 N2 O2 | [M + H]+1 | 7.659 | −0.16 | 198.1368 | 199.1441 | 57.58 | 72.08077, 126.12772, 171.14919, 199.14410 | PDB, R5M |
| Pentamycin | C35 H58 O12 | [M + H]+1 | 12.335 | −0.85 | 670.39225 | 671.3996 | 46 | 79.05448, 95.04921, 121.06478, 635.37848 | EtOH, ISP2 |
| Cyclo (4 hydroxy-D-Pro-D-Trp) | C16 H17 N3 O3 | [M + H]+1 | 7.342 | −0.12 | 299.12695 | 300.1342 | 12.5 | 114.05464, 282.12396, 300.13428 | PDB |
| Deferrioxamine E | C27 H48 N6 O9 | [M + H]+1 | 20.793 | 1.75 | 600.34933 | 601.3566 | 22.22 | 601.3562 | OMB, ISP5 |
| Cyclo ((D)-Pro-(D)-Leu) | C11 H18 N2 O2 | [M + H]+1 | 7.798 | −1.77 | 210.13646 | 211.1437 | 76.19 | 86.09640, 98.06000, 211.14415, 183.14903 | PDB |
| Actinozine A | C11 H18 N2 O4 | [M + H]+1 | 6.9 | −0.74 | 242.12648 | 243.1337 | 73.58 | 100.03922, 165.10231, 201.12343, 243.13400 | PDB |
| Diterpene derivative | C24 H30 O7 | [M+NH4]+1 | 11.749 | 0.22 | 430.19925 | 448.2331 | 33.33 | 151.07553, 431.20493 | DMSO |
| Streptazone E | C12 H13 N O | [M + H]+1 | 13.092 | −0.09 | 187.0997 | 188.107 | Nil | CcSr |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Das, V.; Chatterjee, N.S.; Pushpakaran, P.U.; Lalitha, K.V.; Joseph, T.C. Exploration of Natural Product Repository by Combined Genomics and Metabolomics Profiling of Mangrove-Derived Streptomyces murinus THV12 Strain. Fermentation 2023, 9, 576. https://doi.org/10.3390/fermentation9060576
Das V, Chatterjee NS, Pushpakaran PU, Lalitha KV, Joseph TC. Exploration of Natural Product Repository by Combined Genomics and Metabolomics Profiling of Mangrove-Derived Streptomyces murinus THV12 Strain. Fermentation. 2023; 9(6):576. https://doi.org/10.3390/fermentation9060576
Chicago/Turabian StyleDas, Vineetha, Niladri Sekhar Chatterjee, Prinetha Uruparambil Pushpakaran, Kuttanappilly V. Lalitha, and Toms C. Joseph. 2023. "Exploration of Natural Product Repository by Combined Genomics and Metabolomics Profiling of Mangrove-Derived Streptomyces murinus THV12 Strain" Fermentation 9, no. 6: 576. https://doi.org/10.3390/fermentation9060576
APA StyleDas, V., Chatterjee, N. S., Pushpakaran, P. U., Lalitha, K. V., & Joseph, T. C. (2023). Exploration of Natural Product Repository by Combined Genomics and Metabolomics Profiling of Mangrove-Derived Streptomyces murinus THV12 Strain. Fermentation, 9(6), 576. https://doi.org/10.3390/fermentation9060576





