Abstract
Biomass residue and waste stream bioconversion is a key pillar for successful transition toward sustainable bioeconomy. Spent microbial biomass (SMB) is a unique type of nutrient-rich residue generated from fermentation. This study addresses the waste–SMB–substrate cycle in fermentation. Data from a range of published fermentation processes using waste and non-waste substrates are analyzed for a variety of fermentation products including alcohols and biofuels, amino acids, polymers (PHA), and organic acids. On average, fermentation of waste substrates produces similar, or up to two–three times higher, amounts of SMB compared to purified substrates. SMB production from waste substrates is further illustrated with data from PHA production. The amino acid composition of SMB from 6 industrially relevant microorganisms is compared and shows relatively low variety (2–8%). The return of SMB as a (co-)substrate in fermentation is then considered by building upon the novel concept of sustainable metabolic engineering (SME). SME incorporates economic, environmental, and social sustainability criteria in its optimization algorithm to select microbial strain designs resulting in the most sustainable products. An example of SME application for SMB amino acid re-use by engineered Escherichia coli is demonstrated and discussed. A design with dual production of succinate and ethanol was found to be the most sustainable.
1. Introduction
To address climate change the production of bioproducts (e.g., chemicals, biopolymers, etc.) from renewable biomass is becoming increasingly significant [1,2,3]. Utilization of biomass residues and waste streams (“bioresidues”) is often desirable as it can reduce the cost of bioproduction, provide higher resource efficiency in circular economy and avoid competition with food for agricultural land [4,5,6]. However, most bioresidues are complex substrates containing a variety of compounds at different concentrations. These can include both beneficial and harmful compounds for microbial growth or product formation [7].
Biotechnology also produces waste streams that are released back into the environment and made available to other industries. Among these wastes is spent microbial biomass (SMB) [8,9], which can be defined as microbial biomass left over after product removal. SMB is an inevitable, commonly protein-rich residue in any biotechnological process that uses and propagates biological organisms and cells as catalysts for biotransformation of substrate into a product. Data on the amount of global industrial production are scarce. For this reason, the only assessment of the global amount of SMB was made using data from 2012, which estimated global SMB to be above 50 million metric tons [8]. Since then, biological production and industrial fermentation has grown and is expected to continue developing rapidly. To illustrate, the global precision fermentation market is expected to grow from USD 1.6 Billion in 2022 to USD 36.3 Billion in 2030 [10]. Raising market value indicates an increase in production volumes, which, in turn, increases SMB generation.
Due to the nature of industrial production, SMB has several favorable properties for SMB re-use. Namely, it has a highly repeatable and reasonably well-defined content (i.e., the contents of a cell grown in a highly controlled environment) and predictable availability (i.e., depends on planned industrial production) [8]. For these reasons, SMB has high potential as a substrate for other bioprocesses.
Metabolic engineering can contribute to circularity through very fine and detailed re-usability strategies enabled by the genome scale metabolic modeling [11,12,13], which considers the entire network of metabolic reactions and all possible steady state metabolic fluxes. Based on the mass conservation law and steady state assumption, the constraint-based modeling approach requires only stoichiometrically balanced reaction equations to capture all possible steady-state metabolism scenarios including biomass production [14]. This enables prediction of the ability to consume different components of substrate (if they are presented in a consumable form) in order to produce a product and/or biomass. A recently proposed sustainable metabolic engineering (SME) concept [15] aims to match the available substrate with an organism for the formation of the most sustainable product. SME considers three components of sustainability: economic, environmental, and social. To improve the sustainability features the organism can be modified by gene deletions and/or insertions to give the best performance in sustainability that correlates to the circularity.
SME can be used as a matching tool between different existing bioresidues in search of re-use opportunities that would not be obvious without genome-scale metabolic modeling, optimization, and engineering. As a result, SMB can be proposed as a valuable substrate for existing productions where the exploited organism and product can benefit from a particular SMB composition.
Here, we aim to contribute to the innovation efforts in resource circulation within bioeconomy by considering a full cycle from fermentation of a waste substrate to SMB, and the return of SMB to fermentation as another waste substrate. To do this, we analyze and compare the available information on SMB formation from waste and non-waste substrates and take the first steps toward exploring sustainable re-use of SMB using the methodology of SME. In this study, we focus on the protein fraction of SMB, which on average composes 40–60% of SMB [8]. For this, the amino acid content of several industrial microorganisms is compared. In addition, we estimate the variability of amino acid composition for an organism growing in various environments using yeast (Saccharomyces cerevisiae) as a model organism, for which the biomass composition has been reported in several different academic studies [16,17]. SME is then used on a test microorganism Escherichia coli to cycle through potential enzyme coding gene deletions and identify the genetic modification designs yielding the most sustainable re-use of SMB as a substrate in subsequent fermentations. In this way, we consider a full cycle from fermentation of a waste substrate to SMB, and the return of SMB to fermentation as another waste substrate. In SME approach, the selection of desired products is a consequence of maximizing toward sustainability in contrast to the traditional approach which selects the target product and optimizes its flux or yield. Amino acid re-use design features are discussed as an illustration for SMB utilization as waste-based substrate. To our knowledge, this is the first study applying in silico modeling tools optimizing for the most sustainable SMB bioconversion strategy.
2. Methods
2.1. Estimating Amounts of SMB from Waste and Non-Waste Substrates
The methodology, general assumptions, and caveats for SMB estimation are described in a previous study [8]. Here, we further explore the coupling between substrate, product, and biomass formation. Namely, substrate is converted into both biomass and product, and with no substrate limitations an increase in biomass results in an increase of cells generating product, thus leading to faster production.
To assess how the choice of purified or waste substrate affects the formation of product and SMB, descriptions of fermentation processes from waste and non-waste substrates were collected using academic databases such as Scopus and Web of Science. Although the activity of bioproduction from waste streams is growing [2,3], it is still a relatively new field, and thus, the amount of suitable data is limited. Reliable comparison requires collection of multiple fermentation processes for the same bioproduct derived from both waste and non-waste substrates. The number of bioproducts that fill this criterion is limited. Additionally, most of the data are derived from early technological development studies (TRL 3–5), where the emphasis is on demonstrating the concept. Thus, the data on by-product formation and cell growth are frequently either left out or reported in study-specific units (for example, optical density) without providing conversion parameters to more comparable units such as dry cell weight (DCW). For these reasons, the scope of this work is framed around a few selected bioproducts representing different small-molecule metabolites.
The information gathered (Supplementary Table S1) was divided into two categories: purified (“laboratory” or non-waste) substrates and waste substrates. This was done on the basis that most traditional bioprocess development studies use well-defined media made from purified substrates, whereas waste substrates have a less well-defined, more complicated chemical makeup. Laboratory substrates mainly include purified carbon sources, such as glucose, fructose, sucrose, pure glycerol (>90%), lactose, etc. Waste substrates, in turn, include a variety of complex substrates such as hemicellulose, lignocellulose, different hydrolysates (e.g., starch, barley straw, wheat straw, etc.), various molasses, kitchen waste, and others. In some cases, data represented fermentation studies where microorganisms were tested on a medium containing the main carbon sources from a waste stream without using the actual waste biomass. For example, a purified xylose or cellulose was added to mimic the main carbon source from lignocellulosic substrates [18]. These data points form a “waste-mimicking” substrate sub-group, which is included within the larger group of waste substrates.
The SMB was defined in a prior study [8] as the amount of microbial biomass as dry cell weight (DCW) at the end of the fermentation. To limit the influence of different process conditions, substrates, and microorganisms, data were grouped according to the product, and the variability visualized using box plot statistical description. For intracellular products such as polyhydroxyalkanoates (PHA), the SMB was calculated as the residual biomass after PHA removal (i.e., “mass of DCW”—"mass of PHA”). Due to the availability of data of PHA production from various waste and purified substrates, PHA were selected for closer examination of the relationship between product and SMB formation.
2.2. Estimating Amino Acid Composition of SMB
SMB is a complex substrate consisting mainly of protein, nucleic acids, lipids, and polysaccharides. SME methodology relies on genome scale metabolic models. To introduce SMB as a substrate for SME, the SMB composition must be broken down and described in detail at the metabolite level. To aid in connecting SMB substrate description with the requirements of SME, we decided to focus this study on the protein and amino acid component of the SMB. Protein fraction generally forms 40–60% of SMB [8].
A search of the literature was performed to estimate and compare the diversity of amino acid composition within various industrially relevant microorganisms forming SMB. They are Escherichia coli, Corynebacterium glutamicum, Streptomyces coelicolor, Saccharomyces cerevisiae, Pichia pastoris, and Aspergillus niger. The estimates of the amino acid composition of an organism were derived from both analytical biomass studies [16,17] and genome-scale metabolic models [19,20,21,22] because these models are frequently created by integrating a wide variety of inputs and their predictive accuracy are evaluated experimentally. To compare data from various sources, the molar amounts of amino acids were calculated and normalized to the sum of all amino acids.
To observe how much the amino acid composition of a single organism can change depending on growth in various growth conditions, we compared data from S. cerevisiae amino acid composition reported in different publications [16,23,24,25]. Data sources where not all amino acids have been measured were excluded from this analysis.
Analysis of the data was performed using the R-software (v4.1.0; [26], the Tidyverse package (v1.3.1; [27]).
2.3. Sustainable Metabolic Engineering (SME) Task Set-Up
SME approach has been implemented according to Muiznieks et al. [28] using COBRA toolbox [29] for calculations and Paint4net [30] and IMFLer [31] for visualization. The designs are ranked according to the integrated sustainability score (ISS), which is a weighted sum of economic, environmental, and social sustainability scores. In this study, all weight coefficients are equal to 1, as all components of sustainability are expressed in monetary units. The values for economic, environmental and social sustainability calculations were consistent with those used in Muiznieks et al. [28].
To reduce the impact of the metabolic flux variability on ISS variability, designs were selected using growth coupling production approach [32,33,34]. This approach is based on the assumption that the strain with the fastest growing phenotype will dominate in the media and genetic engineering is applied to the strain in such a way that produces sufficient amount of product at its maximal growth. The wild type and engineered strain solution space is represented by growth/production envelopes with ISS value on “y” axis. ISS values at maximal growth and at no growth conditions have been used to rank different growth/production envelopes as per Equation (1):
where ISSAtZeroGR is the integrated sustainability score at minimal growth-rate, ISSAtMaxGR is the integrated sustainability score at maximal growth-rate, and nrOfKO is the number of knockouts in a design.
The design generation using the constrained model was performed with optGene [35]. optGene was modified to accept the constrained model and the sustainability indicator vector as global variables. The fitness function was used as described in Muiznieks et al. [28]. A maximum of 10 gene knockouts were allowed.
Several constraints were introduced in the SME task set-up to optimize the utilization of amino acids from SMB. E. coli was used as the only organism to model SMB uptake because of the availability of data about amino acid consumption [36] and high-quality GSMMs [37]. In the study by Zampieri et al. [36], amino acid uptake rates of E. coli were measured and modeled at different time points using a novel constraint-based metabolic modeling approach. To simplify and generalize this process, in our study we introduced additional constraints into the iML1515 GSMM of E. coli [38] based on the data of Zampieri et al. [36], as shown in Figure 1. Firstly, the maximum amino acid uptake rate was calculated for each amino acid and multiplied by 1.5. This was used as the maximum uptake rate for each amino acid, assuming that the maximum measured uptake rate could change between different experimental conditions but should be sufficiently close to the ones measured by Zampieri et al. [36]. Secondly, a constraint for the total amino acid uptake rate of 14.2 mmol/gDW/h was introduced based on the experimental data by Zampieri et al. [36]. The glucose uptake rate was set to 7.5 mmol/gDW/h in the same way. Lastly, the oxygen uptake rate was set to 0 to simulate anaerobic conditions.
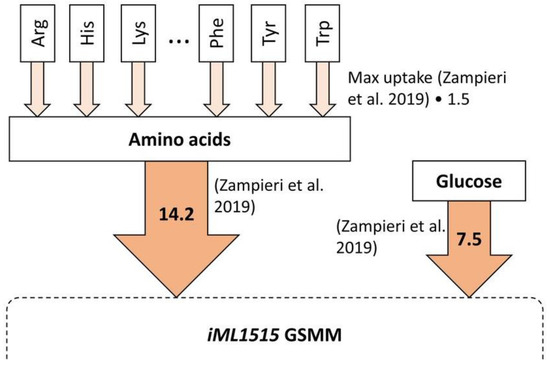
Figure 1.
The constraints added to the genome-scale metabolic model (GSMM) iML1515 of E. coli, total uptake rate of amino acids and glucose. Flux unit—mmol/gDW/h. Additional constraints based on experimental data by Zampieri et al. [36].
The sustainability indicators for various products were the same as in Muiznieks et al. [28]; they are included in the Supplementary Materials (SI2). The SME implementation code in MATLAB (R2020a) is available online at https://github.com/lv-csbg/SME_code (accessed on 30 April 2023) and in the Supplementary Materials SI3.
2.4. Environmental, Economic, and Social Impact of Biomass Extract for SME
The environmental, economic, and social impact of spent microbial biomass (SMB) had to be considered in SME because SMB is a substrate in this study. Although the composition of SMB can vary, the determination of environmental, economic, and social parameters for every variety of SMB was beyond the scope of this study. Therefore, sustainability parameters of biomass extract were simplified and assumed to be the same as for yeast extract, which is already an established substrate and has available data on its economic and environmental impact. The parameters of these impacts were calculated using the same principle as in Muiznieks et al. [28]. The economic impact of biomass extract for SME was assumed to be a cost of 4190 dollars per ton and a price of 5020 dollars per ton [39]. The environmental impact of biomass consisted of two factors: land use (ha/t) and CO2 equivalent (kg CO2 eq/kg). CO2 equivalent was 0.44 kg CO2 eq/kg [40]; however, land use was not factored in, but set to 0. This was done to avoid double counting, as the biomass originates from substrates, and the land impact of substrates is already considered. Additionally, to the best of our knowledge, no data regarding the land use for production of yeast extract are publicly available. The product land use impacts were taken from the literature [41]. Social component was calculated the same way as in Muiznieks et al. [28].
3. Results
3.1. The Comparison of SMB Formation Using Purified and Waste Substrates
Considering the growing demands for bioproduction from various bioresidues, this study examines and estimates how the choice of a waste or non-waste substrate affects SMB formation relative to the product. SMB formation varies depending on the bioproduct as well as the fermentation process (e.g., producer organism, strain, environmental parameters, etc.). Considerations regarding suitable data, bioproduct selection, and grouping of substrates into “pure” or “waste” substrates are described in Section 2.1. In short, data were divided on the basis of whether the fermentation substrate was media containing well-defined, purified carbon sources (e.g., glucose) or substrates with less well-defined, more complicated chemical makeup (e.g., straw). The results for the selected bioproducts from various product groups are summarized in Figure 2.
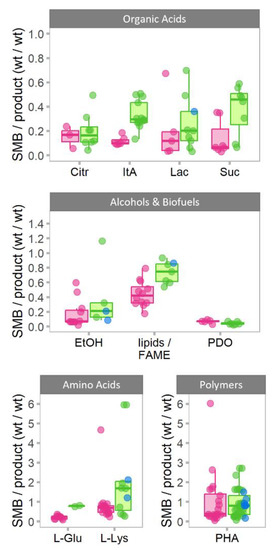
Figure 2.
SMB formation in bioproduction of various products using pure (“lab”) substrates (pink) or substrates derived from waste streams (green) or mimicking main carbon sources from a waste stream (blue). SMB formation is represented as SMB to product–weight ratio. The median value is shown as the bold line in the middle of the box. The lower and upper bounds of box show the first and third quartiles, respectively, and whiskers indicate ±1.5× the interquartile range (IQR). The scattered dots overlaid represent the corresponding data points from individual production processes. Citr—citrate, ItA—itaconic acid, Lac—lactate, Suc—succinate, EtOH—bioethanol, FAME—fatty acid methyl esters, PDO—propanediol, L-Glu—glutamate, L-Lys—lysine, PHA—polyhydroxyalkanoates. For simplified data visualization, several outlying datapoints with significantly higher SMB/product ratios have been excluded. These were data points from pure substrates (i.e., 4× citrate, 1× FAME), and wastes (i.e., 2× bioEOH and 1× PHA). The full dataset and references are shown in Supplementary Table S1.
Overall, for most products the average distribution of SMB/product ratio from waste substrates is higher or extend above that of data from purified substrates. The only exceptions—when the average SMB/product ratio was lower or identical for waste substrates—were for PDO and PHA, respectively. This indicates that biological conversion of waste streams commonly, but not always, generates more SMB for the same amount of product formation. However, most products also have bioprocesses where SMB/product ratio from waste are on par with ones from purified substrates.
Interestingly, for all metabolites, except L-Lys and ItA, the variability of SMB/product ratios (i.e., the size of the boxplot) was similar for both purified and waste streams. We attribute this variability to differences in non-optimal growth conditions and use of different microorganisms. If this hypothesis is true, then this data suggests that variability of growth conditions cause similar variability of SMB/product ratios despite substrate being grouped either as waste or purified.
The lack of variability for PDO and the lower average SMB/product ratio for waste compared to purified substrates can be explained by the fact that glycerol is the only substrate reported for PDO production within this dataset. In this case, the distinction between “waste” and “pure” glycerol lies in the level of purity for each substrate category (above 90% for “pure” grade and 70–90% for “waste” grade).
The Case of SMB Formation in PHA Production from Wastes and Purified Substrates
Many studies have explored PHA production from various waste and purified substrates, providing generous amounts of data on the biomass and PHA produced. Therefore, PHA was selected to provide a more detailed view on the relationship between product (i.e., PHA) and SMB formation. Typically, PHA is produced within the cell (i.e., as a part of biomass) [42], and in this case, SMB was calculated as the residual biomass after PHA removal (i.e., “mass of DCW”—“mass of PHA”); therefore, SMB/product can be expressed as (DCW-PHA)/PHA.
We realize that the best way to compare productivity of these very different processes would be to compare PHA yields (i.e., amount of PHA per g of substrate). However, given the diverse range of substrates, the PHA yield should be calculated relative to the amount of available carbon in each substrate. This information, unfortunately, is often unavailable. Therefore, we rely here on PHA titer as a pointer for PHA productivity, with the caveat that some datapoints might have higher or lower titer due to the higher or lower amount of substrate provided. This lack of data normalization with respect to the available substrate is somewhat offset by using the SMB/product ratio as an indicator as it indirectly shows how much of the carbon is diverted from substrate to product or SMB. To avoid substrate availability differences due to the differences in fermentation set-up, data from batch or fed-batch processes were compared separately. The data representing fermentation in fed-batch processes are shown in Figure 3 and data from batch processes are in Figure S1.
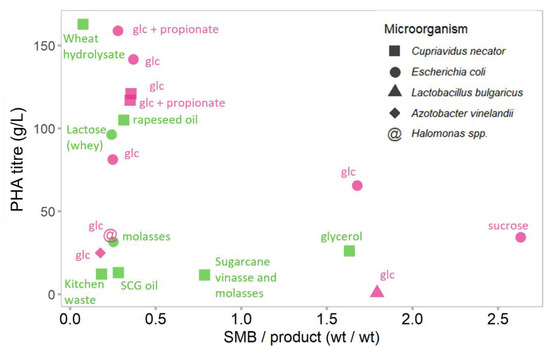
Figure 3.
SMB and PHA formation in PHA bioproduction (fed-batch set-up) using purified (“laboratory”) substrates (pink) or substrates derived from waste streams (green). The shape of the points indicates which microorganism was used for PHA production. The carbon substrate is specified for selected points. glc—glucose, SCG oil—spent coffee grounds oil. Further details and references for data sources are shown in Supplementary Table S2.
The results shown in Figure 3 and Figure S1 summarize the relationship between the reported PHA titers and the SMB/product ratios. Data from both fed-batch and batch studies show a general trend that processes with lower SMB/product ratio correlate with better PHA titer. A small SMB/product ratio shows that most of the substrate is anabolized to produce PHA instead of biomass. An SMB/product ratio lower than 1, means that more than 50% of biomass consists of PHA. Many processes utilizing waste streams and different microbial hosts fall into this bracket. The best titer represents Cupriavidus necator grown on a waste stream (wheat hydrolysate), showing that waste valorization can achieve comparable results. Fermentation using rapeseed oil and lactose (whey) as substrates also showed high product titers. Nevertheless, most processes reporting highest PHA titers used purified substrates. The waste substrates with lower SMB/product ratio and higher PHA titer tend to represent substrates with more readily available carbon and energy content (e.g., different oils and sugars). However, many agricultural residues such as straw and xylose (which mimics main sugars from hydrolyzed lignocellulosic feedstocks) show quite low titer and SMB/product ratio around 1 (Figure S1).
The highest PHA titer outcomes have been reported by C. necator or genetically modified Escherichia coli, while other organisms showed lower concentrations of PHA. Among the other organisms explored, Loktanella spp. is an interesting example, which was able to produce PHA from such lignocellulosic residues as pine and miscanthus with very low SMB/product ratio (Figure S1) [43].
3.2. Amino Acid Composition of SMB
To use SMB as a substrate for SME approach, the SMB composition must be expressed in the form of metabolites. Previous research indicates that, while slightly varying between different organisms, SMB mainly consist of proteins (40–60%), polysaccharides (3–40%), nucleic acids (4–20%), and lipids (2–15%) [8]. To develop and test the application of SME methodology for SMB valorization, we decided to focus on the protein fraction and investigate the amino acid composition of various industrial microbial strains.
Figure 4 shows the estimates of the amino acid composition in different microorganisms, as well as a comparison with casein, the main proteinaceous component of milk [44], which is used as substrate for E. coli in experimental data [36] used in this study to constrain GSMM for SME. Overall, the amino acid distributions among all microorganisms are similar, where Glx (Glu and Gln), Asx (Asp and Asn), Gly, and Ala are more abundant and Cys, Tyr, Trp, His, Phe, and Met are less abundant.
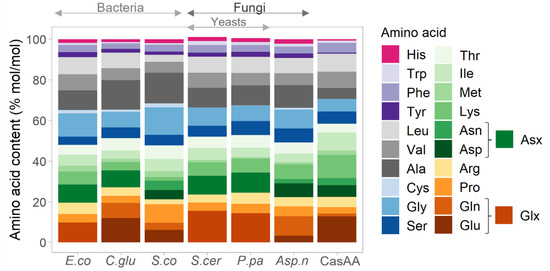
Figure 4.
Estimated amino acid composition of SMB protein fraction in selected microorganisms and a comparison to composition of casamino acids (CasAA) [36]. Data based on the published literature on E. coli (E.co) [19], Corynebacterium glutamicum (C.glu) [20], Streptomyces coelicolor (S.co) [21], Saccharomyces cerevisiae (S.cer) [16], Pichia pastoris (P.pa) [17], and Aspergillus niger (Asp.n) [22]. Due to the experimental limitations, some data sources do not distinguish between Asp and Asn (pooled fraction is named Asx). Similarly, Glx represents pool of Gln and Glu. The different shades of the same color represent amino acids from a common biosynthesis pathway.
As illustrated in Figure 4 and Figure S2, the amino acid composition varies between different organisms by up to around 5%. Most of the data for these amino acid composition estimates come from genome scale metabolic models, which in turn are often based on measurements from a single experimental setting. Depending on growth conditions, the distribution of amino acids within a single organism can vary. This is illustrated in Figure 5, comparing amino acid variability in S. cerevisiae grown in different growth conditions, which shows maximum variability of around 6–7% for such amino acids as Met and Glx.
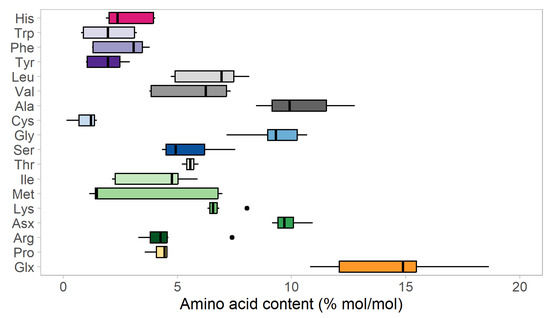
Figure 5.
Variability of amino acid composition of S. cerevisiae grown in various substrates and conditions. The median value is shown as the bold line in the middle of the box. The left and right bounds of box show the first and third quartiles, respectively, and whiskers indicate ± 1.5× the interquartile range (IQR). Outliers in the dataset are indicated as individual points. Data based on S. cerevisiae grown on glucose [16,23], the biomass compositions adjusted for carbon limitation and N-limitation as used for metabolic model iIN800 [24] and the consensus metabolic model of S. cerevisiae Yeast 8.5 [25]. Due to the experimental limitations, some data sources do not distinguish separately Asp and Asn (pooled fraction is named Asx). Similarly, Glx represents pool of Gln and Glu.
The variable composition of amino acids in SMB must be considered when setting up SME. SME does not directly incorporate the composition, nor concentrations of available amino acids, because the availability of substrate does not correlate with its uptake rate. This is due to the fact that different amino acids can have variable affinity toward their membrane transporters [45,46] and different organisms can have varying amounts of transport proteins in different environmental conditions [47]. For these reasons, SME is constrained using data from experimental measurements of amino acid uptake in a relevant setting. For the most precise SME results, one would need to perform metabolomics experiments of amino acid uptake from the specific microorganism growing on the specific amino acid substrate, which was beyond the scope of this study. Instead, we relied on previously published data of Zampieri et al. 2019 [36], where E. coli was grown on casein with glucose. Figure 4 and Figure S2 compares amino acid composition of various SMB with that of casein (CasAA) described in Zampieri et al. 2019 [36]. Although overall the amino acid distribution is similar, there are several notable differences. Cysteine (Cys) is absent (or below detection minimum) from casein, and casein contained about 1–5% less Ala, Gly, Tyr, Thr, Pro, and His and around 1–5% more Lys, Val, Leu, and Phe.
3.3. Sustainability Optimisation of SMB Re-Use as a Substrate
Designs for a constrained model that utilizes glucose and amino acids were created using the sustainable metabolic engineering (SME) approach [15]. Usually, constrained models are used to optimize maximum production of a product. In contrast, SME approach allows optimizing for sustainability taking into account all metabolite exchange fluxes between the cell and the environment. In other words, the calculated sustainability is used to choose the desired product. Consequently, the main product becomes a feature of metabolic engineering design (here—a list of gene deletions for E. coli and the selected substrate composition). The top 5000 designs together with more details can be found in Supplementary Materials (Supplementary Table S3). These designs were made by optimizing the genome-scale metabolic model iML1515 of E. coli [38], which was constrained to allow the uptake of amino acids (Figure 1). The best 5000 designs are remarkably similar to each other. This is shown by the minimum integrated sustainability scores (ISS) at maximum and no growth rate, which are almost identical between designs. The reason for the similarity in designs is the fact that deletion of the same metabolic reactions can be achieved by a large number of different combinations of gene deletions. In addition, the same effect can be achieved by deleting different reactions in a linear metabolic pathway, further increasing the number of gene combinations resulting in the same effect. The opportunity to select different gene combinations is very useful, because it gives a choice to select which genes to delete or to select from available strains with existing gene deletions. Significant variation in designs start only from design #7600 (Supplementary Table S4).
A comparison of different parameters between designs ranked number 1 and 7776 by fitness score and wild-type E. coli can be seen in Table 1. Design #1 has a higher succinate production flux at maximal growth rate and a lower maximal growth rate. The minimum sustainability at maximum and no growth is higher for design #1. Although design #7776 has a slightly higher succinate production rate at maximal growth rate, the integrated sustainability score is smaller, because design #1 produces both succinate and ethanol at high rates.

Table 1.
Comparison of designs ranked #1, #7776, and wild type (control) using the constrained iML1515 GSMM of E. coli. ISS—integrated sustainability score, SS—sustainability score, GR—growth rate.
A closer look at the exchange reactions active for design #1 at maximal growth rate shows that succinate is not the only generated product. Table 2 shows the main exchange reactions active for design #1 that have an associated sustainability coefficient. Although succinate contributes the most to the integrated sustainability score (ISS), ethanol is produced more than succinate—18.93 and 7.98 mmol/gDW/h, respectively. Therefore, this design can be thought of as a dual production design if both ethanol and succinate can be extracted during the downstream process. This dual production is not seen in wild-type at maximal growth rate (not shown). It can also be noted that biomass contributes a small amount to ISS of 1.3 × 10−4 USD/gDW, which can be used as a substrate for later fermentations.

Table 2.
Exchange reactions active for design #1 at the maximum growth rate and minimum sustainability.
A useful tool for design analysis is the so-called production envelope, where the formation of a certain product is plotted at different biomass growth rates. This allows assessing the product growth coupling of a design. A similar envelope can be plotted using the integrated sustainability score obtained by SME (see Section 2.2). The sustainability envelope of design #1 is shown in Figure 6. Design #1 consists of 5 gene deletions, namely, b3844, b0010, b4115, b3927, b1380. In Figure 6, these deletions are introduced in the constrained model that allows both glucose and amino acid uptake. This is compared to the default iML1515 settings, where only glucose is allowed as a substrate. Figure 6 shows that using both glucose and amino acids allows a higher growth rate as well as a higher maximal ISS. Although the minimal ISS at the maximal growth rate for design #1 seems close to the wild-type ISS at maximal growth rate, the difference still is notable at 35.2 and 23.27 (increase by 51%) for design #1 and wild-type, respectively. Moreover, the wild-type envelope shows that positive sustainability is obtained only near maximal growth rate, as opposed to design #1 where the sustainability increases more gradually.
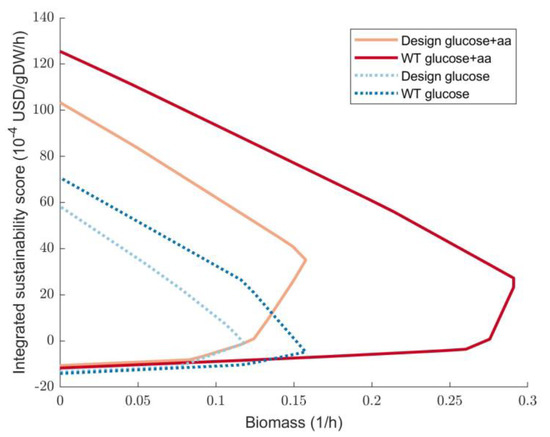
Figure 6.
Sustainability envelopes for design #1 using the iML1515 GSMM with and without amino acids as substrates. WT—wild-type, aa—amino acids.
To show the need for optimization using models that simulate growth on SMB, Figure 7 shows sustainability envelopes for a design that was created for growth on glucose. While this design works well when glucose is the only substrate, using amino acids together with glucose impairs the ISS at maximal growth rate. If only glucose is used, the design improves the minimal sustainability of the wild-type from −4.77 to 33.04 1 × 10−4 USD/gDW/h. However, when amino acids are used as a substrate, the ISS improvement is insignificant at 26.95 and 23.27 1 × 10−4 USD/gDW/h for the design and wild-type, respectively.
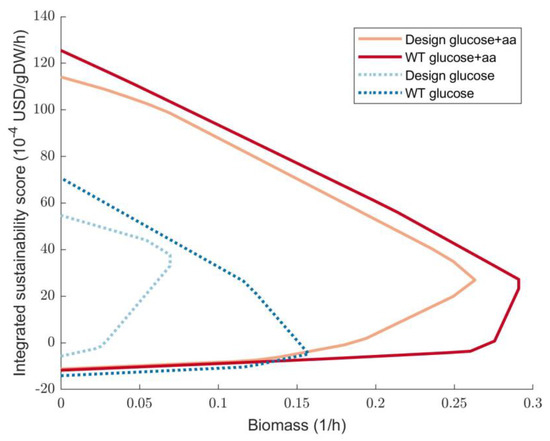
Figure 7.
Sustainability envelopes for a design with 2 gene deletions (b2415 and b3731) for the E. coli model iML1515 with and without amino acids as substrates. WT—wild-type, aa—amino acids.
4. Discussion
This study considers a full cycle from fermentation of a waste substrate to product and SMB, and then the return of SMB to fermentation as another waste substrate.
4.1. SMB Formation from Waste Substrates
Fermentation is becoming increasingly appealing for valorizing diverse organic waste streams such as whey, straw, etc. In this study, we analyze how the use of waste substrate versus purified substrate affects the development of product or SMB. Fermentation of waste substrates produces comparable or higher amount of SMB to that of purified substrates. This indicates potential space to optimize product formation from waste substrates.
The main difference between purified and waste substrates is the complexity of substrate composition (i.e., mostly one carbon source, or a range of related different molecules). This complexity can cause several additional hurdles limiting scope for bioproduction optimization from waste streams. These can broadly be summarized in two groups: (1) variability of chemical composition due to seasonal changes and weather (e.g., for agricultural wastes) [48], and (2) limited nutrient bioavailability to the microbe (e.g., cellulose in lignocellulosic waste substrates [49]). In the latter case, a pre-treatment is required (e.g., thermal, pressure, or enzymatic hydrolysis), which also influences the final composition of the waste substrate provided to the microbe [5]. The variability of the composition of the waste substrate influences the ability to optimize any single microbial strain for efficient bioconversion of bioresidues into product (and SMB).
In addition, the complex content of the waste stream can include both beneficial and harmful compounds for microbial growth or product formation. For example, waste stream might contain beneficial amino acids and vitamins (e.g., brewery spent yeast) [50]. In other cases, waste streams can contain microbe-inhibiting substrates such as furfural and vanillin, which are commonly created during hydrolysis of lignocellulosic substrates [43]. Growth on such substrates might influence the balance of substrate conversion into product and biomass due to the environmental stress applied to the cells.
Given the projections of the increasing availability of SMB, its valorization options should be considered. As discussed in our previous work, several options are available and commonly include use as agricultural fertilizer, biogas production, food, and feed applications [8]. In this study we explore the amino acid content of SMB as a potential substrate for SMB valorization into a fermentation (co-)substrate. The comparison of amino acid content from 5 different industrially relevant organisms and S. cerevisiae growing in various environmental conditions indicate a relatively low variation of the relative abundance of individual amino acids (i.e., 1–8%). This suggests that various SMB can form relatively comparable substrates, and a valorization strategy working on one type of SMB might be applicable to others.
4.2. Optimizing Metabolic Designs for Sustainable SMB Use as a Substrate in Fermentation
In this study, a conceptual example is presented demonstrating the use of SME to identify strain designs for sustainable re-use of SMB focusing on the amino acid fraction of SMB. Furthermore, amino acids were presented as co-substrate together with glucose. The use of amino acids as a sole C-source was not considered due to the thermodynamic limitations associated with amino acid deamination [51], which may limit amino acid consumption, and thus, also limit cell growth and production of any products. In addition, the SME was limited to one organism (E. coli) and did not consider any insertions of reactions. Still, the proposed designs show some interesting features of optimization for sustainability in metabolic engineering.
SME application generated many identical designs in terms of the estimated ISS value. This is due to the combinatorial explosion of different gene set deletions that result in the same metabolic alteration. For every enzyme composed of several proteins, deleting one of the protein coding genes is sufficient to render the enzyme inactive. It becomes possible to choose from several alternative gene deletion sets to implement the same targeted metabolism. This allows for the selection of the most appropriate set of deletions (e.g., the simplest and/or safest).
To set up SME, it is advantageous to have a detailed composition of the substrate, e.g., amino acids from SMB. For many complex bioresidues, detailed composition might not be available and most likely will require either (1) experimental analysis or (2) an approximate estimate using available data. An advantage of the SMB is that if it comes from a tightly controlled industrial process, and the content of SMB might be relatively stable, despite the use of different waste streams, as these will be already partially converted by the microbe. This is supported by analysis of amino acid content variation among different organisms and for the same organism growing on various substrates/conditions is relatively low at 1–8%.
Another hurdle identified for SME adaptation to substrates like SMB, is the requirement to constrain metabolic fluxes for realistic growth simulations on the complex substrate. In this study, the genome-scale model was constrained using mass conservation and substrate uptake constraints. These constraints do not consider physiological limitations, such as gene expression and enzyme kinetics, which therefore lead to high metabolic flux variability. This means that certain production envelopes can include points that are unlikely to happen in vivo. Different metabolic modeling approaches—for example, enzyme kinetic constraints based on omics experimental data such as those from the GECKO toolbox [52,53]—could be introduced to further constrain the model. This could reduce flux variability when amino acids are used as a substrate. This would allow for more precise metabolic modeling and potentially result in higher diversity of strain designs. However, implementing such complex constraints is difficult, because the assumptions made from wild-type organisms might not hold true when certain metabolic engineering interventions are performed.
To further develop the SME use for designing a strain growing on a particular waste substrate, SME application should be expanded to allow consideration of more than one candidate organism. In addition, insertions of reactions within the metabolic network should be enabled to better use the potential of SME guided strain design. Looking even further in future, SME could be used to improve sustainability of a cluster of biotechnological productions suggesting selection of a strain that can be sustainably re-used in another factory. This way one can utilize the fact that strains with equally good productivity features can be different in the re-use features of the SMB. As a result, a cluster of biotechnological companies could improve sustainability by forming synergy without reduction in productivity.
5. Conclusions
This study addresses the waste–SMB–substrate cycle in fermentation. We draw from published data and in silico modeling to explore (1) formation of waste bioresidues (i.e., Spent Microbial Biomass, SMB) from waste substrates and (2) SMB re-use as a fermentation co-substrate by identifying genetic designs leading to sustainable products.
The key findings are: (1) on average, waste fermentation produces more or similar amounts of SMB per product (at times reaching 2–3× more SMB), contributing to the increasing availability of SMB as waste bioresidues; (2) the amino acid composition of various SMB has relatively low variety (2–8%), illustrating the foreseeable nature of SMB as a fermentation (co-)substrate; (3) SME indicates that sustainable co-fermentation of glucose and amino acids using E. coli produces succinate together with ethanol; and (4) use of different substrates (e.g., glucose or additional amino acids) changes the size and shape of the sustainability envelope, reflecting the variety of possible outcomes and the accompanying technological risk for not attaining the desired results.
To conclude, this work demonstrates that with the expected increase of industrial bioproduction, it is worth considering and directing innovation efforts toward valorization of the increasing amounts of SMB within bioeconomy. SMB features such as nutrient availability, low variability, and predictable supply make SMB suitable (co-)substrate for fermentation. Furthermore, such in silico tools as SME can help to design and increase sustainable paths for SMB re-circulation within bioeconomy.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fermentation9060531/s1, Figure S1: SMB formation in PHA bioproduction (batch fermentation); Figure S2: Variability of amino acid composition selected microorganisms and compared to composition of casamino acids (CasAA); Table S1: SMB data from purified and waste substrates; Table S2: SMB data from PHA bioproduction from purified and waste substrates; Table S3: First 5000 SME generated strain designs; Table S4: SME generated strain designs ranked from 7600 to 8000. Tables in SI2: Indicator values used to calculate economic, environmental, and social sustainability in SME. Files in SI3: MATLAB code for SME implementation.
Author Contributions
Conceptualization, A.S. and E.S.; methodology, A.S. and R.M.; software, R.M.; investigation, A.S., M.R.B. and R.M; resources, E.S.; data curation, A.S., M.R.B. and R.M.; writing—original draft preparation, A.S., M.R.B. and R.M.; writing—review and editing, A.S., M.R.B., R.M. and E.S.; visualization, A.S. and R.M.; supervision, A.S. and E.S.; project administration, A.S. and E.S.; funding acquisition, A.S. and E.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by European Regional Development Fund grant number 1.1.1.2/VIAA/4/20/610 and University of Latvia project “Optimization of biotechnological processes for effective utilization of renewable resources” (Nr. Y5-AZ20-ZF-N−270).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data is contained within the article or Supplementary Material.
Acknowledgments
The authors thank Elina Dace for sharing helpful suggestions.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Lee, S.Y.; Kim, H.U.; Chae, T.U.; Cho, J.S.; Kim, J.W.; Shin, J.H.; Kim, D.I.; Ko, Y.-S.; Jang, W.D.; Jang, Y.-S. A Comprehensive Metabolic Map for Production of Bio-Based Chemicals. Nat. Catal. 2019, 2, 18–33. [Google Scholar] [CrossRef]
- Alonso, S.; Rendueles, M.; Díaz, M. Microbial Production of Specialty Organic Acids from Renewable and Waste Materials. Crit. Rev. Biotechnol. 2015, 35, 497–513. [Google Scholar] [CrossRef] [PubMed]
- Tomás-Pejó, E.; Morales-Palomo, S.; González-Fernández, C. Microbial Lipids from Organic Wastes: Outlook and Challenges. Bioresour. Technol. 2021, 323, 124612. [Google Scholar] [CrossRef] [PubMed]
- Karthick, C.; Nanthagopal, K. A Comprehensive Review on Ecological Approaches of Waste to Wealth Strategies for Production of Sustainable Biobutanol and Its Suitability in Automotive Applications. Energy Convers. Manag. 2021, 239, 114219. [Google Scholar] [CrossRef]
- Sirohi, R.; Prakash Pandey, J.; Kumar Gaur, V.; Gnansounou, E.; Sindhu, R. Critical Overview of Biomass Feedstocks as Sustainable Substrates for the Production of Polyhydroxybutyrate (PHB). Bioresour. Technol. 2020, 311, 123536. [Google Scholar] [CrossRef]
- Kircher, M. Bioeconomy—Present Status and Future Needs of Industrial Value Chains. New Biotechnol. 2021, 60, 96–104. [Google Scholar] [CrossRef]
- Alibardi, L.; Cossu, R. Effects of Carbohydrate, Protein and Lipid Content of Organic Waste on Hydrogen Production and Fermentation Products. Waste Manag. 2016, 47, 69–77. [Google Scholar] [CrossRef]
- Stikane, A.; Dace, E.; Stalidzans, E. Closing the Loop in Bioproduction: Spent Microbial Biomass as a Resource within Circular Bioeconomy. New Biotechnol. 2022, 70, 109–115. [Google Scholar] [CrossRef]
- Paramasivam, P.; Kanagesan, K.; Bhuyar, P.; Govindan, N.; Ab. Rahim, M.H.; Maniam, G.P. Biomass and Lipid Production from Indigenous Nannochloropsis sp. by Employing Stress Factors for Improved Biodiesel Production. Environ. Dev. Sustain. 2021. [Google Scholar] [CrossRef]
- Research and Markets Ltd. Global Precision Fermentation Market by Ingredient (Whey & Casein Protein, Egg White, Collagen Protein, Heme Protein), Microbe (Yeast, Algae, Fungi, Bacteria), Application (Meat & Seafood, Dairy Alternatives, Egg Alternatives), and Region—Forecast to 2030; Research and Markets: Dublin, Ireland, 2022. [Google Scholar]
- Thiele, I.; Palsson, B.Ø. A Protocol for Generating a High-Quality Genome-Scale Metabolic Reconstruction. Nat. Protoc. 2010, 5, 93–121. [Google Scholar] [CrossRef]
- Stephanopoulos, G. Metabolic Engineering. Curr. Opin. Biotechnol. 1994, 5, 196–200. [Google Scholar] [CrossRef]
- Otero, J.M.; Nielsen, J. Industrial Systems Biology. Biotechnol. Bioeng. 2010, 105, 439–460. [Google Scholar] [CrossRef]
- Stalidzans, E.; Seiman, A.; Peebo, K.; Komasilovs, V.; Pentjuss, A. Model-Based Metabolism Design: Constraints for Kinetic and Stoichiometric Models. Biochem. Soc. Trans. 2018, 46, 261–267. [Google Scholar] [CrossRef]
- Stalidzans, E.; Dace, E. Sustainable Metabolic Engineering for Sustainability Optimisation of Industrial Biotechnology. Comput. Struct. Biotechnol. J. 2021, 19, 4770–4776. [Google Scholar] [CrossRef]
- Lange, H.C.; Heijnen, J.J. Statistical Reconciliation of the Elemental and Molecular Biomass Composition of Saccharomyces Cerevisiae. Biotechnol. Bioeng. 2001, 75, 334–344. [Google Scholar] [CrossRef]
- Carnicer, M.; Baumann, K.; Töplitz, I.; Sánchez-Ferrando, F.; Mattanovich, D.; Ferrer, P.; Albiol, J. Macromolecular and Elemental Composition Analysis and Extracellular Metabolite Balances of Pichia Pastoris Growing at Different Oxygen Levels. Microb. Cell Factories 2009, 8, 65. [Google Scholar] [CrossRef]
- Ilmén, M.; Koivuranta, K.; Ruohonen, L.; Suominen, P.; Penttilä, M. Efficient Production of L-Lactic Acid from Xylose by Pichia Stipitis. Appl. Environ. Microbiol. 2007, 73, 117–123. [Google Scholar] [CrossRef]
- Orth, J.D.; Conrad, T.M.; Na, J.; Lerman, J.A.; Nam, H.; Feist, A.M.; Palsson, B.Ø. A Comprehensive Genome-Scale Reconstruction of Escherichia Coli Metabolism—2011. Mol. Syst. Biol. 2011, 7, 535. [Google Scholar] [CrossRef]
- Kjeldsen, K.R.; Nielsen, J. In Silico Genome-Scale Reconstruction and Validation of the Corynebacterium Glutamicum Metabolic Network. Biotechnol. Bioeng. 2009, 102, 583–597. [Google Scholar] [CrossRef]
- Amara, A.; Takano, E.; Breitling, R. Development and Validation of an Updated Computational Model of Streptomyces Coelicolor Primary and Secondary Metabolism. BMC Genom. 2018, 19, 519. [Google Scholar] [CrossRef]
- Upton, D.J.; McQueen-Mason, S.J.; Wood, A.J. In Silico Evolution of Aspergillus Niger Organic Acid Production Suggests Strategies for Switching Acid Output. Biotechnol. Biofuels 2020, 13, 27. [Google Scholar] [CrossRef] [PubMed]
- Albers, E.; Larsson, C.; Lidé, N.G.; Niklasson, C.; Gustafsson, L. Influence of the Nitrogen Source on Saccharomyces Cerevisiae Anaerobic Growth and Product Formation. Appl. Environ. Microbiol. 1996, 62, 3187–3195. [Google Scholar] [CrossRef] [PubMed]
- Nookaew, I.; Jewett, M.C.; Meechai, A.; Thammarongtham, C.; Laoteng, K.; Cheevadhanarak, S.; Nielsen, J.; Bhumiratana, S. The Genome-Scale Metabolic Model iIN800 of Saccharomyces Cerevisiae and Its Validation: A Scaffold to Query Lipid Metabolism. BMC Syst. Biol. 2008, 2, 71. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Li, F.; Sánchez, B.J.; Zhu, Z.; Li, G.; Domenzain, I.; Marcišauskas, S.; Anton, P.M.; Lappa, D.; Lieven, C.; et al. Cerevisiae Metabolic Model Yeast8 and Its Ecosystem for Comprehensively Probing Cellular Metabolism. Nat. Commun. 2019, 10, 3586. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 30 April 2023).
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Muiznieks, R.; Dace, E.; Stalidzans, E. Sustainable Metabolic Engineering Design Development Employing Complex Objective Function. Preprints.org 2023, 2023050094. [Google Scholar] [CrossRef]
- Heirendt, L.; Arreckx, S.; Pfau, T.; Mendoza, S.N.; Richelle, A.; Heinken, A.; Haraldsdóttir, H.S.; Wachowiak, J.; Keating, S.M.; Vlasov, V.; et al. Creation and Analysis of Biochemical Constraint-Based Models Using the COBRA Toolbox v.3.0. Nat. Protoc. 2019, 14, 639–702. [Google Scholar] [CrossRef]
- Kostromins, A.; Stalidzans, E. Paint4Net: COBRA Toolbox Extension for Visualization of Stoichiometric Models of Metabolism. Biosystems 2012, 109, 233–239. [Google Scholar] [CrossRef]
- Petrovs, R.; Stalidzans, E.; Pentjuss, A. IMFLer: A Web Application for Interactive Metabolic Flux Analysis and Visualization. J. Comput. Biol. 2021, 28, 1021–1032. [Google Scholar] [CrossRef]
- Klamt, S.; Mahadevan, R. On the Feasibility of Growth-Coupled Product Synthesis in Microbial Strains. Metab. Eng. 2015, 30, 166–178. [Google Scholar] [CrossRef]
- von Kamp, A.; Klamt, S. Growth-Coupled Overproduction Is Feasible for Almost All Metabolites in Five Major Production Organisms. Nat. Commun. 2017, 8, 15956. [Google Scholar] [CrossRef]
- Motamedian, E.; Berzins, K.; Muiznieks, R.; Stalidzans, E. OptEnvelope: A Target Point Guided Method for Growth-Coupled Production Using Knockouts. 2023.03.10.532079. Available online: https://www.biorxiv.org/content/10.1101/2023.03.10.532079v1 (accessed on 29 April 2023).
- Patil, K.R.; Rocha, I.; Förster, J.; Nielsen, J. Evolutionary Programming as a Platform for in Silico Metabolic Engineering. BMC Bioinform. 2005, 6, 308. [Google Scholar] [CrossRef]
- Zampieri, M.; Hörl, M.; Hotz, F.; Müller, N.F.; Sauer, U. Regulatory Mechanisms Underlying Coordination of Amino Acid and Glucose Catabolism in Escherichia coli. Nat. Commun. 2019, 10, 3354. [Google Scholar] [CrossRef]
- King, Z.A.; Lu, J.; Dräger, A.; Miller, P.; Federowicz, S.; Lerman, J.A.; Ebrahim, A.; Palsson, B.O.; Lewis, N.E. BiGG Models: A Platform for Integrating, Standardizing and Sharing Genome-Scale Models. Nucleic Acids Res. 2016, 44, D515–D522. [Google Scholar] [CrossRef]
- Monk, J.M.; Lloyd, C.J.; Brunk, E.; Mih, N.; Sastry, A.; King, Z.; Takeuchi, R.; Nomura, W.; Zhang, Z.; Mori, H.; et al. iML1515, a Knowledgebase That Computes Escherichia Coli Traits. Nat. Biotechnol. 2017, 35, 904–908. [Google Scholar] [CrossRef]
- Misailidis, N.; Petrides, D. Yeast Extract Production—Process Modeling and Techno-Economic Assessment (TEA) Using SuperPro Designer. August 2020. preprint. Available online: https://www.researchgate.net/publication/343922223_Yeast_Extract_Production_-_Process_Modeling_and_Techno-Economic_Assessment_TEA_using_SuperPro_Designer (accessed on 30 April 2023).
- Adom, F.; Dunn, J.B. (Eds.) Material and Energy Flows in the Production of Macro and Micronutrients, Buffers, and Chemicals Used in Biochemical Processes for the Production of Fuels and Chemicals from Biomass; Energy Systems Division, Argonne National Laboratory: Argonne, IL, USA, 2015; p. 16. [Google Scholar]
- Patel, M.; Crank, M.; Dornburg, V.; Hermann, B.; Roes, A.L.; Hüsing, B.; Overbeek, L.; Terragni, F.; Recchia, E. Medium and Long-Term Opportunities and Risks of the Biotechnological Production of Bulk Chemicals from Renewable Resources—The BREW Project. Utrecht University, Utrecht, The Netherlands. 2006. Project Report. Available online: https://dspace.library.uu.nl/handle/1874/21824 (accessed on 30 April 2023).
- Philip, S.; Keshavarz, T.; Roy, I. Polyhydroxyalkanoates: Biodegradable Polymers with a Range of Applications. J. Chem. Technol. Biotechnol. 2007, 82, 233–247. [Google Scholar] [CrossRef]
- Lee, S.M.; Cho, D.-H.; Jung, H.J.; Kim, B.; Kim, S.H.; Bhatia, S.K.; Gurav, R.; Jeon, J.-M.; Yoon, J.-J.; Kim, W.; et al. Finding of Novel Polyhydroxybutyrate Producer Loktanella Sp. SM43 Capable of Balanced Utilization of Glucose and Xylose from Lignocellulosic Biomass. Int. J. Biol. Macromol. 2022, 208, 809–818. [Google Scholar] [CrossRef]
- Wang, J.; Su, Y.; Jia, F.; Jin, H. Characterization of Casein Hydrolysates Derived from Enzymatic Hydrolysis. Chem. Cent. J. 2013, 7, 62. [Google Scholar] [CrossRef]
- Burkovski, A.; Krämer, R. Bacterial Amino Acid Transport Proteins: Occurrence, Functions, and Significance for Biotechnological Applications. Appl. Microbiol. Biotechnol. 2002, 58, 265–274. [Google Scholar] [CrossRef]
- Bianchi, F.; van’t Klooster, J.S.; Ruiz, S.J.; Poolman, B. Regulation of Amino Acid Transport in Saccharomyces Cerevisiae. Microbiol. Mol. Biol. Rev. 2019, 83, e00024-19. [Google Scholar] [CrossRef]
- Brown, C.J.; Todd, K.M.; Rosenzweig, R.F. Multiple Duplications of Yeast Hexose Transport Genes in Response to Selection in a Glucose-Limited Environment. Mol. Biol. Evol. 1998, 15, 931–942. [Google Scholar] [CrossRef] [PubMed]
- Gontard, N.; Sonesson, U.; Birkved, M.; Majone, M.; Bolzonella, D.; Celli, A.; Angellier-Coussy, H.; Jang, G.W.; Verniquet, A.; Broeze, J.; et al. A Research Challenge Vision Regarding Management of Agricultural Waste in a Circular Bio-Based Economy. Crit. Rev. Environ. Sci. Technol. 2018, 48, 614–654. [Google Scholar] [CrossRef]
- Comelli, R.N.; Seluy, L.G.; Benzzo, M.T.; Isla, M.A. Combined Utilization of Agro-Industrial Wastewaters for Non-Lignocellulosic Second-Generation Bioethanol Production. Waste Biomass Valorization 2020, 11, 265–275. [Google Scholar] [CrossRef]
- Jaeger, A.; Arendt, E.K.; Zannini, E.; Sahin, A.W. Brewer’s Spent Yeast (BSY), an Underutilized Brewing By-Product. Fermentation 2020, 6, 123. [Google Scholar] [CrossRef]
- Wernick, D.G.; Liao, J.C. Protein-Based Biorefining: Metabolic Engineering for Production of Chemicals and Fuel with Regeneration of Nitrogen Fertilizers. Appl. Microbiol. Biotechnol. 2013, 97, 1397–1406. [Google Scholar] [CrossRef]
- Sánchez, B.J.; Zhang, C.; Nilsson, A.; Lahtvee, P.J.; Kerkhoven, E.J.; Nielsen, J. Improving the Phenotype Predictions of a Yeast Genome-Scale Metabolic Model by Incorporating Enzymatic Constraints. Mol. Syst. Biol. 2017, 13, 935. [Google Scholar] [CrossRef]
- Domenzain, I.; Sánchez, B.; Anton, M.; Kerkhoven, E.J.; Millán-Oropeza, A.; Henry, C.; Siewers, V.; Morrissey, J.P.; Sonnenschein, N.; Nielsen, J. Reconstruction of a Catalogue of Genome-Scale Metabolic Models with Enzymatic Constraints Using GECKO 2.0. Nat. Commun. 2022, 13, 3766. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).