The Effect of Lactobacillus planturum YQM48 Inoculation on the Quality and Microbial Community Structure of Alfalfa Silage Cultured in Saline-Alkali Soil
Abstract
1. Introduction
2. Materials and Methods
2.1. Site Description and Alfalfa Production
2.2. LAB Strains
2.3. Silage Preparation
2.4. Characterization of the Alfalfa and the Silage
2.5. Culture-Dependent Bacterial Enumeration
2.6. High-Throughput Sequencing of the Bacterial Populations
2.7. Statistical Analysis
3. Results
3.1. Characteristics of Fresh Alfalfa Grown on Saline–Alkaline Soil
3.2. Effects of L. Plantarum YQM48 on Chemical Composition of Alfalfa Silage
3.3. Effects of L. plantarum YQM48 on Fermentation Characteristics of Alfalfa Silage
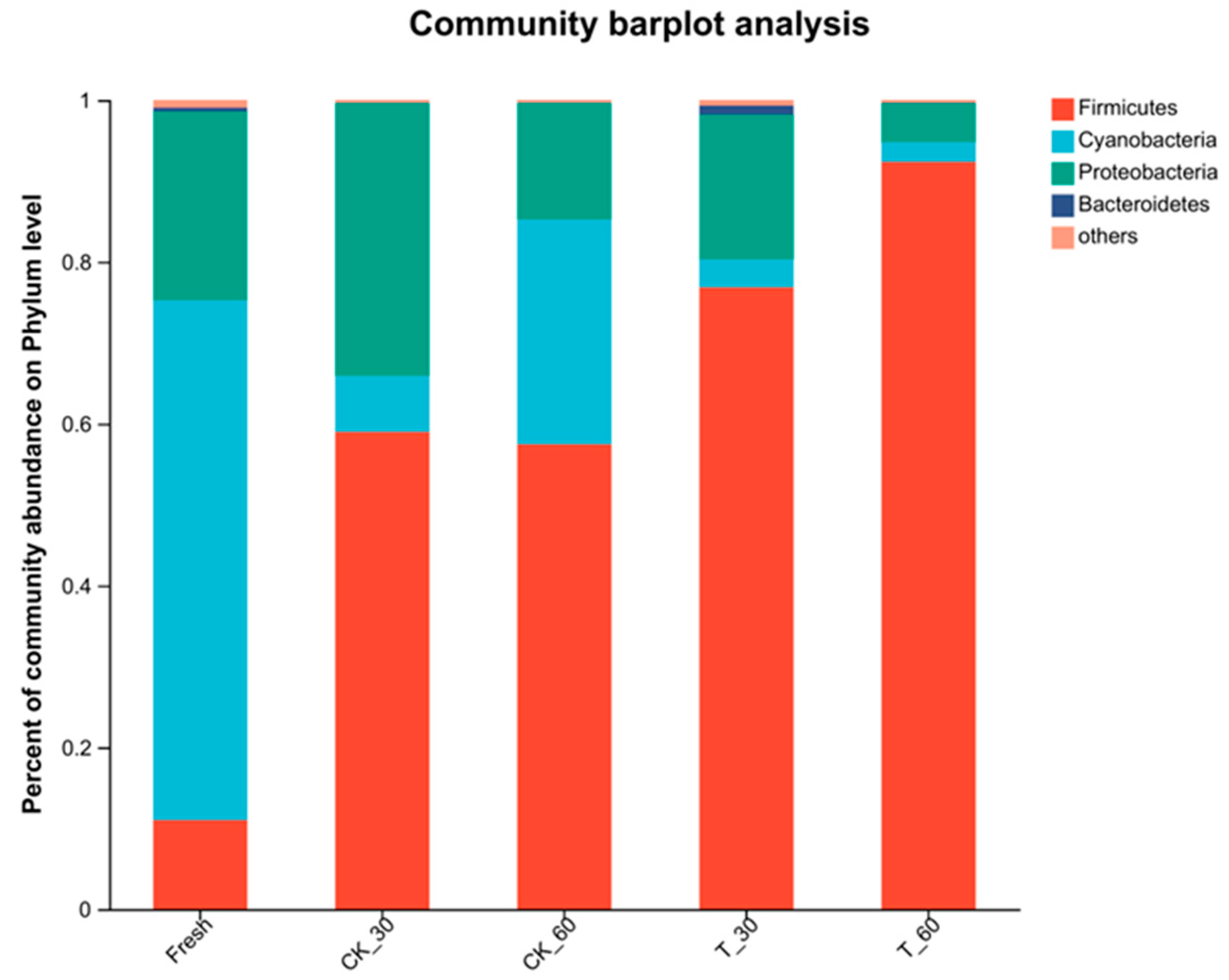
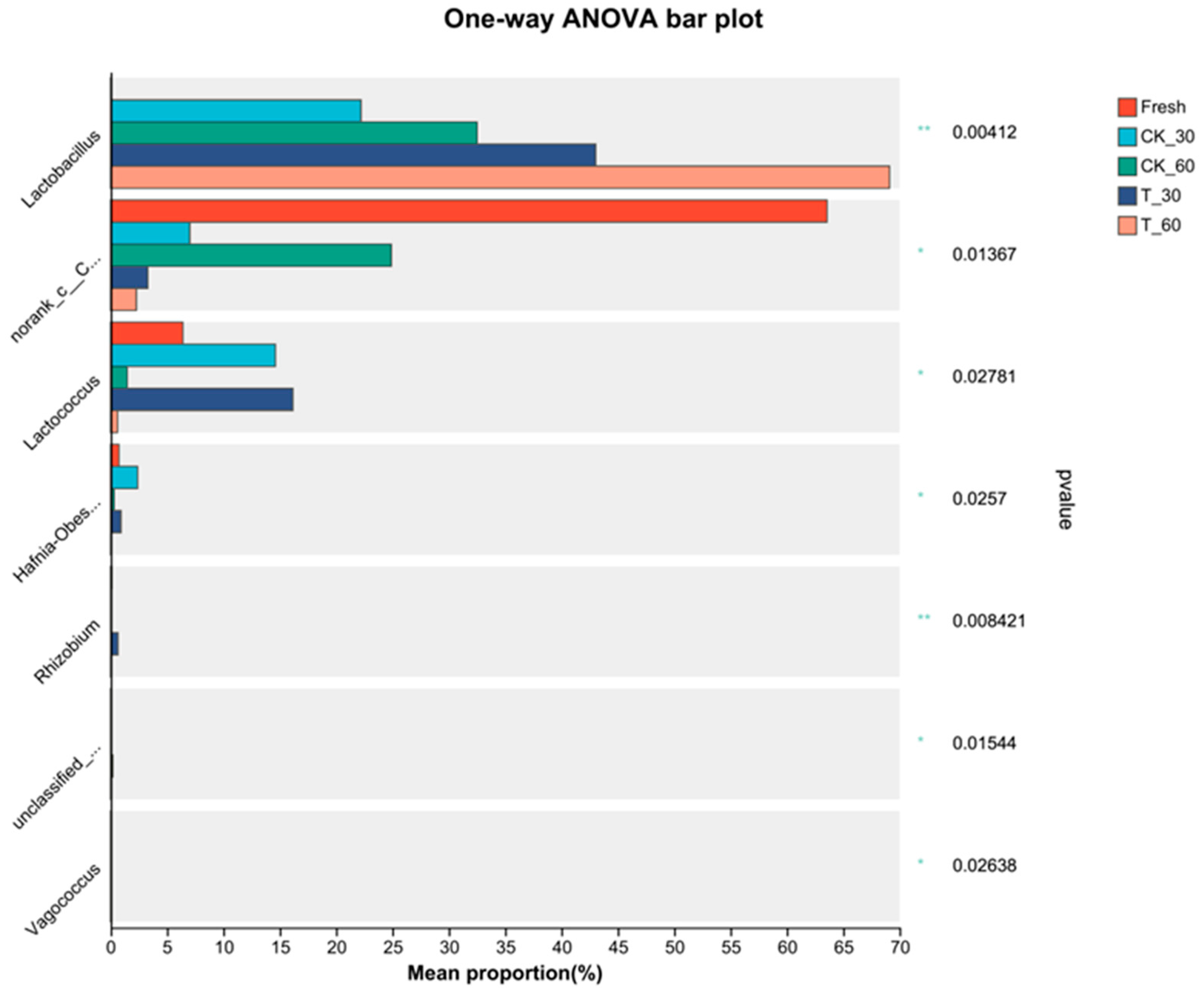
3.4. Characterization of the Bacterial Communities in Fresh Alfalfa and in the Silage
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shao, H.; Chu, L.; Lu, H.; Qi, W.; Chen, X.; Liu, J.; Kuang, S.; Tang, B.; Wong, V. Towards sustainable agriculture for the salt-affected soil. Land Degrad. Dev. 2018, 30, 574–579. [Google Scholar] [CrossRef]
- Li, C.; Wang, Z.; Xu, Y.; Sun, J.; Ruan, X.; Mao, X.; Hu, X.; Liu, P. Analysis of the effect of Modified biochar on saline-alkali soil remediation and crop growth. Sustainability 2023, 15, 5593. [Google Scholar] [CrossRef]
- Zhang, L.; Ge, A.; An, F.; Guo, L.; Nie, Z.; Liu, J.; Yang, F.; Wang, Z. Soil bacterial microbiota predetermines rice yield in reclaiming saline-sodic soils leached with brackish ice. J. Sci. Food Agric. 2021, 101, 6472–6483. [Google Scholar] [CrossRef]
- He, F.; Wang, G.; Wang, L.; Li, Z.; Tong, Z.; Wang, Y.; Li, X. Effects of organic base fertilizer and inorganic topdressing on alfalfa productivity and the soil bacterial community in saline soil of the huanghe river delta in China. Agronomy 2022, 12, 2811. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, Z.; Li, Z.; Yang, B.; Li, B.; Tang, X.; Lai, Y. Comprehensive study on saline-alkali soil amelioration with sediment of irrigation area in northeast China. Arab. J. Chem. 2023, 16, 104608. [Google Scholar] [CrossRef]
- Lu, Q.; Zhen, W.; Duo, W.S. The Potential effects on microbiota and silage fermentation of alfalfa under salt stress. Front. Microbiol. 2021, 12, 688–695. [Google Scholar] [CrossRef]
- Liu, H.; Lu, X.; Li, Z.; Tian, C.; Song, J. The role of root-associated microbes in growth stimulation of plants under saline conditions. Land Degrad. Dev. 2021, 32, 3471–3486. [Google Scholar] [CrossRef]
- Elgharably, A.; Benes, S. Alfalfa biomass yield and nitrogen fixation in response to applied mineral nitrogen under saline soil conditions. J. Soil Sci. Plant Nutr. 2021, 21, 744–755. [Google Scholar] [CrossRef]
- Yang, C.; Wang, X.; Miao, F.; Li, Z.; Tang, W.; Sun, J. Assessing the effect of soil salinization on soil microbial respiration and diversities under incubation conditions. Appl. Soil Ecol. 2020, 155, 103671. [Google Scholar] [CrossRef]
- Zhang, H.; Gao, S.; Zhang, X.; Meng, N.; Chai, X.; Wang, Y. Fermentation characteristics and the dynamic trend of chemical components during fermentation of massa medicata dermentata. Arab. J. Chem. 2021, 15, 103472. [Google Scholar] [CrossRef]
- Dong, Z.; Yuan, X.; Wen, A.; Desta, S.T.; Shao, T. Effects of calcium propionate on the fermentation quality and aerobic stability of alfalfa silage. Asian-Australas. J. Anim. Sci. 2017, 30, 1278–1284. [Google Scholar] [CrossRef] [PubMed]
- Van, S.; Robertson, J.; Lewis, B. Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J. Dairy Sci. 1991, 74, 83–97. [Google Scholar]
- Lei, C.; Gang, G.; Cheng, Q.; Jie, Z.; Masataka, S.; Tao, S. The effects of replacement of whole-plant corn with oat and common vetch on the fermentation quality, chemical composition and aerobic stability of total mixed ration silage in Tibet. Anim. Sci. 2015, 86, 69–76. [Google Scholar]
- Gao, W.; Ynag, G.; Gao, X.; Yu, Z.; Xu, Q.; Yuan, X. Effects of nitrogen, phosphorus and potassium fertilizer on yield and quality of silage maize. Crop J. 2018, 21, 144–149. [Google Scholar]
- Hao, J.; Yu, H.; Jia, Y.; Wang, Z.; Ge, G.; Sun, L. Effects of silage density and silage time on fermentation quality and nutrient composition of alfalfa. J. Grassl. Sci. 2022, 30, 2492–2496. [Google Scholar]
- Si, Q.; Wang, Z.; Liu, W.; Liu, M.; Ge, G.; Jia, Y.; Du, S. Influence of cellulase or Lactiplantibacillus plantarum on the ensiling performance and bacterial community in mixed silage of alfalfa and leymus chinensis. Microorganisms 2023, 11, 426. [Google Scholar] [CrossRef]
- Hao, W.; Tian, P.; Zheng, M.; Wang, H.; Xu, C. Characteristics of proteolytic microorganisms and their effects on proteolysis in total mixed ration silages of soybean curd residue. J. Anim. Sci. 2020, 33, 100–110. [Google Scholar] [CrossRef] [PubMed]
- Silva, V.P.; Pereira, O.G.; Leandro, E.S.; Da Silva, T.C.; Ribeiro, K.G.; Mantovani, H.C.; Santos, S.A. Effects of lactic acid bacteria with bacteriocinogenic potential on the fermentation profile and chemical composition of alfalfa silage in tropical conditions. J. Dairy Sci. 2016, 99, 1895–1902. [Google Scholar] [CrossRef]
- Hisham, M.B.; Hashim, A.M.; Hanafi, N.M.; Rahman, N.A.; Tan, C.K.; Nazli, M.H. Bacterial communities associated with silage of different forage crops in Malaysian climate analysed using 16S amplicon metagenomics. Sci. Rep. 2022, 12, 7107. [Google Scholar] [CrossRef]
- Huo, W.; Zhang, Y.; Zhang, L.; Shen, C.; Zhang, S.; Wang, C.; Guo, G. Effect of Lactobacilli inoculation on protein and carbohydrate fractions, ensiling characteristics and bacterial community of alfalfa silage. Front. Microbiol. 2022, 13, 1070175. [Google Scholar] [CrossRef]
- Fan, X.; Xie, Z.; Cheng, Q.; Li, M.; Long, J.; Lei, Y.; Jia, Y.; Wang, Z. Fermentation quality, bacterial community, and predicted functional profiles in silage prepared with alfalfa, perennia ryegrass and their mixture in the karst region. Front. Microbiol. 2022, 13, 1062515. [Google Scholar] [CrossRef]
- Bao, X.; Feng, H.; Guo, G.; Huo, W.; Li, Q.; Xu, Q.; Liu, Q.; Wang, C.; Chen, L. Effects of laccase and lactic acid bacteria on the fermentation quality, nutrient composition, enzymatic hydrolysis, and bacterial community of alfalfa silage. Front. Microbiol. 2022, 13, 1035942. [Google Scholar] [CrossRef] [PubMed]
- Si, H.; Liu, H.; Li, Z.; Nan, W.; Jin, C.; Sui, Y.; Li, G. Effect of Lactobacillus plantarum and Lactobacillus buchneri addition on fermentation, bacterial community and aerobic stability in lucerne silage. Anim. Prod. Sci. 2018, 59, 1528–1536. [Google Scholar] [CrossRef]
- Yang, F.; Wan, Y.; Zhao, S.; Wang, Y. Lactobacillus plantarum inoculants delay spoilage of high moisture alfalfa silages by regulating bacterial community composition. Front. Microbiol. 2020, 11, 1989. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Xie, Y.; Yu, Z.; Meng, G.; Wu, Z. Effect of Lactobacillus plantarum expressing multifunctional glycoside hydrolases on the characteristics of alfalfa silage. Appl. Microbiol. Biotechnol. 2019, 103, 7983–7995. [Google Scholar] [CrossRef]
- Chen, Q.; Li, M.; Fan, X.; Chen, Y.; Sun, H.; Xie, Y.; Zheng, Y.; Chen, C.; Li, P. Effects of epiphytic and exogenous lactic acid bacteria on fermentation quality and microbial community compositions of paper mulberry silage. Front. Microbiol. 2022, 13, 973500. [Google Scholar] [CrossRef]
- Su, R.; Ni, K.; Wang, T.; Yang, X.; Zhang, J.; Liu, Y.; Shi, W.; Yan, L.; Jie, C. Effects of ferulic acid esterase-producing Lactobacillus fermentum and cellulase additives on the fermentation quality and microbial community of alfalfa silage. PeerJ 2019, 7, e7712. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; He, L.; Xing, Y.; Zhou, W.; Yang, F.; Chen, X.; Zhang, Q. Fermentation quality and microbial community of alfalfa and stylo silage mixed with moringa oleifera leaves. Bioresour. Technol. 2019, 3, 129. [Google Scholar] [CrossRef]
- Bai, B.; Qiu, R.; Wang, Z.; Liu, Y.; Bao, J.; Sun, L.; Liu, T.; Ge, G.; Jia, Y. Effects of cellulase and lactic acid bacteria on ensiling performance and bacterial community of Caragana korshinskii silage. Microorganisms 2023, 11, 337. [Google Scholar] [CrossRef]
- Fan, X.; Zhao, S.; Yang, F.; Wang, Y. Effects of lactic acid bacterial inoculants on fermentation quality, bacterial community, and mycotoxins of alfalfa silage under vacuum or nonvacuum treatment. Microorganisms 2021, 9, 2614. [Google Scholar] [CrossRef]
- He, L.; Wang, Y.; Guo, X.; Chen, X.; Zhang, Q. Evaluating the effectiveness of screened lactic acid bacteria in improving crop residues silage: Fermentation parameter, nitrogen fraction, and bacterial community. Front. Microbiol. 2022, 13, 680988. [Google Scholar] [CrossRef] [PubMed]
- Na, N.; Moge, Q.; Wu, N.; Sun, L.; Xu, H.; Zhao, Y.; Wei, X.; Xue, Y.; Tao, Y. Bacterial community and fermentation quality of ensiling alfalfa with commercial lactic acid bacterial additives. Front. Microbiol. 2022, 13, 836899. [Google Scholar] [CrossRef] [PubMed]
- Silva, V.P.; Odilon, G.P.; Eliana, S.L.; Rosinea, A.P.; Mariele, C.N.; Karina, G.R. Selection of lactic acid bacteria from alfalfa silage and its effects as inoculant on silage fermentation. Agriculture 2020, 10, 518. [Google Scholar] [CrossRef]
- Xie, Y.; Bao, J.; Li, W.; Sun, Z.; Gao, R.; Wu, Z.; Yu, Z. Effects of applying lactic acid bacteria and molasses on the fermentation quality, protein fractions and in vitro digestibility of baled alfalfa silage. Agronomy 2021, 11, 91. [Google Scholar] [CrossRef]
- Sikora, M.C.; Hatfield, R.D.; Kalscheur, K.F. Impact of long-term storage on alfalfa leaf and stem silage characteristics. Agronomy 2021, 11, 2505. [Google Scholar] [CrossRef]
- Mu, L.; Wang, Q.; Cao, X.; Zhang, Z. Effects of fatty acid salts on fermentation characteristics, bacterial diversity and aerobic stability of mixed silage prepared with alfalfa, rice straw and wheat bran. J. Sci. Food Agric. 2021, 102, 1475–1487. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Wang, Y.; Yang, F.; Wang, Y.; Zhang, H. Screening a Lactobacillus plantarum strain for good adaption in alfalfa ensiling and demonstrating its improvement of alfalfa silage quality. J. Appl. Microbiol. 2020, 129, 233–242. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Li, J.; Dong, Z.; Shao, T. The reconstitution mechanism of napier grass microiota during the ensiling of alfalfa and their contributions to fermentation quality of silage. Bioresour. Technol. 2019, 297, 122391. [Google Scholar] [CrossRef]


| Items | CK30 | T30 | CK60 | T60 |
|---|---|---|---|---|
| Dry matter (DM) (weight %) | 27.5 ± 0.24 b | 28.7 ± 0.12 a | 27.3 ± 0.12 b | 28.6 ± 0.22 a |
| Crude protein (CP, % DM) * | 19.5 ± 0.05 b | 21.2 ± 0.17 a | 19.0 ± 0.17 c | 21.9 ± 0.12 a |
| Acid detergent fiber (ADF, % DM) * | 31.6 ± 0.12 a | 31.7 ± 0.09 a | 31.2 ± 0.21 b | 31.5 ± 0.05 ab |
| Neutral detergent fiber (NDF, % DM) * | 35.7 ± 0.05 b | 36.3 ± 0.08 a | 35.2 ± 0.12 c | 36.3 ± 0.08 a |
| Water soluble carbohydrates (WSC, % DM) * | 5.03 ± 0.21 c | 5.15 ± 0.05 a | 5.01 ± 0.25 c | 5.07 ± 0.05 b |
| Items | CK30 | T30 | CK60 | T60 |
|---|---|---|---|---|
| pH | 5.1 ± 0.05 a | 4.4 ± 0.09 c | 4.7 ± 0.05 b | 4.3 ± 0.17 c |
| Lactic acid (LA, % DM) * | 3.7 ± 0.05 c | 4.5 ± 0.12 b | 3.6 ± 0.12 c | 4.9 ± 0.05 a |
| Acetic acid (AA, % DM) * | 3.1 ± 0.05 b | 3.5 ± 0.05 b | 3.3 ± 0.05 c | 3.8 ± 0.01 a |
| Butyric acid (BA, % DM) * | 1.5 ± 0.14 ab | 1.3 ± 0.18 bc | 1.7 ± 0.09 a | 1.2 ± 0.01 c |
| NH4+-N (% DM) | 3.0 ± 0.13 a | 1.9 ± 0.12 b | 3.1 ± 0.05 a | 1.8 ± 0.1 b |
| Treatment | OTU | Shannon | Ace | Chao 1 | Coverage |
|---|---|---|---|---|---|
| Fresh | 114.7 a | 1.6 b | 125.4 a | 127.7 a | 0.99 a |
| T30 | 94 ab | 2.6 a | 108.6 ab | 105.1 ab | 0.99 a |
| CK30 | 94 ab | 2.1 ab | 95.1 b | 90.8 b | 0.99 a |
| T60 | 106.3 ab | 2.2 ab | 116.6 ab | 115.5 ab | 0.99 a |
| CK60 | 81.3 b | 1.9 ab | 98.8 b | 99.5 ab | 0.99 a |
| Items | Fresh | CK30 | CK60 | T30 | T60 |
|---|---|---|---|---|---|
| Carbohydrate Metabolism | 1,827,729 ab | 2,595,879 a | 1,708,199 ab | 1,366,396 b | 878,991 b |
| Amino Acid Metabolism | 1,734,546 ab | 2,025,808 a | 1,331,435 abc | 1,096,366 bc | 648,071 c |
| Cellular Processes and Signaling | 880,958 ab | 1,337,523 a | 713,491 bc | 496,637 bc | 272,306 c |
| Xenobiotics Biodegradation and Metabolism | 501,238 a | 522,642 a | 384,554 ab | 345,977 ab | 204,920 b |
| Glycan Biosynthesis and Metabolism | 429,512 ab | 574,628 a | 329,849 bc | 254,812 bc | 150,012 c |
| Enzyme Families | 366,165 ab | 513,575 a | 311,147 bc | 237,307 bc | 147,550 c |
| Metabolism of Other Amino Acids | 334,661 ab | 422,054 a | 274,368 abc | 222,434 bc | 135,539 c |
| Biosynthesis of Other Secondary Metabolites | 144,487 ab | 151,372 a | 106,654 abc | 92,316 bc | 53,808 c |
| Environmental Adaptation | 25,369 a | 26,743 a | 18,606 ab | 13,691 b | 8913 b |
| Signaling Molecules and Interaction | 22,710 a | 19,672 a | 28,117 a | 31,392 a | 30,907 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Wang, Y.; Zhang, L.; Liu, L.; Cai, T.; Chang, C.; Sa, D.; Yin, Q.; Jiang, X.; Li, Y.; et al. The Effect of Lactobacillus planturum YQM48 Inoculation on the Quality and Microbial Community Structure of Alfalfa Silage Cultured in Saline-Alkali Soil. Fermentation 2023, 9, 511. https://doi.org/10.3390/fermentation9060511
Liu Y, Wang Y, Zhang L, Liu L, Cai T, Chang C, Sa D, Yin Q, Jiang X, Li Y, et al. The Effect of Lactobacillus planturum YQM48 Inoculation on the Quality and Microbial Community Structure of Alfalfa Silage Cultured in Saline-Alkali Soil. Fermentation. 2023; 9(6):511. https://doi.org/10.3390/fermentation9060511
Chicago/Turabian StyleLiu, Yinghao, Yongjie Wang, Lianyi Zhang, Ling Liu, Ting Cai, Chun Chang, Duowen Sa, Qiang Yin, Xiaowei Jiang, Yuyu Li, and et al. 2023. "The Effect of Lactobacillus planturum YQM48 Inoculation on the Quality and Microbial Community Structure of Alfalfa Silage Cultured in Saline-Alkali Soil" Fermentation 9, no. 6: 511. https://doi.org/10.3390/fermentation9060511
APA StyleLiu, Y., Wang, Y., Zhang, L., Liu, L., Cai, T., Chang, C., Sa, D., Yin, Q., Jiang, X., Li, Y., & Lu, Q. (2023). The Effect of Lactobacillus planturum YQM48 Inoculation on the Quality and Microbial Community Structure of Alfalfa Silage Cultured in Saline-Alkali Soil. Fermentation, 9(6), 511. https://doi.org/10.3390/fermentation9060511




