Screening of a Novel Fibrinolytic Enzyme-Producing Streptomyces from a Hyper-Arid Area and Optimization of Its Fibrinolytic Enzyme Production
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Morphological and Molecular Identification of Strains
2.3. Preparation of the Crude Enzyme Solution and Assay of Enzyme Activity
2.4. Determination of Fibrinolytic and Protease Activities
2.5. In Vitro Anticoagulant and Thrombolytic Activity
2.6. Thrombolysis and Casein Degradation through SDS-PAGE
2.7. Single-Factor Condition Optimization of Fibrinolytic Enzyme Production
2.8. Experimental Design
2.9. Response Surface Model Validation
2.10. Determination of Fermentation Process Curve
2.11. Statistical Analysis
3. Results
3.1. Screening of Fibrinolytic Enzyme-Producing Actinomycetes
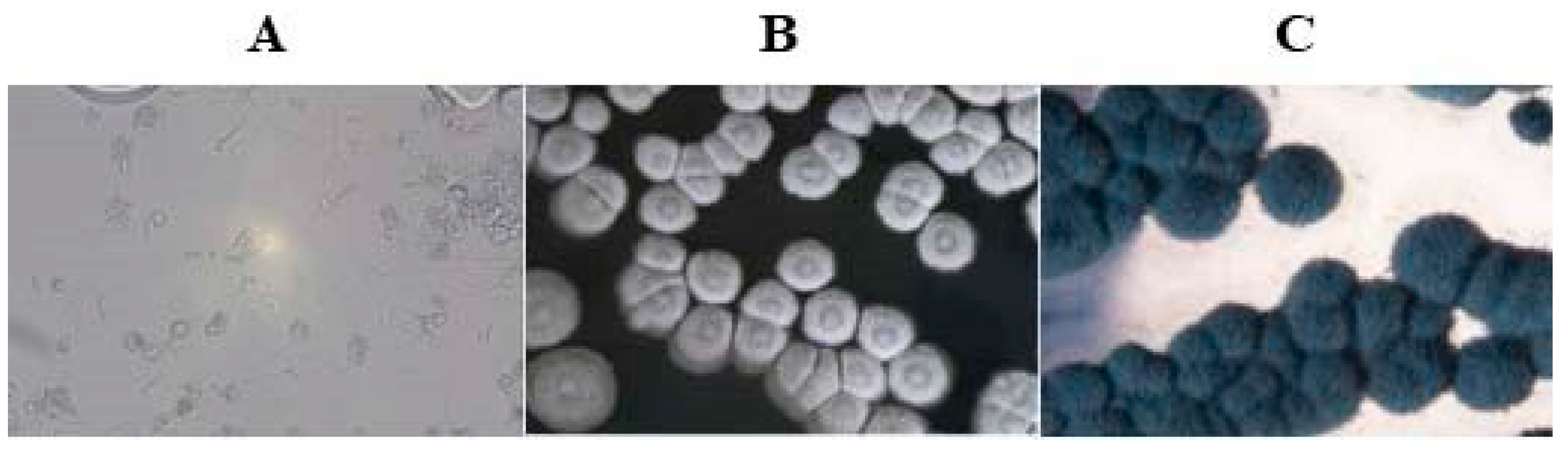
3.2. Morphological and Phylogenetic Analysis of the Strain
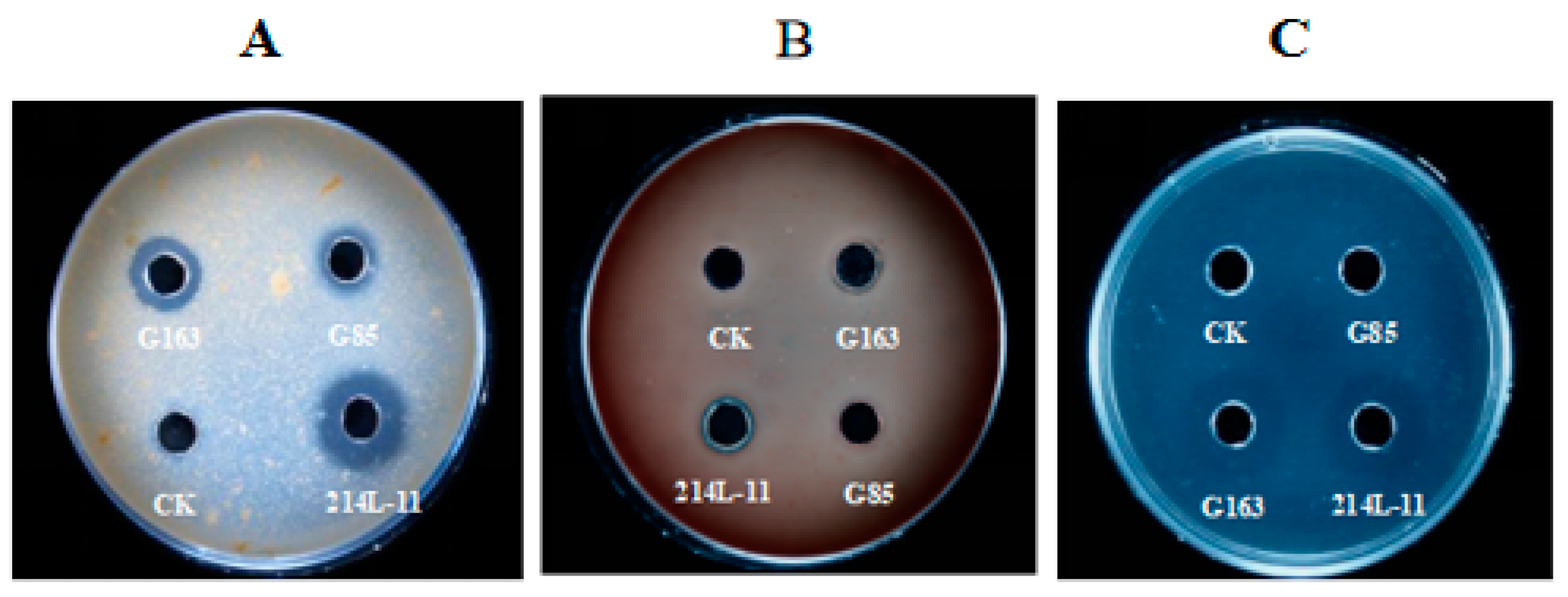
3.3. In Vitro Fibrinolytic Effect on Anticoagulant Activity and Blood Clots
3.4. Fibrinolytic and Casein Degradation Activity through SDS-PAGE Analysis
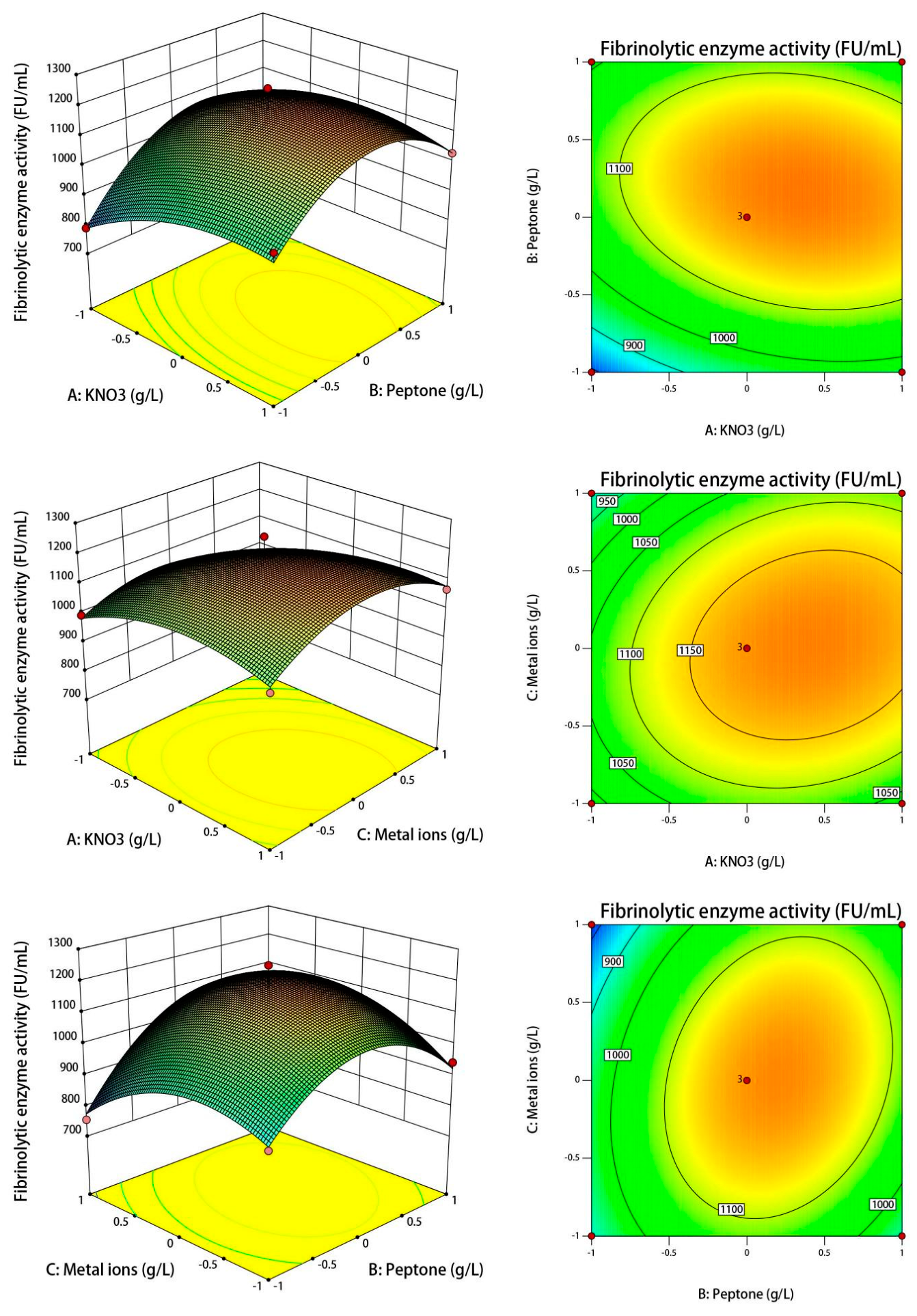
3.5. Optimization of Fermentation Conditions
3.6. Validation of the Models
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, W.; Geng, X.; Chen, W.; Wang, H.; Ng, T.B. Purification and characterization of a novel protease from the inky cap mushroom, coprinopsis atramentaria (agaricomycetes). Int. J. Med. Mushrooms 2018, 20, 349–358. [Google Scholar] [CrossRef]
- Barros, P.; Silva, P.; Nascimento, T.P.; Costa, R.; Bezerra, R.P.; Porto, A. Fibrinolytic enzyme from arthrospira platensis cultivated in medium culture supplemented with corn steep liquor. Int. J. Biol. Macromol. 2020, 164, 3446–3453. [Google Scholar] [CrossRef]
- Blann, A.D.; Landray, M.J.; Lip, G.Y. Abc of antithrombotic therapy: An overview of antithrombotic therapy. Bmj-Br. Med. J. 2002, 325, 762–765. [Google Scholar] [CrossRef]
- Li, G.; Liu, X.; Cong, S.; Deng, Y.; Zheng, X. A novel serine protease with anticoagulant and fibrinolytic activities from the fruiting bodies of mushroom agrocybe aegerita. Int. J. Biol. Macromol. 2021, 168, 631–639. [Google Scholar] [CrossRef]
- Medcalf, R.L. Plasminogen activation-based thrombolysis for ischaemic stroke: The diversity of targets may demand new approaches. Curr. Drug Targets 2011, 12, 1772–1781. [Google Scholar] [CrossRef]
- Murray, V.; Norrving, B.; Sandercock, P.A.; Terent, A.; Wardlaw, J.M.; Wester, P. The molecular basis of thrombolysis and its clinical application in stroke. J. Intern. Med. 2010, 267, 191–208. [Google Scholar] [CrossRef]
- Lu, M.; Gao, Z.; Xing, S.; Long, J.; Li, C.; He, L.; Wang, X. Purification, characterization, and chemical modification of bacillus velezensis sn-14 fibrinolytic enzyme. Int. J. Biol. Macromol. 2021, 177, 601–609. [Google Scholar] [CrossRef]
- Thokchom, S.; Joshi, S.R. Screening of fibrinolytic enzymes from lactic acid bacterial isolates associated with traditional fermented soybean foods. Food Sci. Biotechnol. 2014, 23, 1601–1604. [Google Scholar] [CrossRef]
- Zhao, L.; Lin, X.; Fu, J.; Zhang, J.; Tang, W.; He, Z. A novel bi-functional fibrinolytic enzyme with anticoagulant and thrombolytic activities from a marine-derived fungus aspergillus versicolor zlh-1. Mar. Drugs 2022, 20, 356. [Google Scholar] [CrossRef] [PubMed]
- Verma, P.; Chatterjee, S.; Keziah, M.S.; Devi, S.C. Fibrinolytic protease from marine streptomyces rubiginosus vitpss1. Cardiovasc. Hematol. Agents Med. Chem. 2018, 16, 44–55. [Google Scholar] [CrossRef] [PubMed]
- Cheng, G.; He, L.; Sun, Z.; Cui, Z.; Du, Y.; Kong, Y. Purification and biochemical characterization of a novel fibrinolytic enzyme from streptomyces sp. P3. J. Microbiol. Biotechnol. 2015, 25, 1449–1459. [Google Scholar] [CrossRef] [PubMed]
- Chitte, R.R.; Dey, S. Potent fibrinolytic enzyme from a thermophilic streptomyces megasporus strain sd5. Lett. Appl. Microbiol. 2000, 31, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Dhamodharan, D.; Naine, J.S.; Keziah, M.S.; Devi, S.C. Novel fibrinolytic protease producing streptomyces radiopugnans vitsd8 from marine sponges. Mar. Drugs 2019, 17, 164. [Google Scholar] [CrossRef]
- Mao, J.; Tang, Q.; Zhang, Z.; Wang, W.; Wei, D.; Huang, Y.; Liu, Z.; Shi, Y.; Goodfellow, M. Streptomyces radiopugnans sp. Nov., A radiation-resistant actinomycete isolated from radiation-polluted soil in china. Int. J. Syst. Evol. Microbiol. 2007, 57, 2578–2582. [Google Scholar] [CrossRef]
- Vijayaraghavan, P.; Arasu, M.V.; Anantha Rajan, R.; Al-Dhabi, N.A. Enhanced production of fibrinolytic enzyme by a new xanthomonas oryzae ind3 using low-cost culture medium by response surface methodology. Saudi J. Biol. Sci. 2019, 26, 217–224. [Google Scholar] [CrossRef]
- He, Z.; Wang, Y.; Bai, X.; Chu, M.; Yi, Y.; Zhu, J.; Gu, M.; Jiang, L.; Zhang, Z. Bacterial community composition and isolation of actinobacteria from the soil of flaming mountain in xinjiang, china. Microorganisms 2023, 11, 489. [Google Scholar] [CrossRef] [PubMed]
- Campos-M, M.; Campos-C, R. Applications of quartering method in soils and foods. Int. J. Eng. Res. Appl. 2017, 7, 35–39. [Google Scholar] [CrossRef]
- Li, Y.; Li, Y.; Wang, L.W.; Bao, J. Streptomyces dengpaensis sp. Nov., An actinomycete isolated from desert soil. Int. J. Syst. Evol. Microbiol. 2018, 68, 3322–3326. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Thompson, J.D.; Gibson, T.J.; Plewniak, F.; Jeanmougin, F.; Higgins, D.G. The clustal_x windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997, 25, 4876–4882. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. Mega7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.G.; Wang, H.F.; Yang, L.L.; Zhou, X.K.; Zhi, X.Y.; Duan, Y.Q.; Xiao, M.; Zhang, Y.M.; Li, W.J. Egibacter rhizosphaerae gen. Nov., Sp. Nov., An obligately halophilic, facultatively alkaliphilic actinobacterium and proposal of egibaceraceae fam. Nov. And egibacterales ord. Nov. Int. J. Syst. Evol. Microbiol. 2016, 66, 283–289. [Google Scholar] [CrossRef]
- Xu, F. Measurement of Proteinase Activity; SB/T 10317-1999; Ministry of Commerce of the People’s Republic of China: Beijing, China, 1987.
- Astrup, T.; Müllertz, S. The fibrin plate method for estimating fibrinolytic activity. Arch. Biochem. Biophys. 1952, 40, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Guo, T. Study on Extraction and Separation and Biological Activity of Poecilobdella manillensis Anticoagulant Active Substance. Master’s Thesis, Soochow University, Suzhou, China, 2020. [Google Scholar]
- Huang, S.; Huang, D.; Wu, Q.; Hou, M.; Tang, X.; Zhou, J. Effect of environmental c/n ratio on activities of lignin-degrading enzymes produced by phanerochaete chrysosporium. Pedosphere 2020, 30, 285–292. [Google Scholar] [CrossRef]
- Chen, W.; Gao, L.; Song, L.; Sommerfeld, M.; Hu, Q. An improved phenol-sulfuric acid method for the quantitative measurement of total carbohydrates in algal biomass. Algal Res. 2023, 70, 102986. [Google Scholar] [CrossRef]
- Kim, J.S.; Kim, J.E.; Choi, B.S.; Park, S.E.; Sapkota, K.; Kim, S.; Lee, H.H.; Kim, C.S.; Park, Y.; Kim, M.K.; et al. Purification and characterization of fibrinolytic metalloprotease from perenniporia fraxinea mycelia. Mycol. Res. 2008, 112, 990–998. [Google Scholar] [CrossRef]
- Mi, Y.; Dong, C.; Hou, Z.; Shi, Y. Purification and characterization of the fibrinolytic enzyme from a marine streptomyces strain. Chin. J. Mar. Drugs 2016, 35, 43–48. [Google Scholar]
- Abdul, R.P.; Rengaswamy, D. Fibrinolytic enzyme-an overview. Curr. Pharm. Biotechnol. 2022, 23, 1336–1345. [Google Scholar] [CrossRef]
- Verma, J.; Sourirajan, A.; Dev, K. Bacterial diversity in 110 thermal hot springs of indian himalayan region (ihr). 3 Biotech 2022, 12, 238. [Google Scholar] [CrossRef]
- Xie, F.; Pathom-Aree, W. Actinobacteria from desert: Diversity and biotechnological applications. Front. Microbiol. 2021, 12, 765531. [Google Scholar] [CrossRef]
- Bay, S.K.; Waite, D.W.; Dong, X.; Gillor, O.; Chown, S.L.; Hugenholtz, P.; Greening, C. Chemosynthetic and photosynthetic bacteria contribute differentially to primary production across a steep desert aridity gradient. Isme J. 2021, 15, 3339–3356. [Google Scholar] [CrossRef]
- Choe, Y.H.; Kim, M.; Lee, Y.K. Distinct microbial communities in adjacent rock and soil substrates on a high arctic polar desert. Front. Microbiol. 2020, 11, 607396. [Google Scholar] [CrossRef]
- Molina-Menor, E.; Gimeno-Valero, H.; Pascual, J.; Pereto, J.; Porcar, M. High culturable bacterial diversity from a european desert: The tabernas desert. Front. Microbiol. 2020, 11, 583120. [Google Scholar] [CrossRef] [PubMed]
- Aini, G.; Aji, D. Climate change in turpan city in recent 57 years. J. Guangzhou Univ. (Nat. Sci. Ed.) 2012, 11, 83–87. [Google Scholar]
- Huo, P.; Mao, J.; Shi, Y. Study on the pilot fermentation technology of the xjt9503 thermostable neutral protease. J. Wuxi Univ. Light Ind. 2003, 22, 89–92. [Google Scholar]
- Budihal, S.R.; Agsar, D.; Patil, S.R. Enhanced production and application of acidothermophilic streptomyces cellulase. Bioresour. Technol. 2016, 200, 706–712. [Google Scholar] [CrossRef]
- Dhanaseeli, P.; Balasubramanian, V. Screening, production and optimization of protease enzyme from streptomyces species. Biosci. Biotechnol. Res. Asia 2014, 11, 369–376. [Google Scholar] [CrossRef]
- Singh, T.A.; Jajoo, A.; Bhasin, S. Production and application of glucose isomerase from streptomyces enissocaesilis and amylase from streptomyces sp. For the synthesis of high fructose corn syrup. Sn Appl. Sci. 2020, 2, 1968. [Google Scholar] [CrossRef]
- Deng, Q.; Zhou, L.; Luo, M.; Deng, Z.; Zhao, C. Heterologous expression of avermectins biosynthetic gene cluster by construction of a bacterial artificial chromosome library of the producers. Synth. Syst. Biotechnol. 2017, 2, 59–64. [Google Scholar] [CrossRef]
- Fang, Q.; Maglangit, F.; Mugat, M.; Urwald, C.; Kyeremeh, K.; Deng, H. Targeted isolation of indole alkaloids from Streptomyces sp. Ct37. Molecules 2020, 25, 1108. [Google Scholar] [CrossRef]
- Martin-Sanchez, L.; Singh, K.S.; Avalos, M.; van Wezel, G.P.; Dickschat, J.S.; Garbeva, P. Phylogenomic analyses and distribution of terpene synthases among streptomyces. Beilstein J. Org. Chem. 2019, 15, 1181–1193. [Google Scholar] [CrossRef] [PubMed]
- Mcculloch, K.M.; Mccranie, E.K.; Smith, J.A.; Sarwar, M.; Mathieu, J.L.; Gitschlag, B.L.; Du, Y.; Bachmann, B.O.; Iverson, T.M. Oxidative cyclizations in orthosomycin biosynthesis expand the known chemistry of an oxygenase superfamily. Proc. Natl. Acad. Sci. USA 2015, 112, 11547–11552. [Google Scholar] [CrossRef]
- Kim, M.; Oh, H.S.; Park, S.C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16s rrna gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef]
- Liu, X.; Kopparapu, N.K.; Li, Y.; Deng, Y.; Zheng, X. Biochemical characterization of a novel fibrinolytic enzyme from cordyceps militaris. Int. J. Biol. Macromol. 2017, 94, 793–801. [Google Scholar] [CrossRef]
- Tang, Z.; Zhu, C.; Zhang, N.; Mo, Y.; Xie, X. Optimization of fermentation conditions for douchi fibrinolytic enzyme production by streptomyces albus hs1. J. Hainan Norm. Univ. (Nat. Sci.) 2010, 23, 308–313+337. [Google Scholar]
- Silva, G.M.D.M.; Bezerra, R.P.; Teixeira, J.A.; Silva, F.O.; Correia, J.M.; Porto, T.S.; Lima-Filho, J.L.; Porto, A.L.F. Screening, production and biochemical characterization of a new fibrinolytic enzyme produced by Streptomyces sp. (Streptomycetaceae) isolated from amazonian lichens. Acta Amazon. 2016, 46, 323–332. [Google Scholar] [CrossRef]
- Khursade, P.S.; Galande, S.H.; Shiva, K.P.; Prakasham, R.S. Stenotrophomonas maltophilia gd2: A potential and novel isolate for fibrinolytic enzyme production. Saudi J. Biol. Sci. 2019, 26, 1567–1575. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Song, Y.; Wang, M.; Chen, J.; Duan, L.; Deng, Y. Comparative study of nattokinase and urokinase. Chin. J. Pharm. 2016, 14, 125–134. [Google Scholar]





| Factors | Codes | Level | |
|---|---|---|---|
| −1 | +1 | ||
| Soluble starch (g/L) | A | 11.25 | 18.75 |
| KNO3 (g/L) | B | 0.375 | 0.625 |
| Peptone (g/L) | C | 0.375 | 0.625 |
| Inoculum size (%) | D | 1.5 | 2.5 |
| Liquid volume (%) | E | 15 | 25 |
| Mn2+ (g/L) | F | 0.15 | 0.25 |
| Initial pH | G | 7.5 | 8.5 |
| Virtual factor | H~L | - | - |
| Factors | Codes | Level | ||
|---|---|---|---|---|
| −1 | 0 | 1 | ||
| KNO3 (g/L) | A | 0.45 | 0.5 | 0.55 |
| Peptone (g/L) | B | 0.45 | 0.5 | 0.55 |
| Mn2+ (g/L) | C | 0.18 | 0.2 | 0.22 |
| Term | Stdized Effect | Sum of Squares | Contribution (%) | Importance |
|---|---|---|---|---|
| A: Soluble starch | 40.92 | 4370.85 | 2.64 | |
| B: KNO3 | 115.58 | 42005.97 | 25.40 | Important |
| C: Peptone | 101.62 | 32679.29 | 19.76 | Important |
| D: Inoculum size | −4.38 | 7.97 | 4.820 × 10−3 | |
| E: Liquid volume | −15.42 | 990.45 | 0.60 | |
| F: Metal ions | −88.58 | 25023.51 | 15.13 | Important |
| G: Initial pH | 87.02 | 21304.30 | 12.88 | |
| H: (Virtual factor) | −14.00 | 15236.14 | 9.21 | |
| I: (Virtual factor) | −21.02 | 1695.04 | 1.03 | |
| J: (Virtual factor) | 54.58 | 9860.19 | 5.96 | |
| K: (Virtual factor) | 60.98 | 12184.54 | 7.37 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, Z.; Sun, Y.; Chu, M.; Zhu, J.; Zhang, Y.; Tang, Q.; Osman, G.; Jiang, L.; Zhang, Z. Screening of a Novel Fibrinolytic Enzyme-Producing Streptomyces from a Hyper-Arid Area and Optimization of Its Fibrinolytic Enzyme Production. Fermentation 2023, 9, 410. https://doi.org/10.3390/fermentation9050410
He Z, Sun Y, Chu M, Zhu J, Zhang Y, Tang Q, Osman G, Jiang L, Zhang Z. Screening of a Novel Fibrinolytic Enzyme-Producing Streptomyces from a Hyper-Arid Area and Optimization of Its Fibrinolytic Enzyme Production. Fermentation. 2023; 9(5):410. https://doi.org/10.3390/fermentation9050410
Chicago/Turabian StyleHe, Zixuan, Yang Sun, Min Chu, Jing Zhu, Yu Zhang, Qiyong Tang, Ghenijan Osman, Ling Jiang, and Zhidong Zhang. 2023. "Screening of a Novel Fibrinolytic Enzyme-Producing Streptomyces from a Hyper-Arid Area and Optimization of Its Fibrinolytic Enzyme Production" Fermentation 9, no. 5: 410. https://doi.org/10.3390/fermentation9050410
APA StyleHe, Z., Sun, Y., Chu, M., Zhu, J., Zhang, Y., Tang, Q., Osman, G., Jiang, L., & Zhang, Z. (2023). Screening of a Novel Fibrinolytic Enzyme-Producing Streptomyces from a Hyper-Arid Area and Optimization of Its Fibrinolytic Enzyme Production. Fermentation, 9(5), 410. https://doi.org/10.3390/fermentation9050410






