Transcriptomics-Guided Investigation of the SLCG_Lrp Regulon Provides New Insights into Its Role for Lincomycin Biosynthesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Strains, Plasmids along with Culture
2.2. Gene Deletion and Overexpression Mutant Establishment
2.3. Fermentation and Lincomycin Determination
2.4. RNA-Seq
2.5. RNA Extraction and RT-qPCR Analysis
2.6. Protein Expression and Purification
2.7. Electrophoretic Mobility Shift Assays (EMSAs)
2.8. Statistical Analysis
3. Results
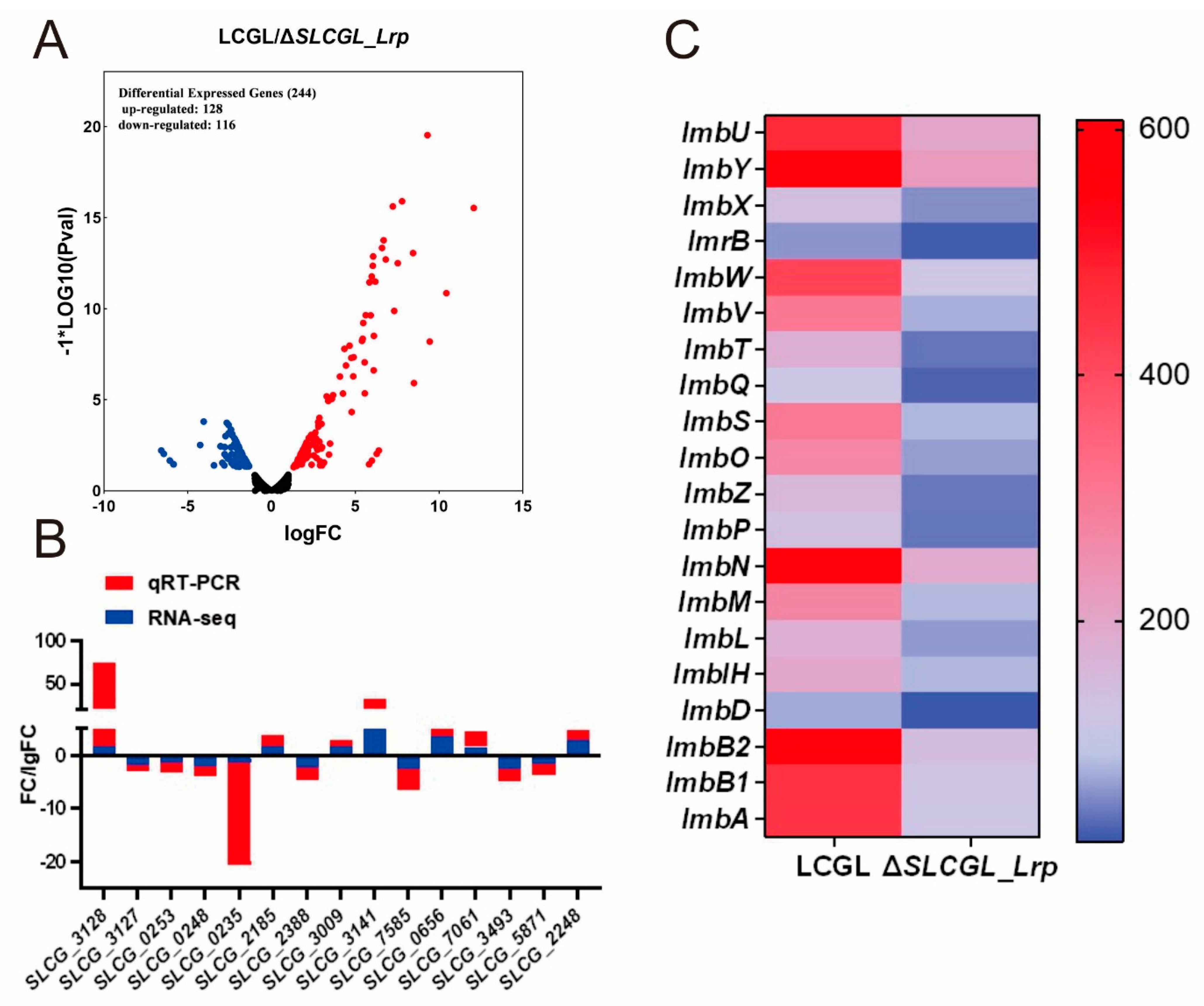
3.1. Transcriptome Analysis of LCGL and ΔSLCGL_Lrp
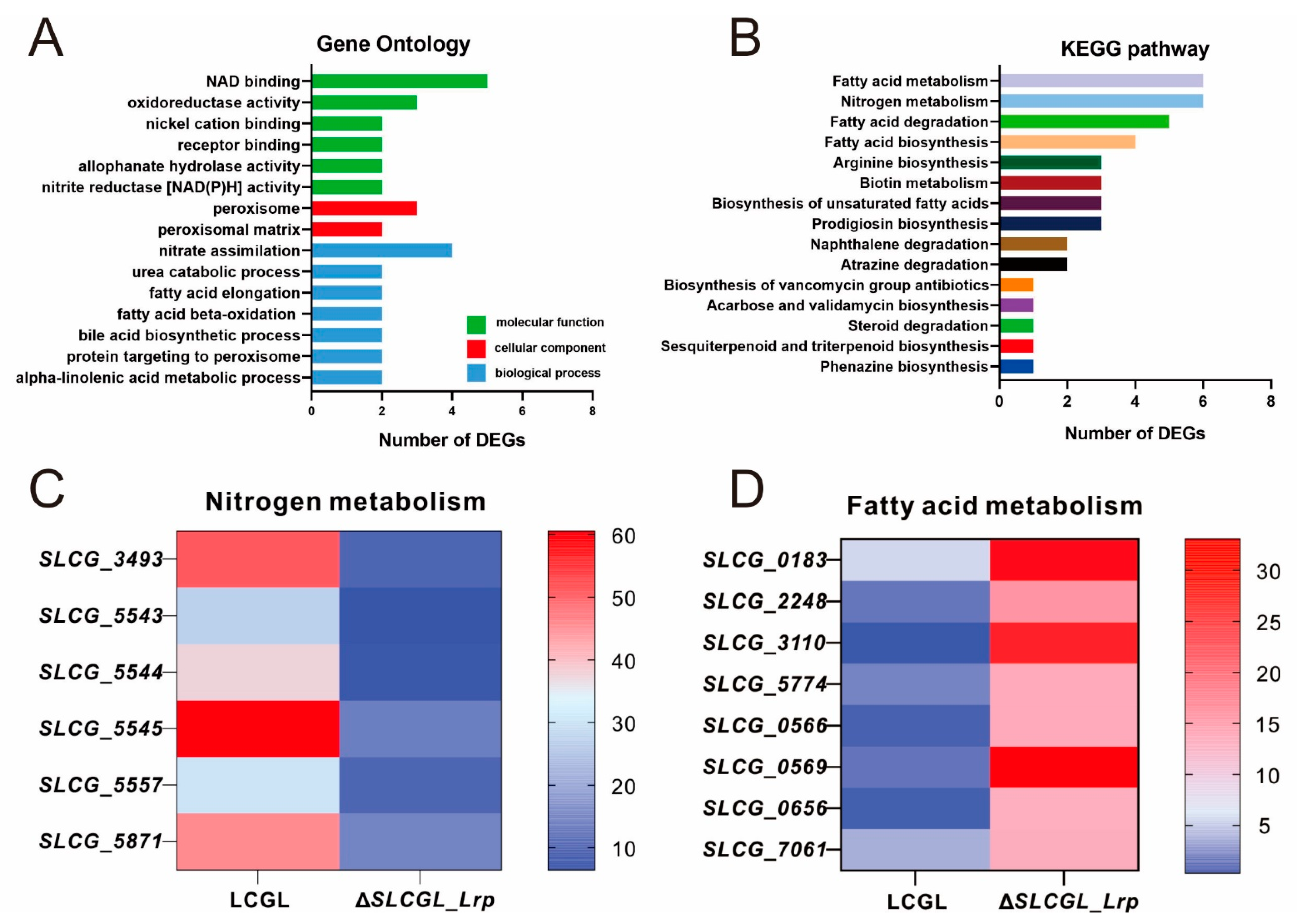
3.2. GO as Well as KEGG Analysis on DEGs
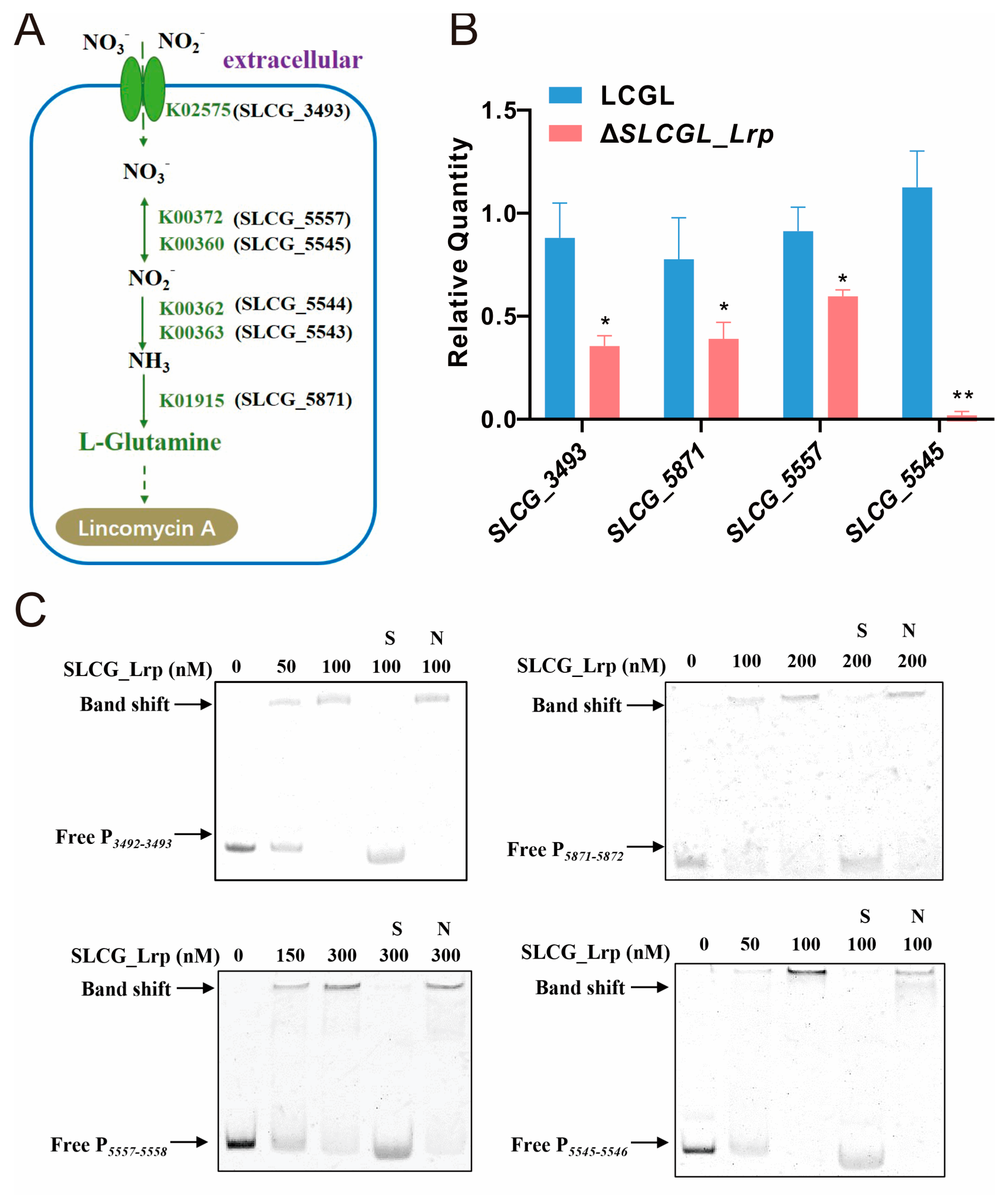
3.3. SLCG_Lrp Directly Promotes the Transcription of Nitrogen Metabolism Genes
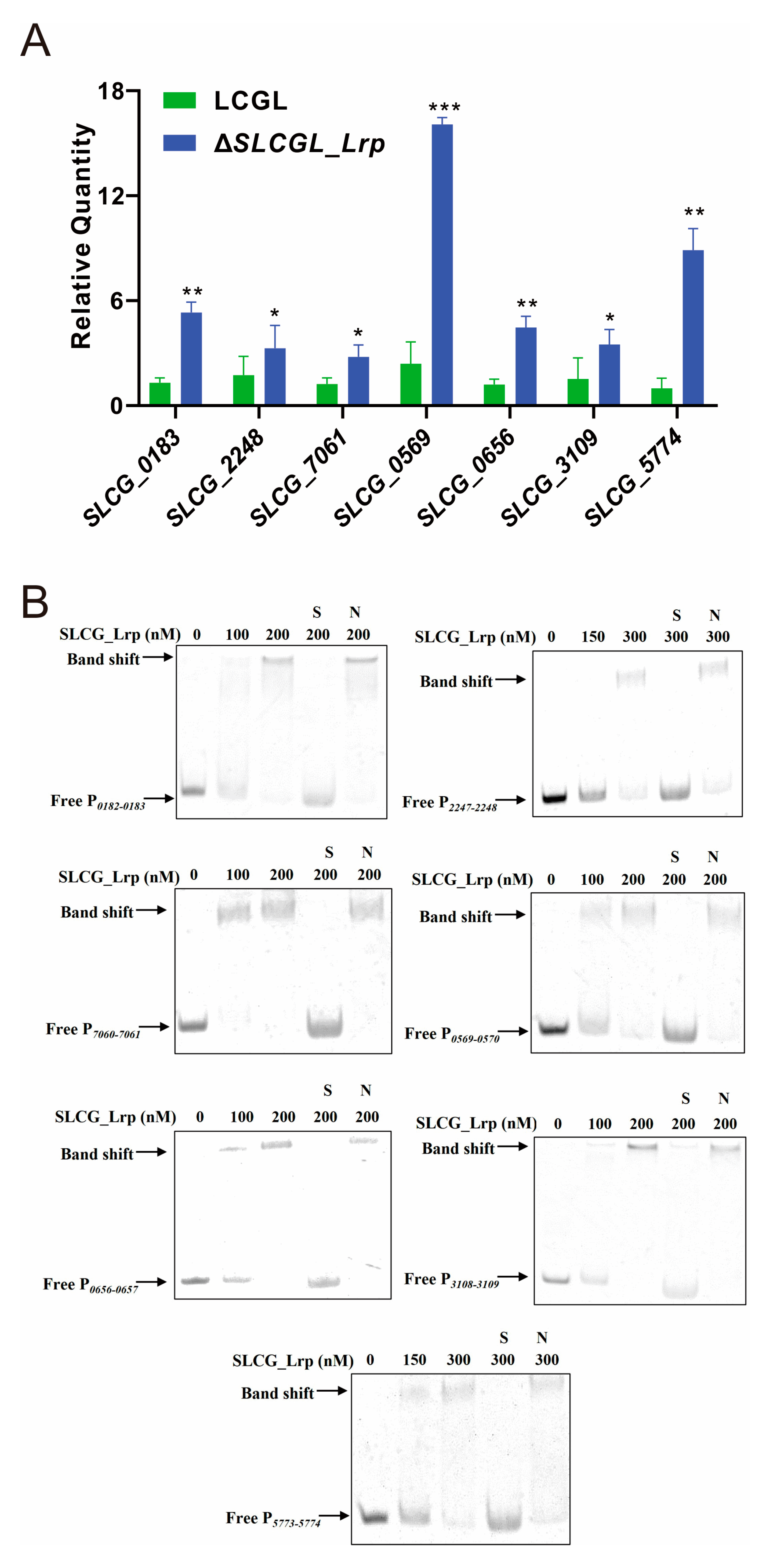
3.4. SLCG_Lrp Directly Represses the Transcription of Fatty Acid Metabolism Genes
3.5. SLCG_Lrp Transcriptionally Modulates Five Regulatory Gene in S. lincolnensis
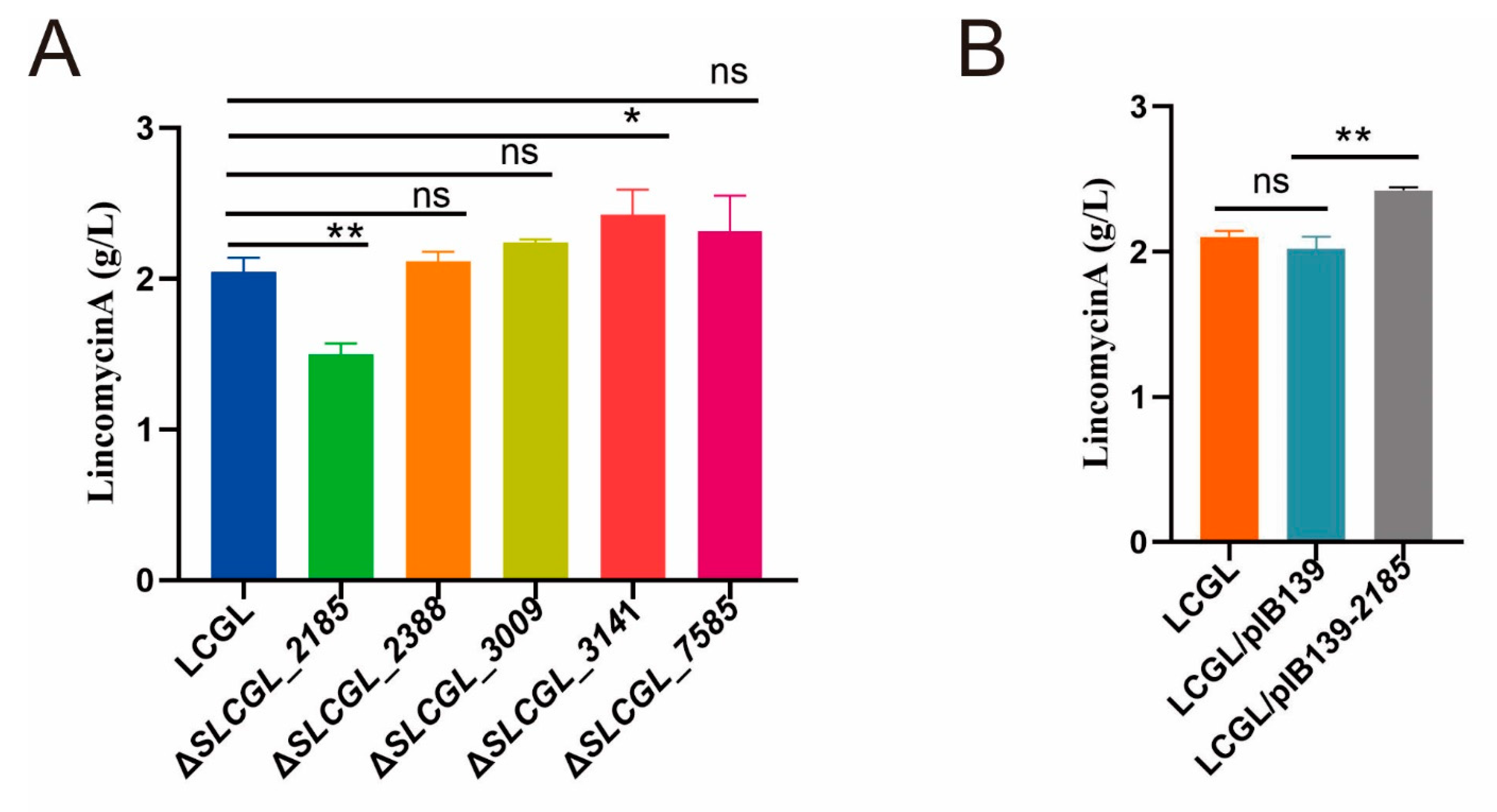
3.6. SLCG_2185 Shows Direct Transcriptional Activation on Lincomycin Biosynthetic Cluster
3.7. Rational Improvement of the Industrial High-Yield S. lincolnensis LA129X Strain
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Spížek, J.; Řezanka, T. Lincomycin, cultivation of producing strains and biosynthesis. Appl. Microbiol. Biotechnol. 2003, 63, 510–519. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Wang, M.; Xu, D.; Zhang, Q.; Liu, W. Metabolic coupling of two small-molecule thiols programs the biosynthesis of lincomycin A. Nature 2015, 518, 115–119. [Google Scholar] [CrossRef]
- Wu, H.; Wang, Y.; Yuan, L.; Mao, Y.; Wang, W.; Zhu, L.; Wu, P.; Fu, C.; Müller, R.; Weaver, D.; et al. Inactivation of SACE_3446, a TetR family transcriptional regulator, stimulates erythromycin production in Saccharopolyspora erythraea. Synth Syst. Biotechnol. 2016, 1, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Koberská, M.; Kopecký, J.; Olsovská, J.; Jelínková, M.; Ulanova, D.; Man, P.; Flieger, M.; Janata, J. Sequence analysis and heterologous expression of the lincomycin biosynthetic cluster of the type strain Streptomyces lincolnensis ATCC 25466. Folia Microbiol. 2008, 53, 395–401. [Google Scholar] [CrossRef]
- Koberska, M.; Vesela, L.; Vimberg, V.; Lenart, J.; Vesela, J.; Kamenik, Z.; Janata, J.; Balikova Novotna, G. Beyond Self-Resistance: ABCF ATPase LmrC Is a Signal-Transducing Component of an Antibiotic-Driven Signaling Cascade Accelerating the Onset of Lincomycin Biosynthesis. mBio 2021, 12, e0173121. [Google Scholar] [CrossRef]
- Hou, B.; Zhu, X.; Kang, Y.; Wang, R.; Wu, H.; Ye, J.; Zhang, H. LmbU, a Cluster-Situated Regulator for Lincomycin, Consists of a DNA-Binding Domain, an Auto-Inhibitory Domain, and Forms Homodimer. Front. Microbiol. 2019, 10, 989. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhao, Q.; Zhang, Q.; Liu, W. Differences in PLP-Dependent Cysteinyl Processing Lead to Diverse S-Functionalization of Lincosamide Antibiotics. J. Am. Chem. Soc. 2016, 138, 6348–6351. [Google Scholar] [CrossRef] [PubMed]
- van der Heul, H.; Bilyk, B.; McDowall, K.; Seipke, R.; van Wezel, G. Regulation of antibiotic production in Actinobacteria: New perspectives from the post-genomic era. Nat. Prod. Rep. 2018, 35, 575–604. [Google Scholar] [CrossRef]
- Wang, R.; Kong, F.; Wu, H.; Hou, B.; Kang, Y.; Cao, Y.; Duan, S.; Ye, J.; Zhang, H. Complete genome sequence of high-yield strain S. lincolnensis B48 and identification of crucial mutations contributing to lincomycin overproduction. Synth Syst. Biotechnol. 2020, 5, 37–48. [Google Scholar] [CrossRef]
- Wu, H.; Chu, Z.; Zhang, W.; Zhang, C.; Ni, J.; Fang, H.; Chen, Y.; Wang, Y.; Zhang, L.; Zhang, B. Transcriptome-guided target identification of the TetR-like regulator SACE_5754 and engineered overproduction of erythromycin in Saccharopolyspora erythraea. J. Biol. Eng. 2019, 13, 11. [Google Scholar] [CrossRef]
- Ziegler, C.; Freddolino, P. The leucine-responsive regulatory proteins/feast-famine regulatory proteins: An ancient and complex class of transcriptional regulators in bacteria and archaea. Crit. Rev. Biochem. Mol. Biol. 2021, 56, 373–400. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Wang, H.; Xie, J. Regulatory and pathogenesis roles of Mycobacterium Lrp/AsnC family transcriptional factors. J. Cell Biochem. 2011, 112, 2655–2662. [Google Scholar] [CrossRef]
- Cho, B.; Barrett, C.; Knight, E.; Park, Y.; Palsson, B. Genome-scale reconstruction of the Lrp regulatory network in Escherichia coli. Proc. Natl. Acad. Sci. USA 2008, 105, 19462–19467. [Google Scholar] [CrossRef] [PubMed]
- Peeters, E.; Charlier, D. The Lrp family of transcription regulators in archaea. Archaea 2010, 2010, 750457. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Chen, Y.; Wang, W.; Ren, M.; Wu, P.; Wang, Y.; Li, C.; Zhang, L.; Wu, H.; Weaver, D.; et al. Engineering of an Lrp family regulator SACE_Lrp improves erythromycin production in Saccharopolyspora erythraea. Metab. Eng. 2017, 39, 29–37. [Google Scholar] [CrossRef]
- Liu, J.; Chen, Y.; Li, L.; Yang, E.; Wang, Y.; Wu, H.; Zhang, L.; Wang, W.; Zhang, B. Characterization and engineering of the Lrp/AsnC family regulator SACE_5717 for erythromycin overproduction in Saccharopolyspora erythraea. J. Ind. Microbiol. Biotechnol. 2019, 46, 1013–1024. [Google Scholar] [CrossRef]
- Liu, J.; Li, J.; Dong, H.; Chen, Y.; Wang, Y.; Wu, H.; Li, C.; Weaver, D.; Zhang, L.; Zhang, B. Characterization of an Lrp/AsnC family regulator SCO3361, controlling actinorhodin production and morphological development in Streptomyces coelicolor. Appl. Microbiol. Biotechnol. 2017, 101, 5773–5783. [Google Scholar] [CrossRef]
- Lu, Z.; Zhang, X.; Dai, J.; Wang, Y.; He, W. Engineering of leucine-responsive regulatory protein improves spiramycin and bitespiramycin biosynthesis. Microb. Cell Fact 2019, 18, 38. [Google Scholar] [CrossRef]
- Liu, J.; Wang, Y.; He, H.; Dong, S.; Tang, L.; Yang, E.; Wang, W.; Zhang, B. The leucine-responsive regulatory protein SCAB_Lrp modulates thaxtomin biosynthesis, pathogenicity, and morphological development in Streptomyces scabies. Mol. Plant Pathol. 2023, 24, 167–178. [Google Scholar] [CrossRef]
- Xu, Y.; Tang, Y.; Wang, N.; Liu, J.; Cai, X.; Cai, H.; Li, J.; Tan, G.; Liu, R.; Bai, L.; et al. Transcriptional regulation of a leucine-responsive regulatory protein for directly controlling lincomycin biosynthesis in Streptomyces lincolnensis. Appl. Microbiol. Biotechnol. 2020, 104, 2575–2587. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Du, L.; Liu, R.; Ying, L.; Zhao, G. An Efficient Intergeneric Conjugation of DNA from Escherichia coli to Mycelia of the Lincomycin-Producer Streptomyces lincolnensis. Int. J. Mol. Sci. 2012, 13, 4797–4806. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Tan, G.; Ke, M.; Li, J.; Tang, Y.; Meng, S.; Niu, J.; Wang, Y.; Liu, R.; Wu, H.; et al. Enhanced lincomycin production by co-overexpression of metK1 and metK2 in Streptomyces lincolnensis. J. Ind. Microbiol. Biotechnol. 2018, 45, 345–355. [Google Scholar] [CrossRef] [PubMed]
- Bierman, M.; Logan, R.; O’brien, K.; Seno, E.; Rao, R.N.; Schoner, B. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene 1992, 116, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Chen, M.; Mao, Y.; Li, W.; Liu, J.; Huang, X.; Zhou, Y.; Ye, B.; Zhang, L.; Weaver, D.; et al. Dissecting and engineering of the TetR family regulator SACE_7301 for enhanced erythromycin production in Saccharopolyspora erythraea. Microb. Cell Fact. 2014, 13, 158. [Google Scholar] [CrossRef]
- Xu, Y.; Ke, M.; Li, J.; Tang, Y.; Wang, N.; Tan, G.; Wang, Y.; Liu, R.; Bai, L.; Zhang, L.; et al. TetR-Type Regulator SLCG_2919 Is a Negative Regulator of Lincomycin Biosynthesis in Streptomyces lincolnensis. Appl. Environ. Microbiol. 2019, 85, e02091-18. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Hellman, L.M.; Fried, M.G. Electrophoretic mobility shift assay (EMSA) for detecting protein–nucleic acid interactions. Nat. Protoc. 2007, 2, 1849–1861. [Google Scholar] [CrossRef] [PubMed]
- Lintner, R.; Mishra, P.; Srivastava, P.; Martinez-Vaz, B.; Khodursky, A.; Blumenthal, R. Limited functional conservation of a global regulator among related bacterial genera: Lrp in Escherichia, Proteus and Vibrio. BMC Microbiol. 2008, 8, 60. [Google Scholar] [CrossRef]
- Fukuda, M.; Takeda, H.; Kato, H.; Doki, S.; Ito, K.; Maturana, A.; Ishitani, R.; Nureki, O. Structural basis for dynamic mechanism of nitrate/nitrite antiport by NarK. Nat. Commun. 2015, 6, 7097. [Google Scholar] [CrossRef]
- Chu, S.; Zhang, D.; Wang, D.; Zhi, Y.; Zhou, P. Heterologous expression and biochemical characterization of assimilatory nitrate and nitrite reductase reveals adaption and potential of Bacillus megaterium NCT-2 in secondary salinization soil. Int. J. Biol. Macromol. 2017, 101, 1019–1028. [Google Scholar] [CrossRef] [PubMed]
- Meng, S.; Wu, H.; Wang, L.; Zhang, B.; Bai, L. Enhancement of antibiotic productions by engineered nitrate utilization in actinomycetes. Appl. Microbiol. Biotechnol. 2017, 101, 5341–5352. [Google Scholar] [CrossRef] [PubMed]
- Neusser, D.; Schmidt, H.; Spizèk, J.; Novotnà, J.; Peschke, U.; Kaschabeck, S.; Tichy, P.; Piepersberg, W. The genes lmbB1 and lmbB2 of Streptomyces lincolnensis encode enzymes involved in the conversion of L-tyrosine to propylproline during the biosynthesis of the antibiotic lincomycin A. Arch. Microbiol. 1998, 169, 322–332. [Google Scholar] [CrossRef] [PubMed]
- Perli, T.; van der Vorm, D.; Wassink, M.; van den Broek, M.; Pronk, J.; Daran, J. Engineering heterologous molybdenum-cofactor-biosynthesis and nitrate-assimilation pathways enables nitrate utilization by Saccharomyces cerevisiae. Metab. Eng. 2021, 65, 11–29. [Google Scholar] [CrossRef]
- Fujihashi, M.; Nakatani, T.; Hirooka, K.; Matsuoka, H.; Fujita, Y.; Miki, K. Structural characterization of a ligand-bound form of Bacillus subtilis FadR involved in the regulation of fatty acid degradation. Proteins 2014, 82, 1301–1310. [Google Scholar] [CrossRef]
- Grau, F.; Burkovski, A.; Muller, Y. Crystal structures of adenylylated and unadenylylated P protein GlnK from Corynebacterium glutamicum. Acta Crystallogr. D Struct. Biol. 2021, 77, 325–335. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Wang, L.; Li, S.; Pan, C.; Cheng, K.; Luo, Y.; Xu, H.; Tian, B.; Zhao, Y.; Hua, Y. Structure and DNA damage-dependent derepression mechanism for the XRE family member DG-DdrO. Nucleic Acids Res. 2019, 47, 9925–9933. [Google Scholar] [CrossRef]
- Zhu, Y.; Lu, T.; Zhang, J.; Zhang, P.; Tao, M.; Pang, X. A novel XRE family regulator that controls antibiotic production and development in Streptomyces coelicolor. Appl. Microbiol. Biotechnol. 2020, 104, 10075–10089. [Google Scholar] [CrossRef]
- Yang, Y.; Luo, M.; Zhou, H.; Li, C.; Luk, A.; Zhao, G.; Fung, K.; Ip, M. Role of Two-Component System Response Regulator bceR in the Antimicrobial Resistance, Virulence, Biofilm Formation, and Stress Response of Group B Streptococcus. Front. Microbiol. 2019, 10, 10. [Google Scholar] [CrossRef] [PubMed]




| Strains and Plasmids | Description | Reference |
|---|---|---|
| E. coli | ||
| DH5α | F recA lacZM15 | [21] |
| BL21 (DE3) | F− ompT hsdSB(rB− mB−) dcm gal λ(DE3) | Novagen |
| S. lincolnensis | ||
| LCGL | A lincomycin producer with artificially integrated attBΦC31 site | [23] |
| ΔSLCGL_Lrp | LCGL derivative with SLCG_Lrp deleted | [20] |
| ΔSLCGL_2185 | LCGL derivative with SLCG_2185 deleted | This study |
| ΔSLCGL_2388 ΔSLCGL_3009 ΔSLCGL_3141 ΔSLCGL_7585 | LCGL derivative with SLCG_2388 deleted LCGL derivative with SLCG_3009 deleted LCGL derivative with SLCG_3141 deleted LCGL derivative with SLCG_7585 deleted | This study This study This study This study |
| LCGL/pIB139 LCGL/pIB139-2185 | LCGL carrying pIB139 LCGL carrying pIB139-2185 | This study This study |
| LA219X LA219X/pIB139-2185 LA219X/pIB139-2185-Lrp | A lincomycin high-yield strain with artificial integrated attBφC31 site LA219X carrying pIB139-2185 LA219X carrying pIB139-2185-Lrp | [23] This study This study |
| Plasmids | ||
| pKC1139 | ori (pSG5), aac(3)IV, lacZ | [24] |
| pKC1139-Δ2185 | pKC1139 derivative containing two 1.8 kb fragments, the upstream and downstream regions of SLCG_2185 | This study |
| pKC1139-Δ2388 | pKC1139 derivative containing two 1.8 kb fragments, the upstream and downstream regions of SLCG_2388 | This study |
| pKC1139-Δ3009 | pKC1139 derivative containing two 1.8 kb fragments, the upstream and downstream regions of SLCG_3009 | This study |
| pKC1139-Δ3141 | pKC1139 derivative containing two 1.8 kb fragments, the upstream and downstream regions of SLCG_3141 | This study |
| pKC1139-Δ7585 | pKC1139 derivative containing two 1.8 kb fragments, the upstream and downstream regions of SLCG_7585 | This study |
| pIB139 pIB139-2185 pIB139-2185-Lrp | φC31 attP-int locus, acc(3)IV, oriT, PermE* pIB139 carrying an extra SLCG_2185 for gene overexpression pIB139 carrying an extra SLCG_2185 and SLCG_Lrp for gene overexpression | [20] This study This study |
| pET28a | T7 promoter, His-tag, kan | Novagen |
| pET28a-Lrp | pET28a-derived plasmid carrying SLCG_Lrp | [20] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, Y.; Xu, W.; Yi, J.; Li, B.; Liu, M.; Zhang, M.; Zheng, Y.; Liu, R.; Wu, H.; Zhang, B. Transcriptomics-Guided Investigation of the SLCG_Lrp Regulon Provides New Insights into Its Role for Lincomycin Biosynthesis. Fermentation 2023, 9, 396. https://doi.org/10.3390/fermentation9040396
Xu Y, Xu W, Yi J, Li B, Liu M, Zhang M, Zheng Y, Liu R, Wu H, Zhang B. Transcriptomics-Guided Investigation of the SLCG_Lrp Regulon Provides New Insights into Its Role for Lincomycin Biosynthesis. Fermentation. 2023; 9(4):396. https://doi.org/10.3390/fermentation9040396
Chicago/Turabian StyleXu, Yurong, Wanlian Xu, Jing Yi, Binglin Li, Meng Liu, Maifei Zhang, Yang Zheng, Ruihua Liu, Hang Wu, and Buchang Zhang. 2023. "Transcriptomics-Guided Investigation of the SLCG_Lrp Regulon Provides New Insights into Its Role for Lincomycin Biosynthesis" Fermentation 9, no. 4: 396. https://doi.org/10.3390/fermentation9040396
APA StyleXu, Y., Xu, W., Yi, J., Li, B., Liu, M., Zhang, M., Zheng, Y., Liu, R., Wu, H., & Zhang, B. (2023). Transcriptomics-Guided Investigation of the SLCG_Lrp Regulon Provides New Insights into Its Role for Lincomycin Biosynthesis. Fermentation, 9(4), 396. https://doi.org/10.3390/fermentation9040396






