Abstract
In recent years, nitrogen pollutants have become one of the main causes of water pollution and eutrophication; thus, it is very important to increase the research on nitrogen removal in wastewater. In this study, a bacterium with outstanding ammonia nitrogen degradation capability was isolated from piggery wastewater and identified as Bacillus tequilensis (designated as A2). The ammonia nitrogen degradation rate of A2 reached the highest level (95%) when the incubation temperature was 42 °C, the initial pH was 7, the seed volume was 5%, the rotation speed was 160 r·min−1, the C/N was 10:1, and the carbon source was sodium citrate. A new nitrite reductase gene was successfully expressed in E. coli BL21 (DE3), and the result showed that the enzyme gene contained 2418 bp and 805 encoding amino acids, the recombinant enzyme was purified through an Ni2+ affinity chromatography column, it had a molecular weight of about 94 kDa, it displayed the maximum enzyme activity at 40 °C and pH 6.0, it exhibited good stability in the range of 25 °C to 35 °C, and it showed a pH of 6.0 to 7.0. A 1 mM concentration of Fe3+ promoted the enzyme activity, followed by a 1 mM concentration of Fe2+ and Mg2+. The kinetic parameters of Km, Kcat, and the Vmax of NiR-A2 were calculated to be 1.37 μmol·mL−1, 4.9 × 102 s−1, and 23.75 μmol·mg−1·min−1, respectively. This strain shows good prospects for wastewater treatment, especially in the treatment of high concentration ammonia nitrogen and nitrite degradation, because of its tolerance to and high degradation rate of high concentrations of ammonia nitrogen and high nitrite.
1. Introduction
In recent years, some small and medium-sized cities have begun discharging aquaculture wastewater with high organic matter, high ammonia nitrogen, excessive odor, and other characteristics, causing serious pollution to surface water, groundwater, etc. The toxicity of ammonia nitrogen in water mainly depends on free ammonia gas. At present, the methods of controlling ammonia gas mainly include physical and chemical management, feed regulation, improved composting, and microbial control [1]. Among these methods, microbial control technology based on nitrification offers the advantages of low cost, high effect, and no secondary pollution [2]; it has been widely used in the field of wastewater treatment. The main form of ammonia gas in wastewater before volatilization is NH4+-N, and the nitrification of microorganisms is an effective way to reduce ammonia volatilization [3], which can convert the NH4+-N in wastewater into nitrate or nitrite and change the dynamic balance between NH4+ and NH3 in wastewater.
There have been many reports on the use of microbial methods to degrade ammonia nitrogen. Klebsiella pneumoniae TN-1 degrades 83.26% of ammonia nitrogen (300 mg·L−1) after being cultured for 7 days [4]. Sphingomonas sp. strain LPN080 can reduce the ammonia nitrogen concentration in aquaculture wastewater from 8 mg·L−1 to 0.3 mg·L−1 within 48 h [5]. Pseudomonas chengduensis BF6 degrades ammonia nitrogen (472 mg·L−1) in the wastewater by 97% within 24 h [6]. Phialemoniopsis curvata TWCC 58054s can almost completely degrade ammonia nitrogen (50 mg·L−1) after 7 days [7]. Bacillus is a type of bacteria that exhibits heterotrophic nitrification–aerobic denitrification [8]: on the one hand, Bacillus degrades ammonia nitrogen by assimilation and converts it into its own nutrients; on the other hand, it can promote water nitrification and convert ammonia nitrogen into nitrate. In addition, Bacillus can secrete large amounts of amylase, protease, and lipase, quickly degrading fish and shrimp residue and excreta, reducing the production of ammonia nitrogen, and solving the problem of high ammonia nitrogen at the source. Nitrite reductase (NiR) acts as a key enzyme in the ammonia nitrogen and nitrite degradation pathway [9], and some types of the nitrite reductase gene (nir) of Bacillus have even been cloned [10,11,12]. So far, there have been many reports of the microbial application of ammonia nitrogen and nitrite degradation in water bodies; however, there is not as much evidence regarding the application of Bacillus to the degradation of ammonia nitrogen and nitrite in water, and even fewer reports on Bacillus in fisheries. According to the reports of Bacillus in the degradation of ammonia nitrogen in water, either the degradation effect of ammonia nitrogen is not ideal, or the production cycle is too long [13,14,15]. Similarly, the application of Bacillus to nitrite degradation in water bodies and in fisheries exhibits the same problem [16,17,18], so it is possible for us to isolate and select one or more types of Bacillus with both ammonia nitrogen and nitrite degradation capacity. In this study, a Bacillus tequilensis A2, with the ability to degrade high concentration ammonia nitrogen, was selected from aquaculture wastewater, and its inhibitory effect on ammonia gas in aquaculture wastewater was studied; moreover, a new NiR (designated as NiR-A2, NCBI gene ID: CP048852.1) from A2 was cloned, expressed, and purified, providing a reference for future theoretical research and practical application.
2. Materials and Methods
2.1. The Source of the Wastewater Sample
The wastewater used in this experiment was obtained from a pig farm in Panyu District, Guangzhou City, Guangdong Province.
2.2. The Main Culture Medium
The Luria–Bertani medium (g·L−1) consisted of agar 15.0, NaCl 10.0, peptone 10.0, and yeast extract 5.0.
The ammonia nitrogen medium (g·L−1) was made up of C6H12O6 5.0, (NH4)2SO4 5.0, K2HPO4 0.5, NaCl 0.85, MgSO4·7H2O 0.25, and pH 7.0.
The trace element solution (g·L−1) was EDTA 50, (NH4)2MoO4 0.05, Fe2(SO4)3 5.0, H3BO3 0.05, CuSO4 1.6, KI 0.01, ZnSO4 2.2, and CoCl2 0.05.
The LB Broth (g·L−1) consisted of peptone 10.0, NaCl 5.0, dextrose 1.0, yeast extract 5.0, and pH 7.0.
The LB Agar (g·L−1) was made up of peptone 10.0, yeast extract 5.0, NaCl 5.0, dextrose 1.0, agar 15.0, and pH 7.0.
2.3. Strains, Plasmids, Reagent, Medium, and Culture Condition
Escherichia coli DH5α, Escherichia coli BL21(DE3), plasmid pUCm-T, plasmid pET-28a(+), Ni-NTA6FF (Pre-Packed Gravity Column, 5 mL), SpeedyCut HindIII, SpeedyCut EcoRI, T4 DNA ligase, and Taq PCR Master Mix (2×, with Blue Dye) were purchased from Sangon Biotech (Shanghai, China); TIANamp Bacteria DNA Kit, TIANprep Mini Plasmid Kit, TIANgel Midi Purification Kit, and TIANquick Midi Purification Kit were provided by TIANGEN (Beijing, China); LB Agar and LB Broth were offer by HuanKai Microbial (Guangzhou, China). All other chemical reagents used in this experiment were of analytical grade. A total of 100 μL of ampicillin (100 mg·mL−1), 200 μL of X-Gal solution (20 mg·mL−1), and 50 μL of isopropyl β-D-1-thiogalactopyranoside (IPTG) (50 mg·mL−1) were added in 100 mL of agar medium for the screening of positive T-A clones (all solutions were sterilized by filtration); LB Broth shake flasks and LB Agar plates were incubated at 37 °C.
2.4. Identification of Strain
2.4.1. Morphology
A2 was cultured on the LB medium plate at 37 °C for 24 h. When a single colony appeared, its shape, size, surface, edge, transparency, colony color, etc. were observed. The morphology of bacteria and spores were observed by Gram staining and spore staining.
2.4.2. Physio-Biochemical Characteristics
Physiological and biochemical experiments, including Gram staining, starch hydrolysis, glucose fermentation, nitrate reduction, (V.P) V.P reaction, citrate utilization, methyl red testing, etc., were conducted according to the Manual for Identification of Common Bacteria Systems.
2.4.3. Molecular Identification
The total genome of the experimental strain was extracted by a bacterial genome DNA extraction kit and used as a PCR amplification template. The 16 S rDNA was amplified by a pair of primers, the upstream primer 27 F: 5′-AGAGTTTGATCCTGGCTCAG-3′ and the downstream primer 1492 R: 5′-TACGGTTACCTTGTTACGACTT-3′. The PCR amplification reaction system (50 μL) consisted of 2 × Easy Taq PCR Super Mix 25 μL, 10 mmol·L−1 upstream primer 1 μL, 10 mmol·L−1 downstream primer 1 μL, template genome DNA 5 μL, and ddH2O 18 μL. The PCR procedure was set as follows: step 1, 94 °C for 4 min; step 2, 30 cycles of 94 °C for 30 s, 54.5 °C for 30 s, and 68 °C for 90 s; step 3, 68 °C for 10 min. The PCR product was detected by 1% agarose gel electrophoresis, recovered from the target stripe, and sequenced. MEGA 5 software was adopted to construct a phylogenetic tree.
2.4.4. In Silico (DDH) Analysis
The relatedness among the genomes was evaluated through in silico DNA–DNA hybridization (DDH) analyses [19], using the server-based genome-to-genome distance calculator (http://ggdc.dsmz.de/distcalc2.php (accessed on 20 October 2022)).
2.4.5. Fatty Acid Analysis
A2 was sent to the Guangdong Provincial Microbial Analysis and Detection Center for analysis of fatty acid components.
2.5. Ammonia Nitrogen Degradation Experiment
A2 was first cultured in LB medium to the logarithmic stage, and the bacterial solution was adjusted to OD600 0.6~0.8 with aseptic water as the seed solution. The seed solution was added to the aquaculture wastewater in a certain proportion, and the resultant wastewater was cultured for 24 h, 48 h, and 72 h. In the single factor experiment, the culture conditions were designed as follows: the carbon sources contained glucose, starch, sodium acetate, mannitol, sodium succinate, and sodium citrate; C/N ratios were set as 0.5, 1.0, 3.0, 5.0, 10.0, 20.0, and 30.0; the inoculum doses were set as 1%, 3%, 5%, 8%, and 10%; the rotary speeds were set as 80, 120, 160, and 200 r·min−1; the temperatures were set as 14, 22, 30, and 38 °C; pH values were set as 5, 6, 7, 8, and 9; the inorganic salts included CuCl2, KCl, NaCl, MgCl2, CaCl2, FeCl3, CoCl2, ZnCl2, and MnCl2, and the concentrations of ammonia nitrogen were set at 100 mg·L−1, 200 mg·L−1, 300 mg·L−1, 400 mg·L−1, and 500 mg·L−1. Based on the results of single factor experiments, the orthogonal experiment was designed.
2.6. Analytical Methods
The content of ammonia nitrogen was determined by Nessler reagent spectrophotometry [20], and the biomass of bacteria was quantified by turbidimetry. All experiments were set with three replicates, and the experimental data were expressed by “mean ± SD”.
2.7. Sequence Analysis and Homology Modeling of NiR-A2
The analysis for the nucleotide sequence of NiR-A2 was conducted using the NCBI ORF Finder tool (ORFfinder Viewer—NCBI). The amino acid sequence of NiR was translated by the online tool EMBOSS Transeq. The theoretical molecular weight (Mw) and isoelectric point (pI) of NiR were predicted by ProtParam. The signal peptide was analyzed by SignalP 4.1. The amino acid sequences of NiR-A2 and other NiRs were searched by the smart blast tool in the NCBI database and aligned using the Clustal Omega tool. The secondary structure of NiR-A2 was analyzed with the SOPMA tool. The three-dimensional structure of the NH2-terminal of NiR-A2 was constructed using SWISS-MODEL server, based on the structure of rubredoxin oxidoreductase from Clostridium acetobutylicum (PDB ID. 3klj.1.A). The rationality of the NH2-terminal structure model was evaluated by the SAVES v6.0 tool, and the Ramachandran plot was drawn using the PROCHECK program. The structure was visualized and analyzed by PyMol software.
2.8. Expression of NiR-A2
The genomic DNA of A2 was extracted according to the specifications of the kit. The nir was obtained by PCR amplification using the genomic DNA as the template and two pairs of cloning primers (5′-ATGGGAAAAAAACAGCTAG-3′ and 5′-TTATGATGTGGTCACGACA-3′). The target gene was inserted into the pUCm-T vector to construct the recombinant plasmid pUCm-T-nir. The pUCm-T-nir was transformed into E. coli DH5α. The positive transformants were selected on the ampicillin-resistant LB plate and further confirmed by enzyme incision and DNA sequencing. The target gene, with two restriction sites, was amplified using pUCm-T-nir (extracted from the positive clone transformants) as the template and two pairs of expression vectors (5′-CGGAATTCATGGGAAAAAAACAGCTAGTTCTTG-3′ and 5′-CCCAAGCTTTCAATGGTGATGGTGATGATGTGATGTGGTCACGACATTTTCAAAT-3′), and then ligated with pET-28 a (+) to construct the expression recombinant vector pET28a (+)-nir. The pET28a (+)-nir was transformed into E. coli BL21(DE3), and the positive transformants were screened and demonstrated by both direct sequencing and restriction endonuclease digestion yield analysis.
The E. coli recombinant BL21(DE3) was grown in LB Broth medium (containing 50 μg·mL−1 kanamycin) at 37 °C and 200 r·min−1 overnight, and then the bacterial fluid was inoculated into the LB broth (containing 50 μg·mL−1 kanamycin) at 37 °C and 200 r·min−1. When the cell biomass (OD600) reached 0.4 up to 0.6, IPTG was added in the medium at a final concentration of 0.8 mM and the cells were cultured overnight.
2.9. Purification and Enzymatic Properties of the Recombinant Enzyme
After IPTG induction and culturing, the cells were collected by centrifugation at 4 °C, then resuspended in lysis buffer (20 mM Tris-HCl, 50 mM NaCl, 1 mM EDTA, pH 8.0) and sonicated on ice. The broken cells were centrifuged at 4 °C, and the precipitate was collected and washed with the inclusion body detergent (20 mM Tris-HCl, 50 mM NaCl, 1 mM EDTA, 1% X-Triton, 2 M Urea, pH 8.0). The inclusion was collected by centrifugation and then dissolved with dissolution buffer (20 mM Tris-HCl, 50 mM NaCl, 1 mM EDTA, 8 M Urea, 20 mM DTT, 5 mM β-mercaptoethanol, pH 8.0).
After the inclusion was dissolved, the resultant solution was purified with Ni-NTA 6FF column affinity chromatography under denaturing conditions [20]. The purification result was visualized using SDS-PAGE gel. The target proteins were collected, and their renaturation was carried out as described in the reference, with slight modifications [21]. The refolded proteins were dialyzed with 50 mM Tris-HCl (pH 7.0) and concentrated by ultrafiltration.
The enzyme activity assay method is described as follows. The assay reaction system (250 μL) consisted of 125 μL phosphate buffer (50 mM, pH 6.5), 15 μL NaCl (0.1 M), 12.5 μL NaNO2 (0.1 M), 7.5 μL methyl viologen (0.1 M), 40 μL Na2S2O4 (0.1 M), and 50 μL Nir-A2; the enzyme reaction was performed at 30 °C for 10 min and terminated by vigorous oscillation, and the nitrite content before and after reaction was determined by spectrophotometry, using the inactivated enzyme instead of the active enzyme as a blank control. The amount of the enzyme required for reducing 1 μmol nitrite per minute was defined as an enzyme viability unit.
The effects of the temperature, pH, metal ion, and EDTA on the enzyme activity were investigated. The kinetic parameters of the enzyme reaction were also measured.
3. Results and Discussion
3.1. Identification of A2
A2 on LB plate medium presented a yellowish, smooth-faced and circular colony morphology, with a tidy margin (Figure 1a). Gram staining was positive (Figure 1b), and spore staining showed that the spores were oval and close to one end of the thallus (Figure 1c).
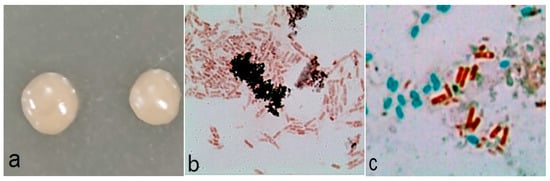
Figure 1.
Morphological characteristics of A2: (a) the colonies of A2; (b) Gram staining; (c) spore staining.
The physiological and biochemical characteristics of A2 were displayed in Table 1. A2 was positive for starch fermentation, sucrose fermentation, glucose fermentation, The physiological and biochemical characteristics of A2 are displayed in Table 1. A2 was positive for starch fermentation, sucrose fermentation, glucose fermentation, D-fructose fermentation, methyl red testing, gelatin hydrolysis, citrate utilization, nitrate reduction, and arginine dihydrolase. It was negative for starch hydrolysis, salicin fermentation, D-mannose fermentation, cellobiose fermentation, indole production, Voges–Proskauer reaction, and catalase. In summary, the characteristics of A2 are consistent with the description for Bacillus tequilensis [22].

Table 1.
The physiological and biochemical characteristics.
The PCR product of 16S rDNA was purified and sequenced, and its sequence size was 1417 bp. The 16S rDNA sequence alignment showed that A2 had the highest homology with Bacillus tequilensis KCTC13622 (accession number: AYTO01000043), and the similarity was 99%. A phylogenetic tree (Figure 2) was constructed based on similar 16S rDNA sequences, which showed that A2 was most closely related to Bacillus tequilensis KCTC13622.

Figure 2.
Phylogenetic analysis using 16S rDNA sequences.
Because in silico DDH analysis has generally been used as a standard method for the delineation of prokaryotic species, in silico DDH value is calculated for the various genomes and used to display the relatedness of the strains. According to reports, DDH technology has been widely used in the identification of microbes, such as the identification of bacteria of asymptomatic periradicular endodontic lesions, the classification of the thermophilic genus Geobacillus and its phylogenomic re-assessment [23], and further identification and taxonomic analysis of the members of the Bacillus subtilis group [24]. The calculated DDH value of A2 and the Bacillus tequilensis genome was 70%, indicating that A2 and Bacillus tequilensis should belong to the same species.
Fatty acid analysis showed that A2 had a near genetic relationship with Bacillus cereus GC subgroup A, followed by Bacillus thuringiensis kurstakii and Bacillus thuringiensis israelensis. Their SIM indexes were 0.777, 0.549, and 0.542, respectively (Table 2 and Figure S1). Each microorganism has its own unique fatty acid intensity profile; through this specific microbial fingerprint profile, Whittaker et al. successfully identified Bacillus anthracis and Bacillus cereus [25]. Combining the above identification results, A2 was finally identified as Bacillus tequilensis.

Table 2.
Fatty acid analysis.
3.2. Optimization of Ammonia Nitrogen Degradation Condition
Many studies have shown that the carbon source is one of the important factors influencing microbial heterotrophic nitrification ability. By controlling the type and concentration of initial organic carbon in the medium, the production and accumulation of ammonia nitrogen can be effectively controlled [26]. Different bacteria have different abilities to degrade ammonia nitrogen when using different types of carbon sources. For Alcaligelies faecalis No.4, only organic acid can be used as a carbon source to degrade ammonia nitrogen [27]; for Bacillus weinstephnisis [28] and Bacillus subtilis Ab03 [13], starch and glucose are the best carbon sources for ammonia nitrogen removal, respectively. In this study, A2 could utilize a variety of carbon sources for growth and metabolism, of which sodium acetate was the best carbon source for ammonia nitrogen degradation, followed by sodium succinate, mannitol, and starch (Figure 3a). The appropriate C/N ratio is of great significance to the efficiency of the bioremediation system; based on microbial metabolic activities, different bacteria require different suitable C/N ratios, generally 6~20 [29]. When the C/N ratio is too low, there is not an adequate electron flow to provide enough energy for strain growth, which causes the strain metabolism to slow down and affects the ammonia nitrogen removal rate, but an excessive C/N ratio can inhibit the growth of bacteria and decrease the degradation rate of ammonia nitrogen [30]. As shown in Figure 3b, whether A2 (3% inoculation volume, v/v) was incubated for 24 h, 48 h, or 72 h in aquaculture wastewater, the most suitable C/N was 0.5, and the highest degradation rate reached 68.9% in 72 h. With the increase in the C/N ratio, the degradation rate showed a downward trend. Inorganic salt is an indispensable substance for microbial life, which mainly functions to constitute the bacterial composition, as the constituent of enzymes, the activator or inhibitor of enzymes, the regulator of the culture medium permeation pressure, the adjustor of the pH value and redox potential, etc. As shown in Figure 3c, CuCl2 inhibited the degradation of ammonia nitrogen. The other eight inorganic salts had no obvious effect on the degradation of ammonia nitrogen at 24 h, but could promote the degradation of ammonia nitrogen to varying degrees at 48 h and 72 h. Among these inorganic salts, KCl and CoCl2 increased the degradation rate by 12.6% and 9.4% at 48 h, and 25.5% and 25.6% at 72 h, respectively. The effect of the initial ammonia nitrogen concentration on the degradation was seen in Figure 3d. The degradation rate decreased with the increase in the initial ammonia nitrogen concentration, whether at 24 h, 48 h, or 72 h. When the concentration of ammonia nitrogen was less than or equal to 200 mg·L−1, the degradation rate of ammonia nitrogen was above 90%. The results showed that A2 could degrade the low and middle concentration of ammonia nitrogen rapidly and completely. When the ammonia nitrogen concentration was 500 mg·L−1, the ammonia nitrogen degradation rate still reached 51.8% at 24 h, 63.6% at 48 h, and 69.2% at 72 h, respectively, indicating that A2 exhibits good tolerance to and a high degradation rate of high concentrations of ammonia nitrogen. The results revealed that A2 showed a high degradation rate for low, medium, and high concentrations of ammonia nitrogen, giving it a wide adaptability and a potential application value in the treatment of wastewater containing ammonia nitrogen.
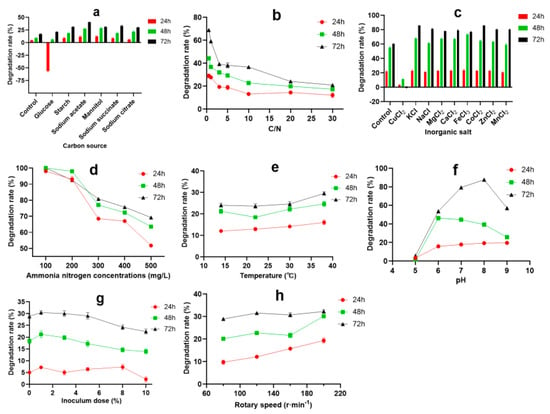
Figure 3.
The effect of different additives in aquaculture wastewater on the degradation of ammonia nitrogen: (a) carbon source; (b) C/N; (c) inorganic salt; (d) initial concentrations of ammonia nitrogen; (e) temperature; (f) pH; (g) inoculum dose; (h) rotary speed.
Temperature is one of the important factors influencing the growth of microorganisms; it influences cell synthesis, mainly through changing the activity of some enzymes, as well as the absorption and utilization of growth substances, thus affecting the growth of microorganisms and the absorption and utilization of growth substances [31]. As shown in Figure 3e, with the increase in culture temperature, the degradation rate of ammonia nitrogen showed an upward trend, as a whole. When the temperature was 38 °C, the degradation rate of A2 in aquaculture wastewater was the highest (29.5%) in 72 h. The initial pH of the culture media affects microbial development, and most species live at a pH 5~9; too high or too low pH is not conducive to cell growth [32,33]. Figure 3f shows that pH values have a great influence on ammonia nitrogen degradation. The degradation rate of ammonia nitrogen increased at first, and then reduced, reaching the maximum (85.3%) at pH 8. The degradation rate of ammonia nitrogen was more than 60% in the range of pH 6 to 9, indicating that A2 shows a good degradation effect on ammonia nitrogen under neutral and alkaline conditions. Whether it was 24 h, 48 h, or 72 h, the degradation rate of ammonia nitrogen was higher when the inoculation amount was in the range of 1% to 5%, of which 1% inoculation amount had the best effect (Figure 3g). Rotary speed reflects the ability of bacteria to obtain oxygen during growth; bacteria will obtain more oxygen at a higher speed, and less oxygen at a lower speed [34]. When the rotation speed was in the range of 80 to 200 r·min−1, the degradation rate of ammonia nitrogen at 24 h and 48 h increased with the increase in the rotation speed, but showed no obvious difference at 72 h (Figure 3h).
The conditions of A2 degrading ammonia nitrogen in aquaculture wastewater were further optimized by orthogonal test, and the results are shown in Table 3.

Table 3.
The orthogonal test result.
The variance analysis indicated that there were differences among carbon sources, C/N, rotation speeds, inoculation doses, and pH values. Their impact on ammonia nitrogen degradation was as follows (from large to small): inoculation dose > pH value > rotating speed > C/N > carbon source. The optimal conditions for degrading ammonia nitrogen were inoculum dose 5%, pH 7, rotary speed 160 r·min−1, C/N 10:1, with sodium citrate as the carbon source. Under the optimal conditions, the actual degradation rate obtained from the validation experiment was 89.9%, which was basically consistent with the results obtained from the orthogonal test.
3.3. Sequence Analysis, Homology Modeling, Expression and Enzyme Characterization of NiR-A2
3.3.1. Sequence Analysis of NiR-A2
The nir of Bacillus tequilensis was successfully amplified by PCR. Its ORF was 2418 bp and encoded 805 amino acids. The theoretical molecular weight (Mw) and isoelectric point (pI) of NiR-A2 were 88.78 kDa and 5.06, respectively. The amino acid sequence of NiR-A2 did not contain a signal peptide. Using the smart blast tool in the NCBI database, five nitrite reductase sequences from Bacillus subtilis subsp. subtilis str.168 (Accession NP_388212.1), Pseudomonas aeruginosa PAO1 (Accession NP_250472.1), Escherichia coli str. K-12 substr.MG1655 (Accession NP_417824.1), Escherichia coli O157:H7 str. Sakai (Accession NP_312243.1), and Streptomyces (Accession WP_011028382.1) were obtained, the amino acid sequences of these NiRs exhibited 100%, 99%, 96%, 96%, and 97% similarity to NiR-A2 (Accession WP_174228931.1), respectively. They were aligned by Clustal Omega, and the results are shown in Figure 4. It could be inferred that the cysteine residues constituted [4Fe-4S] clusters in NiR-A2, and were conserved in Cys418, Cys420, Cys453, Cys456, and Cys482, Cys484, Cys519, Cys522, which is consistent with the reports on some NiRs containing Fe-S motifs [35,36].


Figure 4.
Multiple sequence alignment of NiRs from different strains. Uncharacterized conserved domains are marked with orange boxes, [Fe-S] domains are marked with black boxes, the possible NADH binding sites are marked with red dashed boxes, and the consensus hydrophobic regions are marked by green boxes. ‘*’ indicates positions which have a single, fully conserved residue.
NiR-A2 belonged to the NiRB superfamily (1 aa–800 aa) and contained some uncharacterized domains (marked with orange boxes). NiR-A2 possessed two bacterioferritin-associated ferredoxin (BFD)-like [4Fe-4S] binding domains with “-CXC-” and “-CXXC-” (Figure 4, marked with orange boxes), three possible NADH binding sites (Figure 4, marked with red dashed boxes), and one consensus hydrophobic region (Figure 4, marked with green boxes). These characteristics of NiR-A2 are similar to those of the reported NADH oxidases.
It has been reported that the NADH binding motif is usually located near the N—terminal of the enzyme and exists in the form of -GXGXXG- [37]. According to the research of Ward et al. [38], the NADH binding site is at the amino acid residues of 153–158. Hirano et al. [39] stated that the thermostable H2O2-forming NADH oxidase contains a consensus hydrophobic region and an NADH binding site at the N terminal of the enzyme; the two sites are closely bonded together, and the hydrophobic region is followed by the NADH region. The secondary structure of NiR-A2 included 43.23% alpha-helix (348 aa), 18.51% extended strand (149 aa), 29.94% random coil (241 aa), and 8.32% beta-turn (67 aa).
3.3.2. Homology Modeling for NH2-Terminal Amino Acid Sequence of NiR-A2
The three-dimensional structure (covering residues 2 to 376) of the NH2-terminal of NiR-A2 was constructed, as shown in the Figure 5a. The rationality of the NH2-terminal structure model was evaluated by the Ramachandran plot (Figure 5b). The result of this model revealed that there were 89.1% of total residues (304 aa) in most favored regions, 9.4% residues (32 aa) in additional allowed regions, 0.9% residues (3 aa) in the generously allowed regions, and 0.6% residues (2 aa) in disallowed regions. The percentage of total residues (304 aa) in the most favored regions was close to 90%, which is the standard for a good quality model [40]. The modeled three-dimensional structure contained 12 α-helixes, 17 β-strands, and a few of loops. Moreover, the modeled structure exhibited a conserved three-domain structure, which is used to bind the NAD (P) H and the FAD, the disulfide bond is not involved in the electron transfer process from NADH to rubredoxin [41], and the modeled three-dimensional structure of NiR-A2 almost completely overlapped the NADH: rubredoxin oxidoreductase (Figure 5c); therefore, we speculate that they may have similar functions.
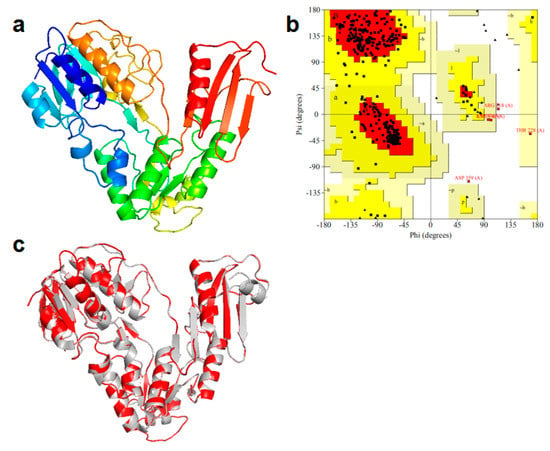
Figure 5.
Homology modeling (NH2-terminal domain) of NiR-A2: (a) the modeled 3D-structure, blue represents the N-terminus, and red represents the C-terminus; (b) the Ramachandran plot of the homology-modeled structure; (c) the overlap between the NADH: rubredoxin oxidoreductase (represented in grey) and the NH2-terminal amino acid sequence of NiR-A2.
3.3.3. Expression and Purification of NiR-A2
The molecular weight of the target protein was estimated to be 88.7 kDa. Because of the addition of 6 × His-Tag, the molecular weight of the recombinant protein was about 90 kDa. The purified recombinant protein was visualized by SDS-PAGE, and it showed a distinct band near the size of 90 kDa (Figure 6a), which is consistent with the inferred molecular weight. The result revealed that the nir gene was successfully expressed in E.coli BL21 (DE3). There have been many reports on microbial NiRs, and their molecular weights are between 20 kDa and 130 kDa; for instance, 127 kDa from Neurospora crassa [42], 120.4 kDa from Hansenula anomala [43], 90 kDa from Rhodobacter capsulatus E1F1 [44], 61.5 kDa from Hydrogenobacter thermophilus TK-6 [45], 55 kDa in soybean nodule [46], 42 kDa from Denitrifying Halophilic Archaeon [47], 35.47 kDa from Geobacillus kaustophilus [6], and 25 kDa from Nitrifier Denitrification [48]. More interestingly, some types of NiRs contain a large and a small subunit, and a portion of these types of NiRs must be in the form of two subunits bound together in order to become catalytically competent [49,50]. Other types of NiR in the form of a single subunit can also undergo the catalysis of chemical reactions [45,47]. The molecular weight of NiR-A2 is close to those of NiRs from Rhodobacter capsulatus E1F1, Bacillus megaterium NCT-2, and Boletus edulis [51], but their amino acid sequences are quite different. Because the recombinant proteins contained six histidine tags, they could be purified by nickel affinity chromatography; the purified proteins were verified by SDS-PAGE (Figure 6b).
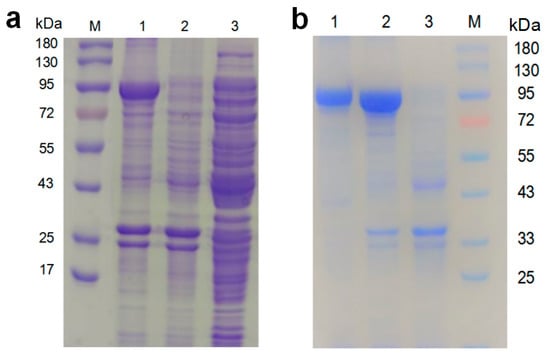
Figure 6.
The expression and purification of the recombinant NiR analyzed by SDS-PAGE: (a) expression of NiR-A2 by the recombinant bacteria. Lane M: molecular mass marker; Lane 1: precipitation of the broken recombinant bacteria (in the presence of IPTG) after centrifugation; Lane 2: precipitation of the broken recombinant bacteria (in the absence of IPTG) after centrifugation; Lane3: suspension of the broken BL21(DE3) harboring pET28a (+) after centrifugation; (b) purification of the recombinant NiR-A2. Lane M: molecular mass marker; Lane 1: purified recombinant NiR-A2; Lane 2: precipitation of the broken recombinant bacteria (in the presence of IPTG) after centrifugation; Lane3: precipitation of the broken BL21(DE3) harboring pET28a (+) after centrifugation.
3.3.4. The Biochemical Characterization of Recombinant NiR-A2
The activities of the recombinant NiR-A2 in the range of 20 °C to 70 °C were investigated. The activity of the recombinant NiR-A2 reached the highest level at 40 °C and maintained a higher level in the range of 30 °C to 50 °C; when the temperature was higher or lower than 40 °C, the enzyme activity presented a downward trend (Figure 7a). According to known reports, the optimum catalytic temperature of NiRs is mostly between 20 °C and 40 °C, i.e., NiRs at 30 °C from Candida utilis, phototrophic bacterium Rhodobacter capsulatus E1F1, Bacillus megaterium NCT-2, and Acidovorax wautersii QZ-4, an NiR at 25~30 °C from Aspergillus oryzae [52], and NiRs at 35 °C from Bacillus cereus LJ01 and Bacillus firmus GY-49, and an NiR at 37 °C from Escherichia coli. Exceptionally, an NiR from Hydrogenobacter thermophilus TK-6 displays the optimum temperature at 70~75 °C.

Figure 7.
The influence of temperature and pH on the activity of NiR-A2: (a) optimum temperature; (b) thermostability; (c) optimum pH; (d) pH stability; (e) metal ions and EDTA; (f) different concentrations of Fe3+.
When the temperature was less than or equal to 35 °C, the purified NiR-A2 was stable, and remained above 80% activity within 60 min and above 50% activity within 120 min (Figure 7b). The activities of the purified NiR-A2 were assayed in the range of pH 4.0 to 10.0. the result indicated that it had the greatest activity at pH 6.0, followed by pH 7.0, and showed a downward trend when the pH was lower or higher than pH 6.0; the activity remained above 65% in the range of pH 5.0 to 9.0, which suggests that NiR-A2 is a neutral enzyme, with a wide pH range (Figure 7c). It has been reported that different types of NiRs have different optimum pH values, and their optimal pH values are mostly between pH 6.0 and pH 9.0 [53,54]. The pH stability tests showed that the purified NiR-A2 exhibited good stability at pH 6.0 and pH 7.0, maintaining more than 75% activity and close to 50% activity, respectively. However, it showed poor stability at pH 5.0 and pH 9.0 (Figure 7d). The effect of different metal ions and EDTA on the enzyme activity is shown in Figure 7e. A total of 1 mM of Fe3+, Fe2+, and Mg2+ significantly increased the enzyme activity by 63.81%, 36.23%, and 12.43%; however, 1 mM of Zn2+, Cu2+ exhibited significant inhibitory effects on the enzyme activity. Except for K+, other ions inhibited enzyme activity when the metal ion concentration was 5 mM. EDTA nearly inactivated the enzyme, implying that NiR-A2 may be a metalloenzyme; there are some reports that NiRs belong to metalloenzyme, such as NiRs from Rhodobacter sphaeroides 2.4.3 [55], Fusarium oxysporum [56], anammox bacterium strain KSU-1 [57], Pseudomonas aureofaciens [58], Sinorhizobium meliloti 2011 [59], Bacillus firmus GY-49 [11], and that Geobacillus kaustophilus [10] belong to the copper-containing type of metalloenzyme. Similarly, NiRs from Pseudomonas aeruginosa [60], Lotus japonicus [61], Orange fifilamentous Beggiatoaceae, Orange Guaymas Basin Beggiatoa spp. [62], and Pseudomonas aeruginosa [63] belong to the iron-containing type. As is shown in Figure 7f, Fe3+ in the range of 0.5 mM to 3 mM obviously enhanced the activity of NiR-A2, and the optimum concentration was 2 mM. Similarly, there have been some reports that Fe3+ can uplift the enzyme activity of NiRs from some different strains, and it is speculated that Fe3+ may be involved in one or more steps of the nitrite reduction process [64,65], or that it may participate in the formation of the active center [66,67]. The Lineweaver–Burk Plot was drawn (Figure S2), and the Km, Kcat, and Vmax of NiR-A2 were calculated to be 1.37 μmol·mL−1, 4.9 × 102 s−1, and 23.75 μmol·mg−1·min−1, respectively. Here, this Km value was close to the gene-type of NiRs reported by Huang et al., Gao et al., Sundermeyer-Klinger et al. [68], Chu et al., and Olmo-Mira et al. Additionally, this Kcat value was similar to the NirK from Bacillus firmus GY-49.
4. Conclusions
A high ammonia nitrogen degrading strain was isolated from aquaculture wastewater and identified as Bacillus tequilensis (A2). Through single-factor optimization and orthogonal tests, we determined the optimal degradation conditions for A2 to treat ammonia nitrogen in aquaculture wastewater, and the experimental results show that the optimal conditions for degrading ammonia nitrogen are inoculum dose 5%, pH 7, rotary speed 160 r·min−1, and C/N 10:1, with sodium citrate as carbon source. Under optimal conditions, the degradation rate reached to 89.9%, and A2 shows high strength ammonia nitrogen tolerance and good wastewater treatment application value. Moreover, a new nitrite reductase gene from A2 was successfully expressed and characterized, and it displays an excellent catalysis capacity for nitrite. Furthermore, A2 provides excellent indigenous species for ammonia nitrogen and nitrite degradation in aquaculture wastewater, which is of great significance for water pollution control.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fermentation9040397/s1, Figure S1: The fatty acid analysis of strain A2; Figure S2: The Lineweaver–Burk Plot.
Author Contributions
This paper was developed based on the collaborative work of the authors. Conceptualization, T.C. and Z.W.; methodology, T.C.; software, Z.W.; validation, Z.W., H.L. and T.C.; formal analysis, Z.W.; investigation, H.L.; resources, H.L.; data curation, Z.W. and H.L.; writing—original draft preparation, Z.W.; writing—review and editing, T.C.; visualization, Z.W.; supervision, T.C.; project administration, T.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data is contained within the article and the Supplementary Material.
Acknowledgments
The authors are thankful to L.X. Luo (South China University of Technology, China) for critically reading and improving the language of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Salim, H.; Patterson, P.; Ricke, S.; Kim, W. Enhancement of microbial nitrification to reduce ammonia emission from poultry manure: A review. World’s Poult. Sci. J. 2014, 70, 839–856. [Google Scholar] [CrossRef]
- Chen, P.; Li, J.; Li, Q.X.; Wang, Y.; Li, S.; Ren, T.; Wang, L. Simultaneous heterotrophic nitrification and aerobic denitrification by bacterium Rhodococcus sp. CPZ24. Bioresour. Technol. 2012, 116, 266–270. [Google Scholar] [CrossRef] [PubMed]
- Sommer, S.; Hutchings, N. Ammonia emission from field applied manure and its reduction—Invited paper. Eur. J. Agron. 2001, 15, 1–15. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, M.; Qu, J.; Zhang, Y.; Liu, H. Characterization and Mechanism Analysis of Tylosin Biodegradation and Simultaneous Ammonia Nitrogen Removal with Strain Klebsiella pneumoniae TN-1. Bioresour. Technol. 2021, 336, 125342. [Google Scholar] [CrossRef]
- Yun, L.; Yu, Z.; Li, Y.; Luo, P.; Jiang, X.; Tian, Y.; Ding, X. Ammonia Nitrogen and Nitrite Removal by a Heterotrophic Sphingomonas Sp. Strain LPN080 and Its Potential Application in Aquaculture. Aquaculture 2019, 500, 477–484. [Google Scholar] [CrossRef]
- Yi, M.; Wang, H.; Ma, X.; Wang, C.; Wang, M.; Liu, Z.; Lu, M.; Cao, J.; Ke, X. Efficient Nitrogen Removal of a Novel Pseudomonas chengduensis Strain BF6 Mainly through Assimilation in the Recirculating Aquaculture Systems. Bioresour. Technol. 2023, 379, 129036. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Xiao, X.; Zhao, Y.; Tu, B.; Zhang, Y. Screening of Efficient Ammonia–Nitrogen Degrading Bacteria and Its Application in Livestock Wastewater. Biomass Convers. Biorefin. 2022, 16, 1–9. [Google Scholar] [CrossRef]
- Zhang, Q.-L.; Liu, Y.; Ai, G.-M.; Miao, L.-L.; Zheng, H.-Y.; Liu, Z.-P. The Characteristics of a Novel Heterotrophic Nitrification-Aerobic Denitrification Bacterium, Bacillus methylotrophicus Strain L7. Bioresour. Technol. 2012, 108, 35–44. [Google Scholar] [CrossRef]
- Canfield, D.E.; Glazer, A.N.; Falkowski, P.G. The Evolution and Future of Earth’s Nitrogen Cycle. Science 2010, 330, 192–196. [Google Scholar] [CrossRef]
- Fukuda, Y.; Tamada, T.; Takami, H.; Suzuki, S.; Inoue, T.; Nojiri, M. Cloning, Expression, Purification, Crystallization and Preliminary X-Ray Crystallographic Study of GK0767, the Copper-Containing Nitrite Reductase from Geobacillus kaustophilus. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011, 67, 692–695. [Google Scholar] [CrossRef]
- Gao, H.; Li, C.; Ramesh, B.; Hu, N. Cloning, Purification and Characterization of Novel Cu-Containing Nitrite Reductase from the Bacillus firmus GY-49. World J. Microbiol. Biotechnol. 2018, 34, 10. [Google Scholar] [CrossRef]
- Huang, Y.; Liang, M.; Zhao, S.; Chen, S.; Liu, J.; Liu, D.; Lu, Y. Isolation, Expression, and Biochemical Characterization: Nitrite Reductase from Bacillus cereus LJ01. RSC Adv. 2020, 10, 37871–37882. [Google Scholar] [CrossRef]
- Wang, Q.; Fu, W.; Lu, R.; Pan, C.; Yi, G.; Zhang, X.; Rao, Z. Characterization of Bacillus subtilis Ab03 for Efficient Ammonia Nitrogen Removal. Syst. Microbiol. Biomanuf. 2022, 2, 580–588. [Google Scholar] [CrossRef]
- Yu, C.-H.; Wang, Y.; Guo, T.; Shen, W.-X.; Gu, M.-X. Isolation and Identification of Ammonia Nitrogen Degradation Strains from Industrial Wastewater. ENG 2012, 04, 790–793. [Google Scholar] [CrossRef]
- Zhang, F.; Xie, F.; Zhou, K.; Zhang, Y.; Zhao, Q.; Song, Z.; Cui, H. Nitrogen Removal Performance of Novel Isolated Bacillus Sp. Capable of Simultaneous Heterotrophic Nitrification and Aerobic Denitrification. Appl. Biochem. Biotechnol. 2022, 194, 3196–3211. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Xie, H.; Yang, E.; Shen, X.; Dai, P.; Zhang, J. Nutrient Removal and Microbial Mechanisms in Constructed Wetland Microcosms Treating High Nitrate/Nitrite Polluted River Water. RSC Adv. 2016, 6, 70848–70854. [Google Scholar] [CrossRef]
- John, E.M.; Krishnapriya, K.; Sankar, T.V. Treatment of Ammonia and Nitrite in Aquaculture Wastewater by an Assembled Bacterial Consortium. Aquaculture 2020, 526, 735390. [Google Scholar] [CrossRef]
- Song, Z.-F.; An, J.; Fu, G.-H.; Yang, X.-L. Isolation and Characterization of an Aerobic Denitrifying Bacillus Sp. YX-6 from Shrimp Culture Ponds. Aquaculture 2011, 319, 188–193. [Google Scholar] [CrossRef]
- Chun, B.H.; Han, D.M.; Kim, K.H.; Jeong, S.E.; Park, D.; Jeon, C.O. Genomic and Metabolic Features of Tetragenococcus halophilus as Revealed by Pan-Genome and Transcriptome Analyses. Food Microbiol. 2019, 83, 36–47. [Google Scholar] [CrossRef]
- Knepp, Z.J.; Ghaner, A.; Root, K.T. Purification and Refolding Protocol for Cold-Active Recombinant Esterase Aa SGNH1 from Aphanizomenon flos-aquae Expressed as Insoluble Inclusion Bodies. Prep. Biochem. Biotechnol. 2022, 52, 394–403. [Google Scholar] [CrossRef]
- Lipničanová, S.; Chmelová, D.; Godány, A.; Ondrejovič, M.; Miertuš, S. Purification of Viral Neuraminidase from Inclusion Bodies Produced by Recombinant Escherichia coli. J. Biotechnol. 2020, 316, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Wintzingerode, F.; Rainey, F.A.; Kroppenstedt, R.M.; Stackebrandt, E. Identification of Environmental Strains of Bacillus Mycoides by Fatty Acid Analysis and Species-Specific 16S RDNA Oligonucleotide Probe. FEMS Microbiol. Ecol. 2006, 24, 201–209. [Google Scholar] [CrossRef]
- Gatti, J.J.; Dobeck, J.M.; Smith, C.; White, R.R.; Socransky, S.S.; Skobe, Z. Bacteria of Asymptomatic Periradicular Endodontic Lesions Identified by DNA-DNA Hybridization. Dent. Traumatol. 2000, 16, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Aliyu, H.; Lebre, P.; Blom, J.; Cowan, D.; De Maayer, P. Phylogenomic Re-Assessment of the Thermophilic Genus Geobacillus. Syst. Appl. Microbiol. 2016, 39, 527–533. [Google Scholar] [CrossRef] [PubMed]
- Whittaker, P.; Fry, F.S.; Curtis, S.K.; Al-Khaldi, S.F.; Mossoba, M.M.; Yurawecz, M.P.; Dunkel, V.C. Use of Fatty Acid Profiles to Identify Food-Borne Bacterial Pathogens and Aerobic Endospore-Forming Bacilli. J. Agric. Food Chem. 2005, 53, 3735–3742. [Google Scholar] [CrossRef]
- Yang, X.; Wang, S.; Zhou, L. Effect of Carbon Source, C/N Ratio, Nitrate and Dissolved Oxygen Concentration on Nitrite and Ammonium Production from Denitrification Process by Pseudomonas stutzeri D6. Bioresour. Technol. 2012, 104, 65–72. [Google Scholar] [CrossRef]
- Joo, H.-S.; Hirai, M.; Shoda, M. Characteristics of Ammonium Removal by Heterotrophic Nitrification-Aerobic Denitrification by Alcaligenes faecalis No. 4. J. Biosci. Bioeng. 2005, 100, 184–191. [Google Scholar] [CrossRef]
- Seenivasagan, R.; Kasimani, R.; Babalola, O.O.; Karthika, A.; Rajakumar, S.; Ayyasamy, P.M. Effect of Various Carbon Source, Temperature and PH on Nitrate Reduction Efficiency in Mineral Salt Medium Enriched with Bacillus weinstephnisis (DS45). Groundw. Sustain. Dev. 2017, 5, 21–27. [Google Scholar] [CrossRef]
- Chen, M.; Wang, W.; Feng, Y.; Zhu, X.; Zhou, H.; Tan, Z.; Li, X. Impact Resistance of Different Factors on Ammonia Removal by Heterotrophic Nitrification–Aerobic Denitrification Bacterium Aeromonas Sp. HN-02. Bioresour. Technol. 2014, 167, 456–461. [Google Scholar] [CrossRef]
- Yang, X.-P.; Wang, S.-M.; Zhang, D.-W.; Zhou, L.-X. Isolation and Nitrogen Removal Characteristics of an Aerobic Heterotrophic Nitrifying–Denitrifying Bacterium, Bacillus subtilis A1. Bioresour. Technol. 2011, 102, 854–862. [Google Scholar] [CrossRef]
- Wang, Y.; Zou, Y.-L.; Chen, H.; Lv, Y.-K. Nitrate Removal Performances of a New Aerobic Denitrifier, Acinetobacter haemolyticus ZYL, Isolated from Domestic Wastewater. Bioprocess Biosyst. Eng. 2021, 44, 391–401. [Google Scholar] [CrossRef]
- Mobarry, B.K.; Wagner, M.; Urbain, V.; Rittmann, B.E.; Stahl, D.A. Phylogenetic Probes for Analyzing Abundance and Spatial Organization of Nitrifying Bacteria. Appl. Environ. Microbiol. 1996, 62, 2156–2162. [Google Scholar] [CrossRef]
- Zumft, W.G. The Biological Role of Nitric Oxide in Bacteria. Arch. Microbiol. 1993, 160, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Fan, X.; Li, J.; Wang, X.; Yuan, Z. Isolation and Identification of Naphthalene Degrading Bacteria and Their Degradation Characteristics under Rainwater Environment in Heavily Polluted Areas. J. Environ. Sci. Health Part A 2021, 56, 434–444. [Google Scholar] [CrossRef] [PubMed]
- Izumi, A.; Schnell, R.; Schneider, G. Crystal Structure of NirD, the Small Subunit of the Nitrite Reductase NirbD from Mycobacterium Tuberculosis at 2.0 Å Resolution: NirD from Mycobacterium tuberculosis. Proteins 2012, 80, 2799–2803. [Google Scholar] [CrossRef] [PubMed]
- Chu, S.; Zhang, D.; Wang, D.; Zhi, Y.; Zhou, P. Heterologous Expression and Biochemical Characterization of Assimilatory Nitrate and Nitrite Reductase Reveals Adaption and Potential of Bacillus megaterium NCT-2 in Secondary Salinization Soil. Int. J. Biol. Macromol. 2017, 101, 1019–1028. [Google Scholar] [CrossRef] [PubMed]
- Yılmaz, H.; Ibici, H.N.; Erdoğan, E.M.; Türedi, Z.; Ergenekon, P.; Özkan, M. Nitrite is reduced by nitrite reductase NirB without small subunit NirD in Escherichia coli. J. Biosci. Bioeng. 2022, 134, 393–398. [Google Scholar] [CrossRef]
- Ward, D.E.; Donnelly, C.J.; Mullendore, M.E.; van der Oost, J.; de Vos, W.M.; Iii, E.J.C. The NADH Oxidase from Pyrococcus furiosus: Implications for the Protection of Anaerobic Hyperthermophiles against Oxidative Stress. Eur. J. Biochem. 2001, 268, 5816–5823. [Google Scholar] [CrossRef]
- Hirano, J.; Miyamoto, K.; Ohta, H. Purification and Characterization of Thermostable H2O2-Forming NADH Oxidase from 2-Phenylethanol-Assimilating Brevibacterium Sp. KU1309. Appl. Microbiol. Biotechnol. 2008, 80, 71. [Google Scholar] [CrossRef]
- Laskowski, R.A.; MacArthur, M.W.; Moss, D.S.; Thornton, J.M. PROCHECK: A Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Crystallogr. 1993, 26, 283–291. [Google Scholar] [CrossRef]
- Nishikawa, K.; Shomura, Y.; Kawasaki, S.; Niimura, Y.; Higuchi, Y. Crystallization and Preliminary X-Ray Analysis of NADH: Rubredoxin Oxidoreductase from Clostridium acetobutylicum. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010, 66, 23–25. [Google Scholar] [CrossRef] [PubMed]
- Colandene, J.D.; Garrett, R.H. Functional Dissection and Site-Directed Mutagenesis of the Structural Gene for NAD(P)H-Nitrite Reductase in Neurospora crassa. J. Biol. Chem. 1996, 271, 24096–24104. [Google Scholar] [CrossRef] [PubMed]
- García-Lugo, P.; González, C.; Perdomo, G.; Brito, N.; Ávila, J.; de la Rosa, J.M.; Siverio, J.M. Cloning, Sequencing, and Expression OfH.a.YNR1 AndH.a.YNI1, Encoding Nitrate and Nitrite Reductases in the Yeast Hansenula anomala. Yeast 2000, 16, 1099–1105. [Google Scholar] [CrossRef]
- Olmo-Mira, M.F.; Cabello, P.; Pino, C.; Martínez-Luque, M.; Richardson, D.J.; Castillo, F.; Roldán, M.D.; Moreno-Vivián, C. Expression and Characterization of the Assimilatory NADH-Nitrite Reductase from the Phototrophic Bacterium Rhodobacter capsulatus E1F1. Arch. Microbiol. 2006, 186, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Hirai, T.; Arai, H.; Ishii, M.; Igarashi, Y. Purification, Characterization, and Gene Cloning of Thermophilic Cytochrome Cd1 Nitrite Reductase from Hydrogenobacter thermophilus TK-6. J. Biosci. Bioeng. 2006, 101, 391–397. [Google Scholar] [CrossRef] [PubMed]
- Hunter, W.J. Purification and Characterization of Soybean Nodule Nitrite Reductase. Physiol. Plant. 1984, 60, 467–472. [Google Scholar] [CrossRef]
- Ichiki, H.; Tanaka, Y.; Mochizuki, K.; Yoshimatsu, K.; Sakurai, T.; Fujiwara, T. Purification, Characterization, and Genetic Analysis of Cu-Containing Dissimilatory Nitrite Reductase from a Denitrifying Halophilic Archaeon, Haloarcula marismortui. J. Bacteriol. 2001, 183, 4149–4156. [Google Scholar] [CrossRef]
- Lawton, T.J.; Bowen, K.E.; Sayavedra-Soto, L.A.; Arp, D.J.; Rosenzweig, A.C. Characterization of a Nitrite Reductase Involved in Nitrifier Denitrification. J. Biol. Chem. 2013, 288, 25575–25583. [Google Scholar] [CrossRef]
- Song, Q.; Wang, B.; Zhao, F.; Han, Y.; Zhou, Z. Expression, Characterization and Molecular Docking of the Assimilatory NaDH-Nitrite Reductase from Acidovorax wautersii QZ-4. Biochem. Eng. J. 2020, 159, 107589. [Google Scholar] [CrossRef]
- Suzuki, S.; Kataoka, K.; Yamaguchi, K. Metal Coordination and Mechanism of Multicopper Nitrite Reductase. Acc. Chem. Res. 2000, 33, 728–735. [Google Scholar] [CrossRef]
- Zhang, W.; Tian, G.; Feng, S.; Wong, J.H.; Zhao, Y.; Chen, X.; Wang, H.; Ng, T.B. Boletus Edulis Nitrite Reductase Reduces Nitrite Content of Pickles and Mitigates Intoxication in Nitrite-Intoxicated Mice. Sci. Rep. 2015, 5, 14907. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, Y.; Zhou, S.; Kim, S.-W.; Fushinobu, S.; Maruyama, J.; Kitamoto, K.; Wakagi, T.; Shoun, H. A Eukaryotic Copper-Containing Nitrite Reductase Derived from a NirK Homolog Gene of Aspergillus oryzae. Biosci. Biotechnol. Biochem. 2010, 74, 984–991. [Google Scholar] [CrossRef] [PubMed]
- Buckley, A.; MacGregor, B.; Teske, A. Identification, Expression and Activity of Candidate Nitrite Reductases From Orange Beggiatoaceae, Guaymas Basin. Front. Microbiol. 2019, 10, 644. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Yang, X.-Y.; Xu, Q.; Cui, H.-L. Characterization of a Novel Cu-Containing Dissimilatory Nitrite Reductase from the Haloarchaeon Halorussus Sp. YCN54. Extremophiles 2020, 24, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, F.; Pistorius, A.; Farkas, D.; De Grip, W.; Hansson, Ö.; Sjölin, L.; Neutze, R. PH Dependence of Copper Geometry, Reduction Potential, and Nitrite Affinity in Nitrite Reductase. J. Biol. Chem. 2007, 282, 6347–6355. [Google Scholar] [CrossRef]
- Matsuoka, M.; Kumar, A.; Muddassar, M.; Matsuyama, A.; Yoshida, M.; Zhang, K.Y.J. Discovery of Fungal Denitrification Inhibitors by Targeting Copper Nitrite Reductase from Fusarium oxysporum. J. Chem. Inf. Model. 2017, 57, 203–213. [Google Scholar] [CrossRef]
- Hira, D.; Toh, H.; Migita, C.T.; Okubo, H.; Nishiyama, T.; Hattori, M.; Furukawa, K.; Fujii, T. Anammox organism KSU-1 Expresses a NirK-Type Copper-Containing Nitrite Reductase Instead of a NirS-Type with Cytochrome Cd1. FEBS Lett. 2012, 586, 1658–1663. [Google Scholar] [CrossRef]
- Gloekner, A.B.; Jiingst, A.; Zumft, W.G. Copper-Containing Nitrite Reductase from Pseudomonas aureofaciens Is Functional in a Mutationally Cytochrome Cdl-Free Background (NirS-) of Pseudomonas stutzeri. Arch. Microbiol. 1993, 160, 18–26. [Google Scholar] [CrossRef]
- Ferroni, F.M.; Guerrero, S.A.; Rizzi, A.C.; Brondino, C.D. Overexpression, Purification, and Biochemical and Spectroscopic Characterization of Copper-Containing Nitrite Reductase from Sinorhizobium meliloti 2011. Study of the Interaction of the Catalytic Copper Center with Nitrite and NO. J. Inorg. Biochem. 2012, 114, 8–14. [Google Scholar] [CrossRef]
- Martí, M.A.; Crespo, A.; Bari, S.E.; Doctorovich, F.A.; Estrin, D.A. QM-MM Study of Nitrite Reduction by Nitrite Reductase of Pseudomonas aeruginosa. J. Phys. Chem. B 2004, 108, 18073–18080. [Google Scholar] [CrossRef]
- Orea, A.; Pajuelo, P.; Pajuelo, E.; Márquez, A.J.; Romero, J.M. Characterisation and Expression Studies of a Root CDNA Encoding for Ferredoxin-Nitrite Reductase from Lotus japonicus. Physiol. Plant. 2001, 113, 193–202. [Google Scholar] [CrossRef] [PubMed]
- MacGregor, B.J.; Biddle, J.F.; Siebert, J.R.; Staunton, E.; Hegg, E.L.; Matthysse, A.G.; Teske, A. Why Orange Guaymas Basin Beggiatoa Spp. Are Orange: Single-Filament-Genome-Enabled Identification of an Abundant Octaheme Cytochrome with Hydroxylamine Oxidase, Hydrazine Oxidase, and Nitrite Reductase Activities. Appl. Environ. Microbiol. 2013, 79, 1183–1190. [Google Scholar] [CrossRef] [PubMed]
- Zennaro, E.; Ciabatti, I.; Cutruzzola, F.; D’Alessandro, R.; Silvestrini, M.C. The Nitrite Reductase Gene of Pseudomonas Aeruginosa: Effect of Growth Conditions on the Expression and Construction of a Mutant by Gene Disruption. FEMS Microbiol. Lett. 1993, 109, 243–250. [Google Scholar] [CrossRef]
- Shahid, S.; Ali, M.; Legaspi-Humiston, D.; Wilcoxen, J.; Pacheco, A.A. A Kinetic Investigation of the Early Steps in Cytochrome c Nitrite Reductase (CcNiR)-Catalyzed Reduction of Nitrite. Biochemistry 2021, 60, 2098–2115. [Google Scholar] [CrossRef] [PubMed]
- Uppal, S.; Khan, M.A.; Kundu, S. Identification and Characterization of a Recombinant Cognate Hemoglobin Reductase from Synechocystis Sp. PCC 6803. Int. J. Biol. Macromol. 2020, 162, 1054–1063. [Google Scholar] [CrossRef]
- Yamazaki, T.; Oyanagi, H.; Fujiwara, T.; Fukumori, Y. Nitrite Reductase from the Magnetotactic Bacterium Magnetospirillum magnetotacticum. A Novel Cytochrome Cd1 with Fe(II): Nitrite Oxidoreductase Activity. Eur. J. Biochem. 1995, 233, 665–671. [Google Scholar] [CrossRef]
- Zeamari, K.; Gerbaud, G.; Grosse, S.; Fourmond, V.; Chaspoul, F.; Biaso, F.; Arnoux, P.; Sabaty, M.; Pignol, D.; Guigliarelli, B.; et al. Tuning the Redox Properties of a [4Fe-4S] Center to Modulate the Activity of Mo-BisPGD Periplasmic Nitrate Reductase. Biochim. Biophys. Acta BBA-Bioenerg. 2019, 1860, 402–413. [Google Scholar] [CrossRef]
- Sundermeyer-Klinger, H.; Meyer, W.; Warninghoff, B.; Bock, E. Membrane-Bound Nitrite Oxidoreductase of Nitrobacter: Evidence for a Nitrate Reductase System. Arch. Microbiol. 1984, 140, 153–158. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).