Abstract
Acid stress is a challenging condition that yeast cells must overcome during fermentation. Enhancing the inherent tolerance of industrial Saccharomyces cerevisiae to organic acid stresses is crucial for increasing fermentation efficiency and reducing economic costs. In a previous study, we constructed a Saccharomyces cerevisiae strain SWY85S with improved tolerance to citric acid stress by modifying the second PEP4-allele. Malic acid is a dominant organic acid in grapefruit, which forms the acidic constituents of wine fermentation mash and finished products. We investigated the malic acid stress tolerance of the strain SWY85S in comparison with that of a strain with one PEP4-allele disrupted and the wild-type strain in this study. Our results revealed that the strain SWY85S demonstrated greater tolerance of malic acid stress, regardless of whether it was cultured with adequate nutrient supplies or under amino acid starvation. Moreover, the strain SWY85S performed remarkably in converting glucose to ethanol during fermentation under malic acid stress. This study provides insights into the role of a vacuolar PEP4-allele coding product in response to environmental stress and the physiological mechanism of yeast to withstand organic acid stress.
1. Introduction
Saccharomyces cerevisiae is commonly used for the fermentation of a variety of industrial products, which includes not only alcoholic beverages and leavened bread but also large-scale ethanol production, such as bioethanol. Generally, the maximum yields of the product can be achieved only when all fermentation conditions are optimal. Yeast cells face multiple environmental stresses simultaneously and sequentially, which can potentially impact cell viability, growth, and fermentation performance [1,2]. These stresses including high osmolarity, fluctuations in dissolved oxygen concentration, increased ethanol concentration, acidic stress, elevated temperature, nutrition starvation and oxidative stresses [2,3,4,5,6,7,8], which can affect cell growth and fermentative behavior, and even potentially hinder cellular metabolism and viability.
Apart from service to industrial product conversions, yeasts utilize various carbon and nitrogen sources to support cell growth and metabolism, leading to the formation of organic acids as byproducts. The optimal pH range for yeast growth is between pH 4.0 and 6.0, which depends on factors such as temperature, the presence of oxygen, and the strain of yeast [9]. The accumulation of organic acids produced during yeast growth and fermentation, along with other environmental stresses, such as sugar osmotic stress, elevated temperature, and high concentrations of ethanol, can cause fermenting cells to face growth inhibition and ultimately limit fermentation rates, or even terminate the fermentation process prematurely [3,10,11]. Normally, when the environmental pH value drops to below 4.0, acidic stress will be formed to affect yeast cells [12]. To better survive and adapt to the ever-changing environment, microorganisms are bound to evolve regulatory strategies to cope with external stresses.
In recent years, research studies have mainly focused on the adaptation and resistance of yeast to acetic acid [13,14,15,16], lactic acid [17,18,19] and weak organic acid-based preservatives [20,21,22]. However, investigations of the effect of intracellular metabolic carboxylic acids that exist in culture and fermentation environments on the growth and metabolism of yeast cells are rarely reported. Malic acid is one of the major organic acids present in industrial fermentation broth [23,24,25], and it is also a potential biomass-derivable “building block” for chemical synthesis in S. cerevisiae as an intermediate in the TCA metabolic cycle (Tricarboxylic acid cycle) [26,27]. Therefore, understanding the factors that affect cellular tolerance to organic acid stress is crucial for the growth of industrial production strains in harsh environments.
L-malic acid is a dominant organic acid in grape must [28] and has a key influence on the fermentation vigor of the yeast in brewing process of yeast and the quality and flavor of the final product, particularly in cold wine-growing regions where grapefruit a has lower respiratory consumption of malic acid at low temperatures. Commercial wine yeast strains of Saccharomyces are not effective in degrading L-malic acid during alcoholic fermentation [28]. High acidity in grape must is helpful in suppressing undesired microbes, and the excess malic acid in grape must can easily be converted to lactic acid by lactobacteria after the main fermentation, which results in a decrease in acidity in winemaking [29]. The use of malic acid-tolerant yeast can effectively reduce the need for acid-reducing agents in cold wine regions and improve the sensory quality of the final product. Therefore, enhancing the tolerance of yeast to malic acid is of utmost importance for winemaking.
Equivalent to the lysosome in higher eukaryotes, the vacuole of S. cerevisiae is involved in several physiological functions, especially proteolysis [30], which is important for the ability of the cell to adapt to changes of environment [31]. Proteolysis is a key regulatory mechanism that is advantageous for controlling protein activity in cells, particularly in response to environmental stress. Eukaryotic cells have evolved various tactics, including selective hydrolysis of specific proteins, to cope with such stress. Hydrolases are the main contributors to intracellular hydrolysis, which can be influenced by changes in proteinase activity [32]. Encoded by PEP4-allele, proteinase A (PrA, EC 3.4.23.25) is a proteolytic enzyme found in vacuoles of S. cerevisiae, and is essential to the S. cerevisiae vacuolar proteolytic system under conditions of nutritional stress, sporulation and vegetative growth, and is implicated in the activities of other hydrolases [33]. Intracellular hydrolysis is primarily governed by the actions of hydrolases, while the variability in proteinase activities gives rise to alterations in intracellular hydrolysis [34]. PrA is also crucial for protein degradation, differentiation, cell survival under stress and expression of other hydrolases [35]. Whilst our comprehension of the structural and functional characteristics of PrA in S. cerevisiae is comprehensive, its involvement in cell physiology remains intricate [36], particularly with respect to its influence on the cell’s response to environmental stresses and changes of its activity under different growing conditions, which are still inadequately understood. In our previous study [32], the PEP4-allele modified strain SWY85S was constructed and showed enhanced tolerance towards environmental citric acid stress. Therefore, we applied L-malic acid stress to investigate the influence of the PEP4-allele on cell tolerance under acid-stressed conditions, and the effects of this PEP4-allele modification on intracellular PrA activity under malic acid stress. The study aimed to further understand the role of primary metabolism in cellular environmental tolerance and to improve the production performance of industrial strains under inevitable stresses.
2. Materials and Methods
2.1. Strains and Culture Conditions
The wild-type industrial S. cerevisiae strain used in this study, SWY85, was kindly provided by Shaoxing Yellow Rice Wine Group Co., Ltd. (Shaoxing, China). Two derived strains were constructed as previously described [32]. Briefly, the strain with one PEP4-allele disrupted was designated SWY85F, and the strain derived from SWY85F with a modified partial ORF (open reading frame) sequence on the second PEP4-allele was designated SWY85S.
Strains were cultured in YPD or SD media, with and without acidification. The cultures were grown in 250 mL glass flasks containing a working volume of 100 mL.
2.2. Chemicals
Bovine hemoglobin was the product of Sigma-Aldrich (St. Louis, MO, USA). L-malic acid of high purity (>99%) was purchased from Hefei BOSF Biotechnology Co., Ltd. (Hefei, China). Folin-phenol was obtained from Shanghai Lida Biological Technology Co., Ltd. (Shanghai, China). Unless otherwise stated, the other chemicals were all of analytical grade and purchased from Hangzhou Kaitai-Biotechnology Co., Ltd. (Hangzhou, China).
2.3. Acidification of Culture Medium
The culture media used for experimental groups were acidified with L-malic acid or hydrochloric acid, while non-acidified culture media were prepared for the control groups, and the corresponding solid media were prepared by adding agar.
2.4. Assay of Cell Growth Inhibitory Rates
An aliquot of yeast cells of each strain on the YPD slope was picked and inoculated into fresh liquid YPD medium to reach the mid-logarithmic phase through shaking it at 180 rpm at 30 °C. The cells were harvested by centrifugation at 5000× g for 3 min and suspended in sterilized distilled water to an OD600 of 0.5. A total of 100 μL of the cell suspensions were then inoculated into the corresponding fresh culture medium (YPD or SD medium, with or without acidification), and cultured at 30 °C and were shaken at 180 rpm. The cell growth inhibitory rates were calculated following the method described previously [32].
2.5. Spot Assay
The overnight growth cells of each strain cultured in YPD liquid medium were collected by centrifugation at 5000× g for 3 min, and subsequently suspended in sterilized distilled water to an OD600 of 6.0, after being rinsed twice. Cell suspensions were 10-fold serially diluted, and 5 µL of each dilution were spotted on the prepared plates. The plates were incubated at 30 °C for 48 to 72 h, and photographed.
2.6. Intracellular PrA Activity Assay
Cell growth and metabolism were rapidly arrested by immersing the culture flasks in crushed ice. Cells were harvested by centrifugation (5000× g, at 4 °C for 3 min), and the intracellular proteinase A (iPrA) enzyme extracts were prepared, followed by measuring iPrA activity using acid-denatured hemoglobin as the substrate [32].
2.7. Measurement of Intracellular Trehalose Accumulation
The cultured yeast cells were quickly chilled with ice-cold sterilized water and then harvested by centrifugation at 5000× g for 3 min at 4 °C, and washed twice with ice-cold sterilized distilled water. The intracellular trehalose accumulation was quantified using the method described by Mahmud et al. [37].
2.8. Anaerobic Fermentations
Strains were pre-cultured in YPD medium until they reached the stationary growth phase before being inoculated or transferred to anaerobic culture media for batch fermentation. The batch anaerobic fermentations were carried out in 250 mL Erlenmeyer flasks containing 100 mL of liquid high-glucose YPD medium. The medium was acidified with L-malic acid or hydrochloric acid to pH 2.7 and pH 2.5, respectively, and also included 1 g yeast extract, 2 g peptone, and 20 g D-glucose. In addition, 100 mL of non-acidified liquid high-glucose YPD medium, with a pH of 6.7, was used, which contained 1 g yeast extract, 2 g peptone, and 20 g D-glucose. Fermentations were carried out at 30 °C and shaken at 120 rpm. A thick silicone rubber stopper, inserted with a polypropylene tube with a loop-trap, was used to seal the mouth of each flask. The air in the headspace of the flask was extracted using a syringe. An equal volume of a glycerol solution (50%, vol/vol) was loaded into the loop-trap of the polypropylene tubes to form a way of releasing the generated CO2, while preventing the dissipation of H2O and entry of air. The fermentation liquor was periodically sampled using syringes, and the residual glucose concentration in fermentation liquor was measured using an enzymatic assay (GOD-POD kit, NZYTech, Lisboa, Portugal). The ethanol yields were measured using an enzymatic method, using an ethanol assay kit (Abcam, Cambridge, MA, USA) according to the manufacturer’s instructions.
2.9. Statistical Analysis
The data were represented as mean±standard deviation of at least three triplicates as data points. An ANOVA and a t-test were performed to determine statistical significance. The results were considered statistically significant at a p-value of less than 0.05 (* p < 0.05, ** p < 0.01, *** p < 0.001, and **** p < 0.0001). Graphs were generated using GraphPad Prism 8.3.0 (GraphPad software LLC, Dotmatics, San Diego, CA, USA) and OriginPro 2021 software (OriginLab Corporation, Northampton, MA, USA). Asterisks were used to indicate statistically significant differences in the figures.
3. Results
3.1. The Second PEP4-Allele Modification Facilitates Cell Growth under Malic Acid Stress
When cultured on YPD agar, with or without acidification with hydrochloric acid (HA) at 30 °C, for the same duration, the colony size of strain SWY85S was smaller than that of the other two strains (Figure 1a–c). However, when cultured in a liquid YPD medium, with or without acidification using hydrochloric acid, the biomass accumulation of strain SWY85S during the mid-late logarithmic phase was significantly higher than that of the strain SWY85F (Figure 1a). The disruption of one PEP4-allele decreased the biomass accumulation of strain SWY85 when cells were cultured to the mid-late logarithmic phase in the liquid YPD medium acidified with hydrochloric acid (Figure 1a). Compared with the other two strains, SWY85S accumulated the highest biomass at pH 3.0 and pH 2.7. Furthermore, hydrochloric acid acidification reduced the biomass accumulation of all three strains during the cultivation period. The stronger acidification (from pH 3.0 to pH 2.7) further reduced the biomass accumulation of the strains SWY85 (from OD600 of 0.5815 to 0.5071, p < 0.005) and SWY85F (from OD600 of 0.5268 to 0.3927, p < 0.001), but not that of the strain SWY85S. Therefore, we speculated that although hydrochloric acid acidification reduced the biomass accumulation of the three strains, the second PEP4-allele modification may be conducive to cellular growth under acidic stress, in contrast to the PEP4-allele disruption.
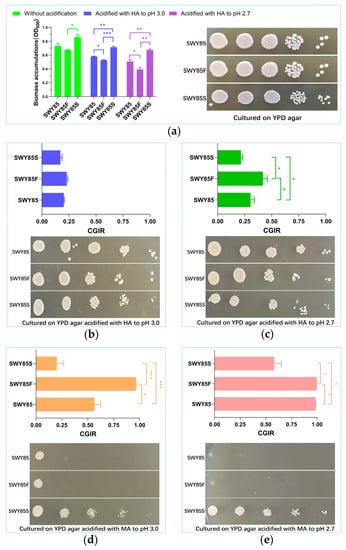
Figure 1.
The effect of malic acid stress on cellular growth of the three strains. The colonies formed on YPD agar and the biomass accumulation of the three strains cultivated in YPD medium with and without acid acidification (a). The colonies formed on acidified YPD agar and the cell growth inhibitory rates (CGIR) of the three strains cultured with an adequate nutrient supply under hydrochloric acid acidification to pH 3.0 (b) and pH 2.7 (c), respectively, and with malic acid stress acidified to pH 3.0 (d) and pH 2.7 (e), respectively. HA, Hydrochloric acid; MA, Malic acid. * p < 0.05, ** p < 0.01, *** p < 0.001.
In order to investigate the effects of disrupting one PEP4-allele and modifying the second PEP4-allele on cellular growth in industrial S. cerevisiae under malic acid stress, the three strains were cultured in YPD medium acidified with L-malic acid and hydrochloric acid, respectively, and the ones cultured without acidification were used as control groups. These control groups were used for calculating the cell growth inhibitory rates (CGIRs). As shown in Figure 1b, no significant difference in CGIRs was found among the three strains when the YPD medium was acidified with hydrochloric acid to pH 3.0. When this acidification was further increased to pH 2.7, the CGIRs of the three strains all showed significant differences from each other, with the CGIR of the strain SWY85F being higher than that of the other two strains, while the strain SWY85S had the lowest CGIR among the three strains (Figure 1c). Moreover, the CGIRs of strains SWY85 and SWY85F were significantly higher when cultured in YPD medium under malic acid acidification compared to hydrochloric acid acidification at the same pH values (p < 0.0001, Figure 1b–e). Notably, the CGIRs of cultivations under acidification with malic acid to pH 3.0 were even higher than those under acidification with hydrochloric acid to pH 2.7 in YPD medium (p < 0.0001, Figure 1c,d). However, the CGIR of strain SWY85S showed no significant change as the acidification with hydrochloric acid increased from pH 3.0 to pH 2.7 (Figure 1b,c). The comparison of the CGIRs caused by malic acid and hydrochloric acid acidification at the same pH value suggested that the cellular growth inhibition effect of malic acid stress comes from the cooperation of protons and malate anions, and that the effect of malate anions was stronger than that of protons. In this study, the tolerance of the industrial strain S. cerevisiae SWY85 to malic acid stress was decreased by disrupting one PEP4-allele (p < 0.05), but was increased by modifying the second PEP4-allele (p < 0.05). As the malic acid acidification was increased from pH 3.0 to pH 2.7, the CGIRs of the strains SWY85 and SWY85F increased to 98.67% and 99.59%, respectively, while the CGIR of the strain SWY85S (58.19%) was significantly lower among the three strains (Figure 1e). Over the same cultivation period under malic acid stress on YPD agar, the strain SWY85S formed more colonies than the strains SWY85 and SWY85F (as shown in Figure 1d,e).
In the current study, we also investigated the effects of PEP4-allele alterations on the tolerance of the strain to malic acid stress under amino acid starvation. It has been recognized for over 30 years that PrA plays a vital role in cells under nutrient stress, especially under nitrogen deficiency, and initiates the activation of a series of vacuolar enzymes [35]. To investigate the impact of pH stress on cell growth, SD culture media were used, and the inhibitory effect of pH stress on cell growth was analyzed. As shown in Figure 2b,c, the inhibitory effect of pH stress on cell growth of the three strains increased as the proton concentration increased (from pH 3.0 to pH 2.7, p < 0.001). However, the CGIR of the second PEP4-allele modified strain caused by the increase in proton concentration was lower than that of the strain with one PEP4-allele disruption (p < 0.05). In addition, the CGIR of the strain SWY85S was significantly lower than that of the strains SWY85 and SWY85F when cultured under acidification with malic acid to pH 3.0 (Figure 2d). However, one PEP4-allele disruption did not significantly impair the cellular tolerance of the strain SWY85 to this intensity of malic acid stress. As the malic acid acidification was increased to pH 2.7, the CGIRs of the three strains showed differences, with the CGIR of strains SWY85, SWY85F and SWY85S being 91.21%, 94.33% and 65.17%, respectively (Figure 2e).
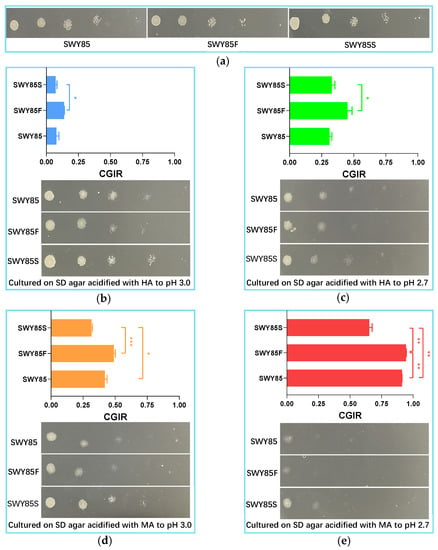
Figure 2.
The inhibitory effect of malic acid stress on cell growth of the three strains under amino acid starvation. The growth status of the strains on SD agar at natural pH (a), and the colonies formed on acidified SD agar and the cell growth inhibitory rates (CGIR) of the three strains cultured with amino acid starvation under acidification with hydrochloric acid to pH 3.0 (b) and pH 2.7 (c), respectively, and under acidification with malic acid to pH 3.0 (d) and pH 2.7 (e), respectively. HA, Hydrochloric acid; MA, Malic acid. * p < 0.05, ** p < 0.01, *** p < 0.001.
Consistent with nutrient-adequate culture conditions, under amino acid starvation, the strength of malic acid anions was the major contributor to the cell growth inhibitory effect on the three strains (p < 0.001).
3.2. The Second PEP4-Allele Modification Stabilized Intracellular PrA Activity under Malate Anions
Based on the aforementioned different malic acid stress resistance between the wild-type and two mutants, we inferred that the cellular tolerance of strain SWY85 against malic acid stress was regulated by the alterations in PEP4 alleles. Therefore, it is necessary to investigate the influence of environmental malic acid stress on the activity of the altered PEP4-allele coding product in the strain. To accomplish this, we inoculated and cultured the three strains according to the cell growth tolerance experiments. For the cultivations with a CGIR greater than 90%, cells were cultured in non-acidified medium for the same period as the cell growth tolerance experiments. Afterwards, the cells were transferred into the acidified culture media and cultivated for 2 h. Finally, crude iPrA enzyme extracts were prepared for analysis.
Cultivations in YPD medium showed that PEP4-allele disruption significantly reduced the iPrA activity of the strain SWY85 (p < 0.05), and this reduction was further extended by the second PEP4-allele modification (p < 0.01) (Figure 3a). When cultivated with adequate nutrients under acidification with malic acid at pH 3.0, the iPrA activity of all three strains increased (by 28.91% for SWY85, by 38.78% for SWY85F, and by 40.75% for SWY85S, Figure 3a). The strengthening acidification (from pH 3.0 to pH 2.7) reduced the iPrA activity of all three strains (under acidification with hydrochloric acid, the reductions in iPrA activity of strains SWY85, SWY85F, and SWY85S were 31.60%, 19.00%, and 49.48%, respectively, p < 0.05; under acidification with malic acid, the reductions in iPrA activity of strains SWY85, SWY85F, and SWY85S were 11.36%, 35.57%, and 84.01%, respectively, p < 0.001, Figure 3a). It was also observed that when cultured in YPD medium at pH 2.7, the iPrA activity of the strain SWY85S showed no significant difference between cultivations under malic acid stress and hydrochloric acid stress (Figure 3a).
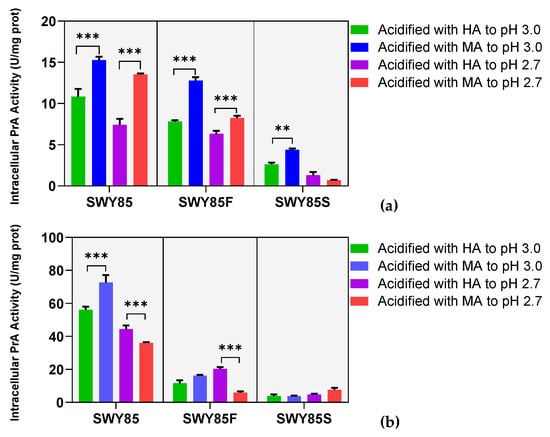
Figure 3.
The iPrA activity of the three strains. Cells were cultured under acid stresses with adequate nutrients (a) and amino acid starvation (b). HA, Hydrochloric acid; MA, Malic acid. ** p < 0.01, *** p < 0.001.
When cultured at pH 3.0, amino acid starvation increased the iPrA activity of the wild-type and strain SWY85F (by 417.72% for SWY85 with p < 0.001, and by 47.41% for SWY85F with p < 0.05), while under malic acid acidification at pH 3.0, it had no significant effect on the iPrA activity of strains SWY85F and SWY85S (p > 0.05), but increased that of strain SWY85 (by 376.02%, p < 0.001). In other words, among the three strains, under amino acid starvation at pH 3.0, the disruption of one PEP4-allele and the modification of the second PEP4-allele both stabilized the iPrA activity under malic acid stress, while the iPrA activity of the wild-type strain was reduced by this strength of malic acid stress (by 20.78% with p < 0.001) (Figure 3b). In addition, the iPrA activity of strains SWY85 and SWY85F was reduced by the strengthening of malic acid stress (from pH 3.0 to pH 2.7), whether cultured under amino acid starvation or with adequate nutrients. Interestingly, at pH 2.7, malate anions significantly attenuated the facilitation effect of amino acid starvation on the iPrA activity of the wild-type and strain SWY85F (p < 0.01). This resulted in the lower iPrA activity of strains SWY85 and SWY85F under malic acid stress compared to hydrochloric acid stress (Figure 3b). However, the iPrA activity of strain SWY85S was not significantly affected by malate anions when cultivated under amino acid starvation (Figure 3b). In summary, the second PEP4-allele modification stabilized the iPrA activity of cells exposed to malate anions under the conditions of amino acid starvation (at both pH 3.0 and pH 2.7) or adequate nutrient supplies (at pH 2.7). It is clear that further research investigating the correlation between iPrA activity and cellular growth tolerance to malic acid stress could provide insights into the regulatory mechanisms of the cellular response to environmental stresses.
3.3. Opposing Effects on Intracellular Trehalose Accumulation between the PEP4-Allele Modification and Disruption
The intracellular trehalose content of three strains cultured under different conditions was assayed and the results are presented in Figure 4 and Figure 5. Among the cultivations, cultured without or with hydrochloric acid acidification to pH 3.0 and pH 2.7, the intracellular trehalose content of strain SWY85S was significantly higher than that of the other two strains. One PEP4-allele disruption significantly reduced the intracellular trehalose content accumulation of cells cultured in non-acidified YPD medium and SD medium acidified with hydrochloric acid to pH 2.7. Under malic acid stress at pH 3.0 in YPD medium or in SD medium, the intracellular trehalose accumulation of strain SWY85 was not affected by the one PEP4-allele disruption but was significantly promoted by the second PEP4-allele modification (Figure 4a and Figure 5a).
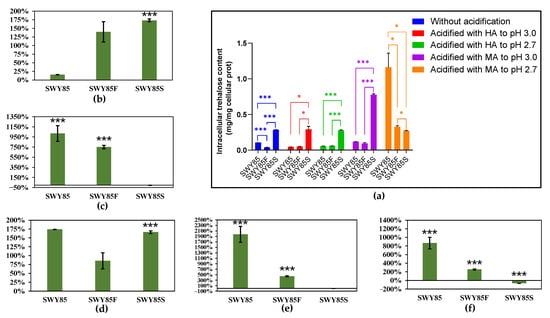
Figure 4.
The intracellular trehalose accumulations of the three strains in YPD medium (a), and the variations of intracellular trehalose content in the cultivations of strains cultured with adequate nutrients between different culture conditions (from non-acidification to the acid stress of pH 3.0 (b) and to the stress of pH 2.7 (c) acidified with malic acid, from the acid stress acidified with hydrochloric acid to the acid stress acidified with malic acid at pH 3.0 (d) and at pH 2.7 (e), and from the acid stress of pH 3.0 acidified with malic acid to the strengthened malic acid stress of pH 2.7 (f), respectively.) HA, Hydrochloric acid; MA, Malic acid. * p < 0.05, *** p < 0.001.
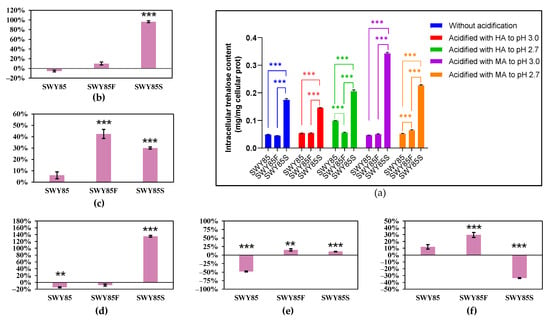
Figure 5.
The accumulated intracellular trehalose in the wild-type strain and the derivatives cultured under different acid stresses combined with amino acid starvation (a), and the variations of intracellular trehalose content in three strains cultured in SD medium between different acid stresses (from non-acidification to acidification with malic acid to pH 3.0 (b) and to pH 2.7(c), from acidification with malic acid to acidification with hydrochloric acid at pH 3.0 (d) and at pH 2.7 (e), and from malic acid stress at pH 3.0 to malic acid stress at pH 2.7 (f), respectively.) HA, Hydrochloric acid; MA, Malic acid. ** p < 0.01, *** p < 0.001.
The impact of malic acid stress on intracellular trehalose accumulation of the three strains was assessed. The results showed that when cultivated in YPD medium at pH 3.0, neither the malate ions nor the combined stress of malate anions and protons altered the trehalose accumulation in the strains SWY85 and SWY85F, but both conditions increased that of the strain SWY85S (by 37.57% and by 173.33, respectively) (Figure 4b,d). Compared to the cultivations cultured without acidification, the acidification of YPD culture medium with malic acid to pH 2.7 increased the intracellular trehalose accumulation of the strains SWY85 and SWY85F (by 1018.53% and by 750.89%, respectively) (Figure 4c). The malate anions at pH 2.7 also increased the accumulated intracellular trehalose of the strains SWY85 and SWY85F (by 1973.19% and by 441.69%, respectively) cultured in YPD medium (Figure 4e). However, malic acid acidification (to pH 2.7) of the YPD culture medium showed no significant effect on the intracellular trehalose accumulation of strain SWY85S (Figure 4c,e).
During amino acid starvation at pH 3.0, malate anions decreased the intracellular trehalose content of the wild-type strain SWY85 by 14.54%, while increasing it by 135.28% in the strain SWY85S (Figure 5d). When combined with proton stress, malate anions had an even greater effect on intracellular trehalose accumulation in strain SWY85S (increasing it by 96.25%) (Figure 5b). In SD culture medium, the intracellular trehalose accumulation of strains SWY85F and SWY85S was increased by malic acid acidification at pH 2.7 (by 42.48% and 30.06%, respectively) compared to cultivations without acidification (Figure 5c). Compared to hydrochloric acid acidification at pH 2.7, the increase in intracellular trehalose accumulation content was 15.30% and 10.40%, respectively (Figure 5e). Furthermore, Figure 5c and e demonstrate that proton stress under amino acid starvation at pH 2.7 significantly increased the intracellular trehalose content of the wild-type strain SWY85 by 53.96%.
Comparing the intracellular trehalose content in the three strains under the two strengths of malic acid stress (pH 3.0 and pH 2.7) revealed that further acidification with malic acid increased the intracellular trehalose accumulation in the strains SWY85 and SWY85F (by 868.26% and 254.88%, respectively), but decreased it in strain SWY85S (by 64.81%) when the cells were cultured with sufficient nutrients (Figure 4f). However, this strengthening of malic acid stress had no significant impact on the intracellular trehalose accumulation in the wild-type strain SWY85, but increased it in strain SWY85F (by 29.59%) and decreased it in strain SWY85S (by 33.73%) when the cells were cultivated under amino acid starvation (Figure 5f). When cultured with sufficient nutrients, the intracellular trehalose content in the wild-type strain and strain SWY85F was increased by 868.26% and 254.88%, respectively (Figure 4f), but was decreased in strain SWY85S by 64.81% (Figure 4f). These results suggest that another factor, in addition to trehalose content, which was caused by the second PEP4-allele modification, may contribute to the enhanced malic acid stress resistance in the strain.
3.4. The PEP4-Allele Modification Maintains Anaerobic Glucose Consumption under Malic Acid Stress
To assess the influence of PEP4-allele alterations on the anaerobic fermentative performance of the strain under malic acid stress, a high-glucose YPD fermentation medium (containing 20% glucose) with and without acidification was prepared and fermented with the three strains anaerobically and shaken at 120 rpm until all fermentations ceased. Yeast fermentation is a process that leads to continuous acidification of the culture medium. Especially in industrial production, the acid stress faced by yeast cells during fermentation is stronger than that during cell growth. Compared with the medium used in the cell growth experiments, the acid stress of the fermentation medium was further strengthened to pH 2.7 and pH 2.5, respectively. Considering that during the fermentation process, acid stress will affect the growth-related metabolism and ethanol production-related metabolism of cells, two methods, namely CIFs (cell-inoculation fermentations) and CTFs (cell-transferring fermentations), were used in the anaerobic fermentation experiments.
As indicated in Figure 6a,b, under the stress of malate anions at pH 2.7, the glucose consumption ratios (GCRs) of strain SWY85S were higher than those of the other two strains after 60 h of culturing in CIFs (60 h–84 h, p < 0.05; 96 h–228 h, p < 0.001). However, no significant differences were observed among the three strains in CTFs (p > 0.05) (Figure 6c,d). We inferred that a positive regulation of the second PEP4-allele modification on the glucose consumption is associated with cell growth-metabolism under malic acid stress at pH 2.7. As the concentration of hydrogen ions increased (to pH 2.5), after 72 h culturing, the GCRs of the strain SWY85S were higher than those of the other two strains in CIFs (p < 0.05) and in CTFs (between SWY85 and SWY85S, p < 0.001; between SWY85F and SWY85S, p < 0.05), whereas one PEP4-allele disruption had no effect on the GCRs of the wild-type strain (Figure 6e,g). Surprisingly, under malic acid stress of pH 2.5, one PEP4-allele disruption boosted the glucose consumption (p < 0.05) of the wild-type strain, whether in CIFs (after culturing for 108 h) or in CTFs (after culturing for 48 h) (Figure 6f,h).
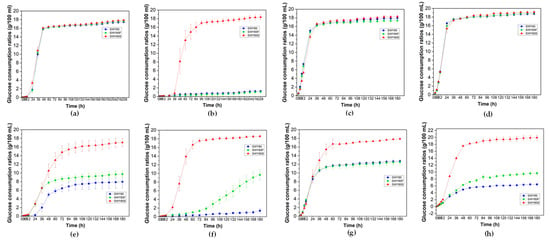
Figure 6.
The glucose consumption ratios of the three strains in high-glucose YPD medium acidified with hydrochloric acid (HA) to pH 2.7 (a,c) and pH 2.5 (e,g), and acidified with malic acid (MA) to pH 2.7 (b,d) and pH 2.5 (f,h) during anaerobic fermentation. Panels (a,b,e,f): cell-inoculation fermentations (CIFs), cells in the stationary growth phase were collected from 100 μL of culture medium, and inoculated into the fermentation medium. Panels (c,d,g,h): cell-transferring fermentations (CTFs), 363 mg of wet cells in the stationary growth phase were transferred into the fermentation medium.
In this study, we observed no inhibitory effect of malic acid stress on the glucose consumption performance of strain SWY85S at pH 2.7 and pH 2.5. As shown in Figure 7, disruption of one PEP4-allele did not affect the final ethanol content produced by the strain SWY85 at different hydrogen ion concentrations (at pH 2.7 and at pH 2.5, p > 0.05; not shown for natural pH). The decrease in pH of the culture medium from pH 2.7 to pH 2.5 inhibited ethanol production in the strains SWY85 and SWY85F (p < 0.0001), but not in the strain SWY85S (p > 0.05). Under malic acid stress of pH 2.5, the final alcohol content produced by the strains SWY85 and SWY85F was lower than that produced by the strain SWY85S (p < 0.0001). In addition, the strain SWY85F produced a higher alcohol content than the wild-type strain (Figure 7c,d). We found no significant inhibition of ethanol production in the strain SWY85S under malic acid stress (acidified to pH 2.7 and pH 2.5, p > 0.05) in both CIFs and CTFs. Notably, the strengthening of malic acid stress (from pH 2.7 to pH 2.5) significantly increased the alcohol production by the strain SWY85F in CIFs (p < 0.0001, Figure 7c). During ethanol fermentation, yeast cells have to confront a combined environmental stress condition composed of acidic stress, hyperosmolarity, oxidative stress, ethanol toxicity, anaerobiosis and nutritional deficiencies [3,38,39,40]. Enhancing the tolerance of S. cerevisiae against multiple stresses is, accordingly, essential for reducing production costs and improving production efficiency [41].
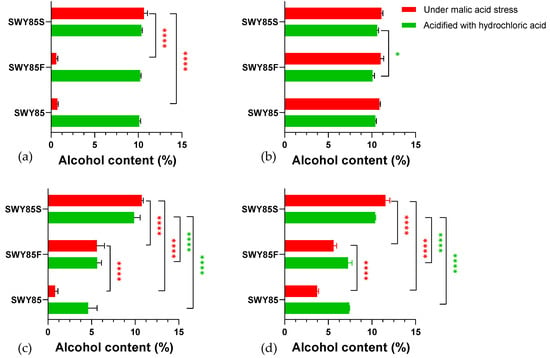
Figure 7.
The final alcohol content in the fermented YPD media, under acid stresses at pH 2.7 (a,b) and pH 2.5 (c,d), respectively. Panel (a,c), cell-inoculation fermentations (CIFs). Panels (b,d), cell-transferring fermentations (CTFs). * p < 0.05, **** p < 0.0001.
4. Discussion
In S. cerevisiae, proteases have been proven to be involved in a wide range of cellular activities, and are crucial requisites for cell survival and metabolism [42]. Among these proteases, proteinase A is believed to play a key role in the activation of other vacuolar zymogens. In the early years, most of the genetic dissection of proteases and protease function concentrated on vacuolar enzymes [42]. Studies have investigated the structure of the S. cerevisiae PEP4 gene and the biological functions of its encoded product on cells [33,43,44,45,46,47].
The insertion of an extraneous DNA fragment into the coding sequence is a powerful technique for reliably eliminating gene function. This technique most often disrupts the ORF and may also replace it entirely [48]. Pleiotropic mutations can affect properties such as levels, electrophoretic mobility, and the processing of PrA [42]. PEP4-allele disruption has been widely used to ameliorate fermentation, improve product quality, or increase the production of a given heterologous protein [49,50,51,52,53,54]. Alugoju et al. [55] reported that quercetin pretreatment alleviated the high sensitivity of yeast pep4 mutant cells to oxidative and apoptotic stress induced by hydrogen peroxide and acetic acid. Chen et al. [56] reported a variant of S. cerevisiae pep4 strain with improved oligotrophic proliferation and cell survival. In our previous study [32], we disrupted one PEP4-allele of the industrial S. cerevisiae strain SWY85. To investigate the phenotype of the strain with vestigial PEP4-allele, we replaced the partial coding sequence in the ORF of another PEP4-allele with the KanMX sequence, resulting in the derived strain with enhanced citric acid stress tolerance during cell growth.
S.cerevisiae cell growth is frequently inhibited by weak acids that contain the organic acids produced during fermentation process [57]. Previous studies have attempted to understand the inhibitory mechanism of some organic acids on the growth of S. cerevisiae [58,59,60]. However, there have been few reports on the relationship between PEP4-allele and yeast cellular tolerance against environmental stress formed with intermediates in the TCA cycle pathway, such as malic acid. The inhibitory effect of malic acid stress on cell growth was found to be stronger than that formed by hydrogen ions at the same pH value (see Figure 1 and Figure 2). Comparisons of cellular tolerance of the three strains against hydrochloric acidification and malic acid stress during cultivation suggested that the second PEP4-allele modification significantly enhanced the malic acid stress tolerance of the S. cerevisiae strain SWY85. With the same pH values, disruption of one PEP4-allele caused increased sensitivity to malic acid stress, as cells cultivated with sufficient nutrients (Figure 1 and Figure 2).
The results of cell growth performance in cultivations indicate that the tolerance of the strain to malic acid stress is correlated with the alterations in the PrA-coding alleles. During cultivation under malic acid stress, iPrA activity may be regulated by the acidification of the culturing environment, which, in turn, affects the cell growth tolerance of strain. The results of iPrA activity assays suggest that the iPrA activity of strain SWY85 was affected by the nutritional status of the culture medium. The increase of iPrA activity under amino acid-limited culture conditions may be an adaptive strategy of cells to cope with nutritional insufficiency in the environment. Regarding the cultivations of the strain with one PEP4-allele disrupted and the wild-type strain, when malic acid acidification was strengthened to pH 2.7 under amino acid starvation, the iPrA activity of these two strains decreased to a level lower than that of the cultivations at the same pH value without malate anions. This result indicates that the decrease in iPrA activity is a specific adaptation of S. cerevisiae to unfavorable nutrient environments [36] and arduous conditions for survival. The strain with one PEP4-allele disrupted showed no decrease in iPrA activity as cells faced the increased hydrogen ion concentrations (Figure 3). We speculated that in the strain SWY85F, the level of iPrA activity expressed by the remaining PEP4-allele was reduced to a level to respond to adverse conditions compared to the wild-type strain. However, this level of iPrA activity was not low enough to enable the cell to tolerate the environmental stress.
As a non-reducing disaccharide, trehalose is thought to be one of the most important molecules for protecting S. cerevisiae against environmental stresses, and the trehalose content is considered a major determinant of stress resistance [61,62,63]. Modification of the second PEP4-allele promoted the accumulation of intracellular trehalose in cells cultured with sufficient nutrients under malic acid stress (at pH 3.0, Figure 4) and in cells cultured under malic acid stress (at pH 3.0 and pH 2.7, Figure 5) with amino acid starvation. However, when cultured in YPD medium acidified with malic acid to pH 2.7, the accumulated trehalose content was positively correlated with the number of intact PEP4 alleles in the strain. Thus, in cells cultured under malic acid acidification with sufficient-nutrient supply (at pH 2.7), trehalose may not be the only molecule that supports cells in tolerating malic acid stress. In addition, the intracellular protein content of the three strains cultivated under different conditions was compared, and the results indicated that the modification of the second PEP4-allele affects the intracellular protein content of the strain cultured under acid stress [32]. Furthermore, some protein molecules regulated by the modified PEP4-allele may play an essential role in the strain’s resistance to the acid stress. However, the mechanism responsible for the large increase in trehalose content in cells of the wild-type strain cultured with sufficient nutrient supply under the strengthened malic acid stress (pH 2.7) requires further investigation.
In the fermentation process, when the ethanol concentration is high, yeast experiences growth inhibition and the loss of cell viability [64]. The cell membrane has been suggested as the primary target site of ethanol toxicity [64]. Based on the foregoing discussion, the modification of the second PEP4-allele may play two roles in explaining the increased ethanol production performance of the strain SWY85S: (i) it promotes the accumulation of trehalose, which has the function of preserving the integrity of the plasma membrane [65], and (ii) it enhances the expression of metabolic pathway-related enzymes involved in ethanol production. However, further research is needed to elucidate the molecular mechanism by which the PEP4-allele modification improves cell tolerance to malic acid stress and enhances cellular ethanol production performance during fermentation.
Author Contributions
Supervision, project administration and funding acquisition, H.Z.; investigation, W.H., H.Y. and X.Y. (Xiaomei Yang); writing—original draft preparation, Y.L.; writing—review and editing, C.S.; formal analysis, X.Y. (Xiaomin Yao); resources, T.L.; critical revision of the manuscript, B.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China, grant number: 31301551.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Attfield, P.V. Stress tolerance: The key to effective strains of industrial baker’s yeast. Nat. Biotechnol. 1997, 15, 1351–1357. [Google Scholar] [CrossRef] [PubMed]
- Betlej, G.; Bator, E.; Oklejewicz, B.; Potocki, L.; Górka, A.; Slowik-Borowiec, M.; Czarny, W.; Domka, W.; Kwiatkowska, A. Long-term adaption to high osmotic stress as a tool for improving enological characteristics in industrial wine yeast. Genes 2020, 11, 576. [Google Scholar] [CrossRef] [PubMed]
- Gibson, B.R.; Lawrence, S.J.; Leclaire, J.P.R.; Powell, C.D.; Smart, K.A. Yeast responses to stresses associated with industrial brewery handling. FEMS Microbiol. Rev. 2007, 31, 535–569. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.M.; Huang, X.W.; Zhang, L.M.; Zhao, N.; Yang, D.M.; Zhang, K.Q. Tolerance and stress response to ethanol in the yeast Saccharomyces cerevisiae. Appl. Microbiol. Biot. 2009, 85, 253–263. [Google Scholar] [CrossRef]
- Stanley, D.; Bandara, A.; Fraser, S.; Chambers, P.J.; Stanley, G.A. The ethanol stress response and ethanol tolerance of Saccharomyces cerevisiae. J. Appl. Microbiol. 2010, 109, 13–24. [Google Scholar] [CrossRef]
- Li, H.; Wang, H.L.; Du, J.; Du, G.; Zhan, J.C.; Huang, W.D. Trehalose protects wine yeast against oxidation under thermal stress. World J. Microb. Biot. 2010, 26, 969–976. [Google Scholar] [CrossRef]
- Kitichantaropas, Y.; Boonchird, C.; Sugiyama, M.; Kaneko, Y.; Harashima, S.; Auesukaree, C. Cellular mechanisms contributing to multiple stress tolerance in Saccharomyces cerevisiae strains with potential use in high-temperature ethanol fermentation. AMB Express 2016, 6, 107. [Google Scholar] [CrossRef]
- Auesukaree, C. Molecular mechanisms of the yeast adaptive response and tolerance to stresses. J. Biosci. Bioeng. 2017, 124, 133–142. [Google Scholar] [CrossRef]
- Narendranath, N.V.; Power, R. Relationship between pH and medium dissolved solids in terms of growth and metabolism of lactobacilli and Saccharomyces cerevisiae during ethanol production. Appl. Environ. Microb. 2005, 71, 2239–2243. [Google Scholar] [CrossRef]
- Attfield, P.V.; Kletsas, S. Hyperosmotic stress response by strains of bakers’ yeasts in high sugar concentration medium. Lett. Appl. Microbiol. 2000, 31, 323–327. [Google Scholar] [CrossRef]
- Mira, N.P.; Teixeira, M.C.; Sá-Correia, I. Adaptive response and tolerance to weak acids in Saccharomyces cerevisiae: A genome-wide view. Omics A J. Integr. Biol. 2010, 14, 525–540. [Google Scholar] [CrossRef] [PubMed]
- Cardona, F.; Carrasco, P.; Pérez-Ortín, J.E.; lí del Olmo, M.; Aranda, A. A novel approach for the improvement of stress resistance in wine yeasts. Int. J. Food. Microbiol. 2007, 114, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Giannattasio, S.; Guaragnella, N.; Ždralević, M.; Marra, E. Molecular mechanisms of Saccharomyces cerevisiae stress adaptation and programmed cell death in response to acetic acid. Front. Microbiol. 2013, 4, 33. [Google Scholar] [CrossRef]
- Meijnen, J.P.; Randazzo, P.; Foulquié-Moreno, M.R.; Van Den Brink, J.; Vandecruys, P.; Stojiljkovic, M.; Dumortier, F.; Zalar, P.; Boekhout, T.; Gunde-Cimerman, N.; et al. Polygenic analysis and targeted improvement of the complex trait of high acetic acid tolerance in the yeast Saccharomyces cerevisiae. Biotechnol. Biofuels 2016, 9, 5. [Google Scholar] [CrossRef]
- Samanfar, B.; Shostak, K.; Moteshareie, H.; Hajikarimlou, M.; Shaikho, S.; Omidi, K.; Hooshyar, M.; Burnside, D.; Márquez, I.G.; Kazmirchuk, T.; et al. The sensitivity of the yeast, Saccharomyces cerevisiae, to acetic acid is influenced by DOM34 and RPL36A. PeerJ 2017, 5, e4037. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Stojiljkovic, M.; Foulquié-Moreno, M.R.; Thevelein, J.M. Polygenic analysis of very high acetic acid tolerance in the yeast Saccharomyces cerevisiae reveals a complex genetic background and several new causative alleles. Biotechnol. Biofuels 2020, 13, 126. [Google Scholar] [CrossRef] [PubMed]
- Mitsui, R.; Yamada, R.; Matsumoto, T.; Yoshihara, S.; Tokumoto, H.; Ogino, H. Construction of lactic acid-tolerant Saccharomyces cerevisiae by using CRISPR-Cas-mediated genome evolution for efficient d-lactic acid production. Appl. Microbiol. Biot. 2020, 104, 9147–9158. [Google Scholar] [CrossRef]
- Yamada, R.; Kumata, Y.; Mitsui, R.; Matsumoto, T.; Ogino, H. Improvement of lactic acid tolerance by cocktail δ-integration strategy and identification of the transcription factor PDR3 responsible for lactic acid tolerance in yeast Saccharomyces cerevisiae. World J. Microb. Biot. 2021, 37, 19. [Google Scholar] [CrossRef]
- Baldi, N.; de Valk, S.C.; Sousa-Silva, M.; Casal, M.; Soares-Silva, I.; Mans, R. Evolutionary engineering reveals amino acid substitutions in Ato2 and Ato3 that allow improved growth of Saccharomyces cerevisiae on lactic acid. FEMS Yeast Res. 2021, 21, foab033. [Google Scholar] [CrossRef]
- Stratford, M.; Nebe-von-Caron, G.; Steels, H.; Novodvorska, M.; Ueckert, J.; Archer, D.B. Weak-acid preservatives: pH and proton movements in the yeast Saccharomyces cerevisiae. Int. J. Food Microbiol. 2013, 161, 164–171. [Google Scholar] [CrossRef]
- Geoghegan, I.A.; Stratford, M.; Bromley, M.; Archer, D.B.; Avery, S.V. Weak acid resistance A (WarA), a novel transcription factor required for regulation of weak-acid resistance and spore-spore heterogeneity in Aspergillus niger. Msphere 2020, 5, e00685-19. [Google Scholar] [CrossRef] [PubMed]
- Ndukwe, J.K.; Aliyu, G.O.; Onwosi, C.O.; Chukwu, K.O.; Ezugworie, F.N. Mechanisms of weak acid-induced stress tolerance in yeasts: Prospects for improved bioethanol production from lignocellulosic biomass. Process Biochem. 2020, 90, 118–130. [Google Scholar] [CrossRef]
- Volschenk, H.; Viljoen, M.; Grobler, J.; Petzold, B.; Bauer, F.; Subden, R.E.; Young, R.A.; Lonvaud, A.; Denayrolles, M.; van Vuuren, H.J. Engineering pathways for malate degradation in Saccharomyces cerevisiae. Nat. Biotechnol. 1997, 15, 253–257. [Google Scholar] [CrossRef]
- Gurban, A.M.; Prieto-Simón, B.; Marty, J.L.; Noguer, T. Malate biosensors for the monitoring of malolactic fermentation: Different approaches. Anal. Lett. 2006, 39, 1543–1558. [Google Scholar] [CrossRef]
- Fletcher, E.; Feizi, A.; Bisschops, M.M.; Hallström, B.M.; Khoomrung, S.; Siewers, V.; Nielsen, J. Evolutionary engineering reveals divergent paths when yeast is adapted to different acidic environments. Metab. Eng. 2017, 39, 19–28. [Google Scholar] [CrossRef]
- Côrte-Real, M.; Leao, C. Transport of malic acid and other dicarboxylic acids in the yeast Hansenula anomala. Appl. Environ. Microb. 1990, 56, 1109–1113. [Google Scholar] [CrossRef]
- Zelle, R.M.; De Hulster, E.; Van Winden, W.A.; De Waard, P.; Dijkema, C.; Winkler, A.A.; Geertman, J.M.A.; Van Dijken, J.P.; Pronk, J.T.; Van Maris, A.J. Malic acid production by Saccharomyces cerevisiae: Engineering of pyruvate carboxylation, oxaloacetate reduction, and malate export. Appl. Environ. Microb. 2008, 74, 2766–2777. [Google Scholar] [CrossRef]
- Volschenk, H.; Van Vuuren, H.J.; Viljoen-Bloom, M. Malic acid in wine: Origin, function and metabolism during vinification. S. Afr. J. Enol. Vitic. 2006, 27, 123–136. [Google Scholar] [CrossRef]
- Kunkee, R.E. Some roles of malic acid in the malolactic fermentation in wine making. FEMS Microbiol. Lett. 1991, 88, 55–71. [Google Scholar] [CrossRef]
- van den Hazel, H.B.; Kielland-Brandt, M.C.; Winther, J.R. Biosynthesis and function of yeast vacuolar proteases. Yeast 1996, 12, 1–16. [Google Scholar] [CrossRef]
- Wolff, A.M.; Din, N.; Petersen, J.G.L. Vacuolar and extracellular maturation of Saccharomyces cerevisiae proteinase A. Yeast 1996, 12, 823–832. [Google Scholar] [CrossRef]
- Zhang, H.B.; Shao, F.F.; Cong, J.H.; Huang, Y.; Chen, M.F.; He, W.X.; Zhang, T.; Liu, L.Y.; Yao, M.Z.; Gwabin, H.; et al. Modification of the second PEP4-allele enhances citric acid stress tolerance during cultivation of an industrial rice wine yeast strain with one PEP4-allele disrupted. LWT Food Sci. Technol. 2021, 152, 112286. [Google Scholar] [CrossRef]
- Parr, C.L.; Keates, R.A.; Bryksa, B.C.; Ogawa, M.; Yada, R.Y. The structure and function of Saccharomyces cerevisiae proteinase A. Yeast 2007, 24, 467–480. [Google Scholar] [CrossRef]
- Callis, J. Regulation of protein degradation. Plant Cell 1995, 7, 845–857. [Google Scholar] [CrossRef] [PubMed]
- Teichert, U.; Mechler, B.; Müller, H.; Wolf, D.H. Lysosomal (vacuolar) proteinases of yeast are essential catalysts for protein degradation, differentiation, and cell survival. J. Biol. Chem. 1989, 264, 16037–16045. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.B.; Zhang, H.F.; Chen, Q.H.; Ruan, H.; Fu, M.L.; He, G.Q. Effects of proteinase A on cultivation and viability characteristics of industrial Saccharomyces cerevisiae WZJ. Zhejiang Univ. Sci. B 2009, 10, 769–776. [Google Scholar] [CrossRef]
- Mahmud, S.A.; Nagahisa, K.; Hirasawa, T.; Yoshikawa, K.; Ashitani, K.; Shimizu, H. Effect of trehalose accumulation on response to saline stress in Saccharomyces cerevisiae. Yeast 2009, 26, 17–30. [Google Scholar] [CrossRef]
- Caspeta, L.; Castillo, T.; Nielsen, J. Modifying yeast tolerance to inhibitory conditions of ethanol production processes. Front. Bioeng. Biotech. 2015, 3, 184. [Google Scholar] [CrossRef]
- Tesnière, C. Importance and role of lipids in wine yeast fermentation. Appl. Microbiol. Biot. 2019, 103, 8293–8300. [Google Scholar] [CrossRef]
- Zhang, M.M.; Xiong, L.; Tang, Y.J.; Mehmood, M.A.; Zhao, Z.K.; Bai, F.W.; Zhao, X.Q. Enhanced acetic acid stress tolerance and ethanol production in Saccharomyces cerevisiae by modulating expression of the de novo purine biosynthesis genes. Biotechnol. Biofuels 2019, 12, 116. [Google Scholar] [CrossRef]
- Xu, K.; Qin, L.; Bai, W.; Wang, X.; Li, F.; Ren, S.; Gao, X.; Chen, B.; Tong, Y.; Li, J.; et al. Multilevel defense system (MDS) relieves multiple stresses for economically boosting ethanol production of industrial Saccharomyces cerevisiae. ACS Energy Lett. 2020, 5, 572–582. [Google Scholar] [CrossRef]
- Jones, E.W. The synthesis and function of proteases in Saccharomyces: Genetic approaches. Annu. Rev. Genet. 1984, 18, 233–270. [Google Scholar] [CrossRef] [PubMed]
- Hemmings, B.A.; Zubenko, G.S.; Hasilik, A.; Jones, E.W. Mutant defective in processing of an enzyme located in the lysosome-like vacuole of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1981, 78, 43–439. [Google Scholar] [CrossRef] [PubMed]
- Jones, E.W.; Zubenko, G.S.; Parker, R.R. PEP4 gene function is required for expression of several vacuolar hydrolases in Saccharomyces cerevisiae. Genetics 1982, 102, 665–677. [Google Scholar] [CrossRef] [PubMed]
- Zubenko, G.S.; Park, F.J.; Jones, E.W. Mutations in PEP4 locus of Saccharomyces cerevisiae block final step in maturation of two vacuolar hydrolases. Proc. Natl. Acad. Sci. USA 1983, 80, 510–514. [Google Scholar] [CrossRef]
- Ammerer, G.; Hunter, C.P.; Rothman, J.H.; Saari, G.C.; Valls, L.A.; Stevens, T.H. PEP4 gene of Saccharomyces cerevisiae encodes proteinase A, a vacuolar enzyme required for processing of vacuolar precursors. Mol. Cell. Biol. 1986, 6, 2490–2499. [Google Scholar] [CrossRef]
- Woolford, C.A.; Daniels, L.B.; Park, F.J.; Jones, E.W.; Van Arsdell, J.N.; Innis, M.A. The PEP4 gene encodes an aspartyl protease implicated in the posttranslational regulation of Saccharomyces cerevisiae vacuolar hydrolases. Mol. Cell. Biol. 1986, 6, 2500–2510. [Google Scholar] [CrossRef]
- Hegemann, J.H.; Heick, S.B. Delete and repeat: A comprehensive toolkit for sequential gene knockout in the budding yeast Saccharomyces cerevisiae. In Strain Engineering Methods and Protocols; Humana Totowa: Paramus, NJ, USA, 2011; pp. 189–206. [Google Scholar] [CrossRef]
- Chen, D.C.; Wang, B.D.; Chou, P.Y.; Kuo, T.T. Asparagine as a nitrogen source for improving the secretion of mouse α-amylase in Saccharomyces cerevisiae protease A-deficient strains. Yeast 2000, 16, 207–217. [Google Scholar] [CrossRef]
- Komeda, T.; Sakai, Y.; Kato, N.; Kondo, K. Construction of Protease-deficient Candida boidinii Strains Useful for Recombinant Protein Production: Cloning and Disruption of Proteinase A Gene (PEP4) and Proteinase B Gen. Biosci. Biotech. Bioch. 2002, 66, 628–631. [Google Scholar] [CrossRef]
- Zhang, H.B.; Ruan, H.; Li, W.F.; Zhang, W.; Su, Z.R.; He, G.Q.; Chen, Q.H. Construction of recombinant industrial S. cerevisiae strain with barley lipid-transfer protein 1 secretion capability and lower PrA activity. Eur. Food Res. Technol. 2011, 233, 707–716. [Google Scholar] [CrossRef]
- Lu, J.; Dong, J.; Wu, D.; Chen, Y.; Guo, X.; Shi, Y.; Sun, X.; Xiao, D. Construction of recombinant industrial brewer’s yeast with lower diacetyl production and proteinase A activity. Eur. Food Res. Technol. 2012, 235, 951–961. [Google Scholar] [CrossRef]
- Wu, M.; Shen, Q.; Yang, Y.; Zhang, S.; Qu, W.; Chen, J.; Sun, H.; Chen, S. Disruption of YPS1 and PEP4 genes reduces proteolytic degradation of secreted HSA/PTH in Pichia pastoris GSJ. Ind. Microbiol. Biot. 2013, 40, 589–599. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Guan, X.; Wang, Y.; Li, L.; Wu, D.; Chen, Y.; Pei, H.; Xiao, D. Reduction of biogenic amines production by eliminating the PEP4 gene in Saccharomyces cerevisiae during fermentation of Chinese rice wine. Food Chem. 2015, 178, 208–211. [Google Scholar] [CrossRef] [PubMed]
- Alugoju, P.; Janardhanshetty, S.S.; Subaramanian, S.; Periyasamy, L.; Dyavaiah, M. Quercetin protects yeast Saccharomyces cerevisiae pep4 mutant from oxidative and apoptotic stress and extends chronological lifespan. Curr. Microbiol. 2018, 75, 519–530. [Google Scholar] [CrossRef]
- Chen, D.C.; Chen, S.Y.; Gee, M.F.; Pan, J.T.; Kuo, T.T. A variant of Saccharomyces cerevisiae pep4 strain with improved oligotrophic proliferation, cell survival and heterologous secretion of α-amylase. Appl. Microbiol. Biot. 1999, 51, 185–192. [Google Scholar] [CrossRef]
- Giannattasio, S.; Guaragnella, N.; Corte-Real, M.; Passarella, S.; Marra, E. Acid stress adaptation protects Saccharomyces cerevisiae from acetic acid-induced programmed cell death. Gene 2005, 354, 93–98. [Google Scholar] [CrossRef]
- Kawahata, M.; Masaki, K.; Fujii, T.; Iefuji, H. Yeast genes involved in response to lactic acid and acetic acid: Acidic conditions caused by the organic acids in Saccharomyces cerevisiae cultures induce expression of intracellular metal metabolism genes regulated by Aft1p. FEMS Yeast Res. 2006, 6, 924–936. [Google Scholar] [CrossRef]
- Abbott, D.A.; Knijnenburg, T.A.; De Poorter, L.M.; Reinders, M.J.; Pronk, J.T.; Van Maris, A.J. Generic and specific transcriptional responses to different weak organic acids in anaerobic chemostat cultures of Saccharomyces cerevisiae. Fems Yeast Res. 2007, 7, 819–833. [Google Scholar] [CrossRef]
- Ullah, A.; Orij, R.; Brul, S.; Smits, G.J. Quantitative analysis of the modes of growth inhibition by weak organic acids in Saccharomyces cerevisiae. Appl. Environ. Microb. 2012, 78, 8377–8387. [Google Scholar] [CrossRef] [PubMed]
- D’Amore, T.; Crumplen, R.; Stewart, G.G. The involvement of trehalose in yeast stress tolerance. J. Ind. Microbiol. 1991, 7, 191–195. [Google Scholar] [CrossRef]
- Van Dijck, P.; Colavizza, D.; Smet, P.; Thevelein, J.M. Differential importance of trehalose in stress resistance in fermenting and nonfermenting Saccharomyces cerevisiae cells. Appl. Environ. Microb. 1995, 61, 109–115. [Google Scholar] [CrossRef] [PubMed]
- Yoshiyama, Y.; Tanaka, K.; Yoshiyama, K.; Hibi, M.; Ogawa, J.; Shima, J. Trehalose accumulation enhances tolerance of Saccharomyces cerevisiae to acetic acid. J. Biosci. Bioeng. 2015, 119, 172–175. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.C. A possible role of trehalose in osmotolerance and ethanol tolerance in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 1997, 152, 11–15. [Google Scholar] [CrossRef] [PubMed]
- Mahmud, S.A.; Hirasawa, T.; Shimizu, H. Differential importance of trehalose accumulation in Saccharomyces cerevisiae in response to various environmental stresses. J. Biosci. Bioeng. 2010, 109, 262–266. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).