Immobilization of Phospholipase D for Production of Phosphatidylserine via Enzyme-Inorganic Hybrid Nanoflower Strategy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials and Reagents
2.2. Preparation and Characterization of saPLD@NFs
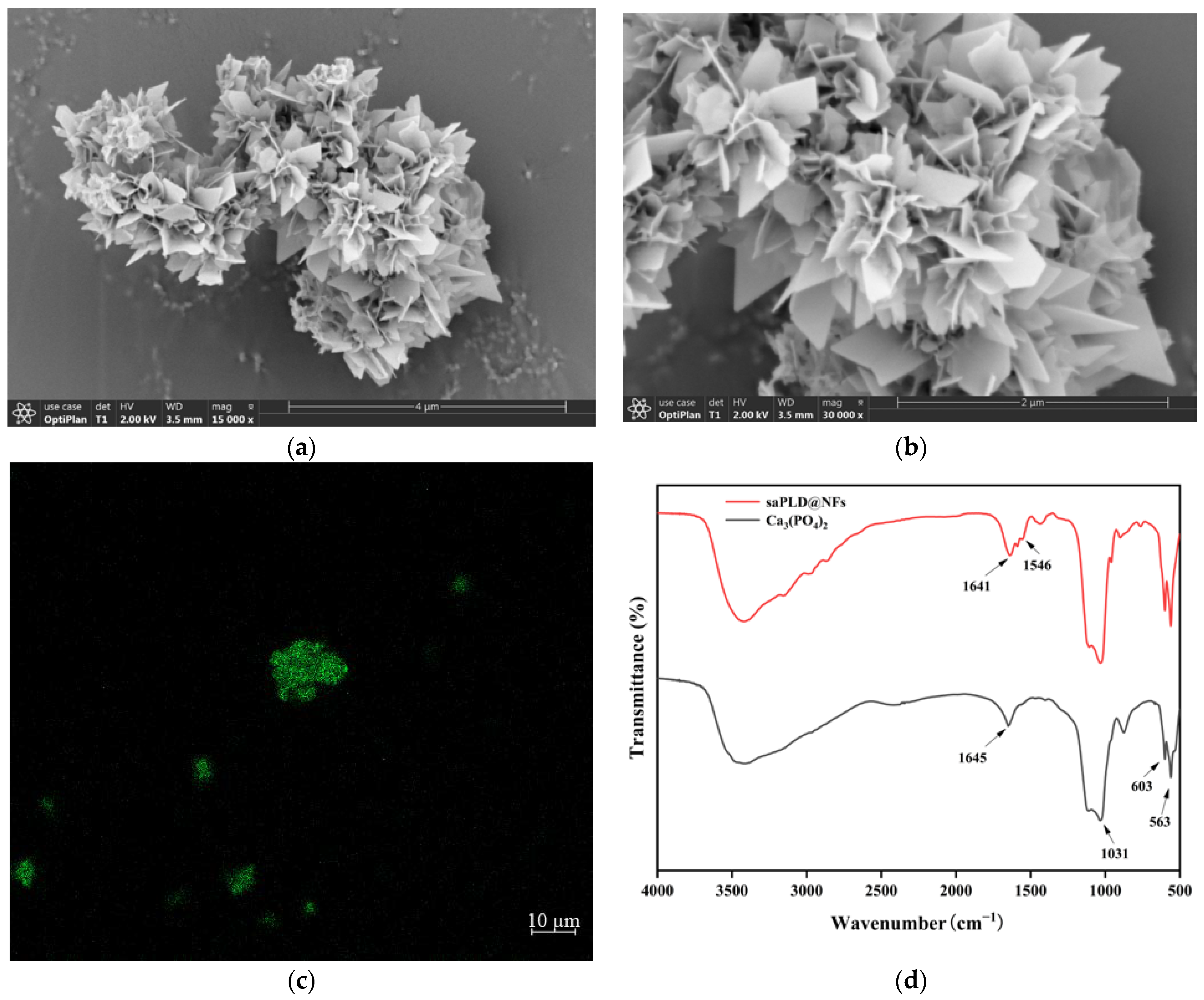
2.2.1. Scanning Electron Microscopy
2.2.2. Confocal Laser Scanning Microscopy
2.2.3. Fourier-Transform Infrared Spectroscopy
2.3. Measurements of saPLD@NFs and Free saPLD Activity
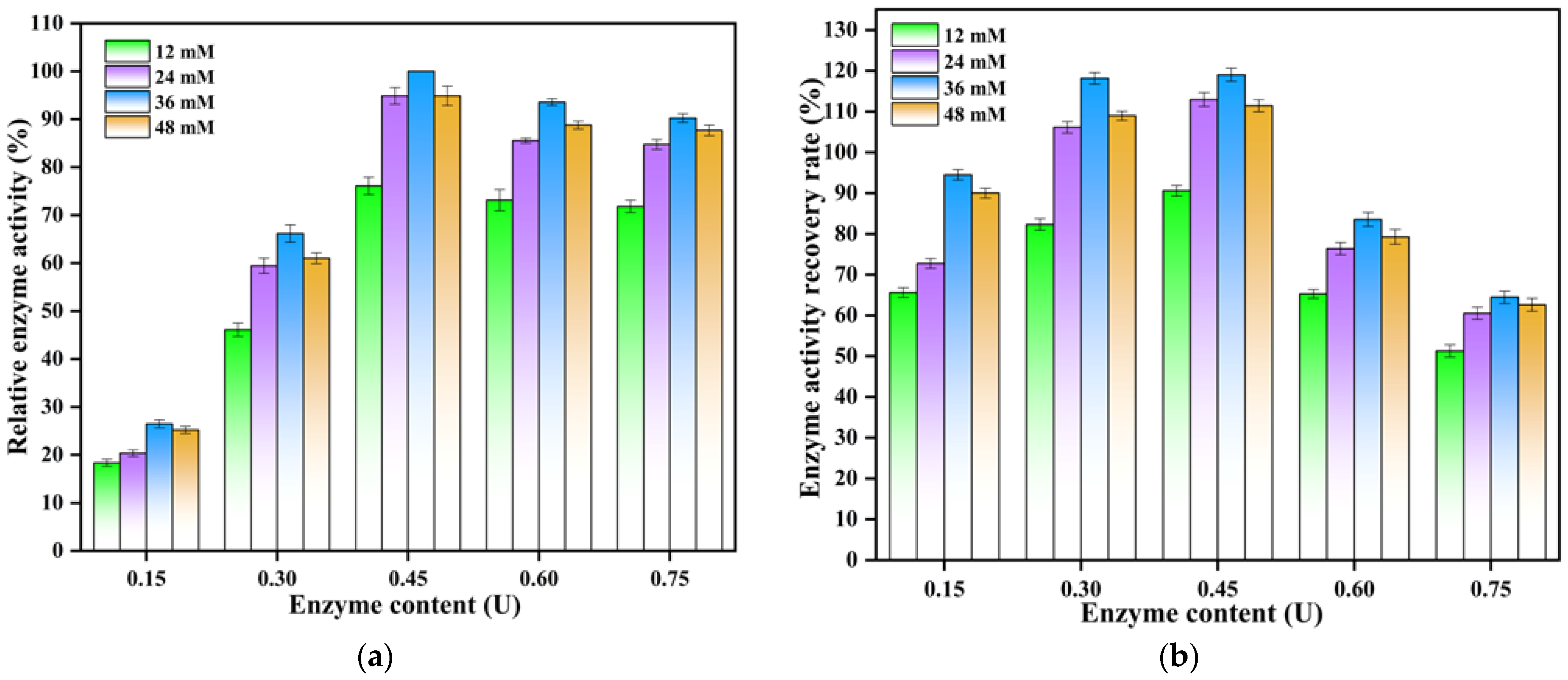
2.4. Optimum Synthesis Conditions of saPLD@NFs
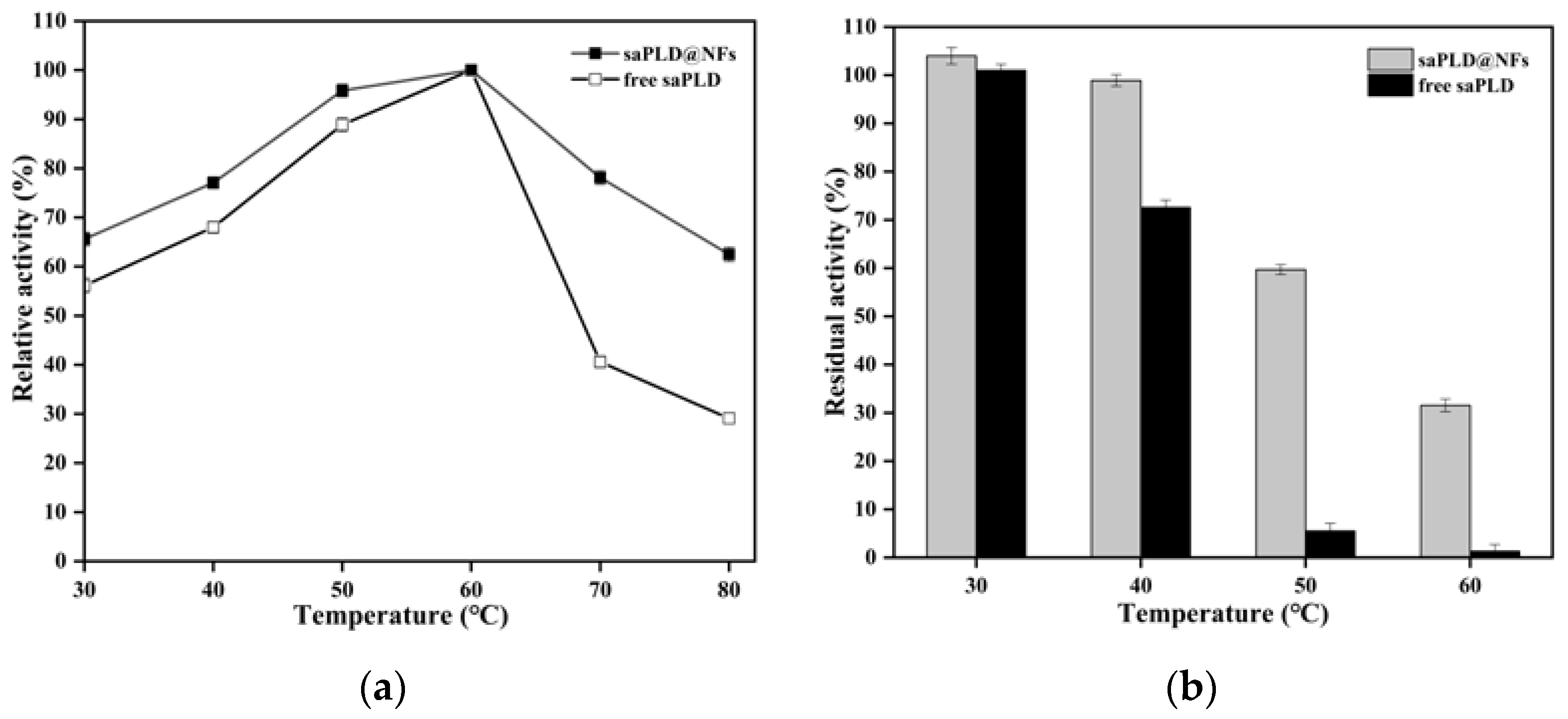
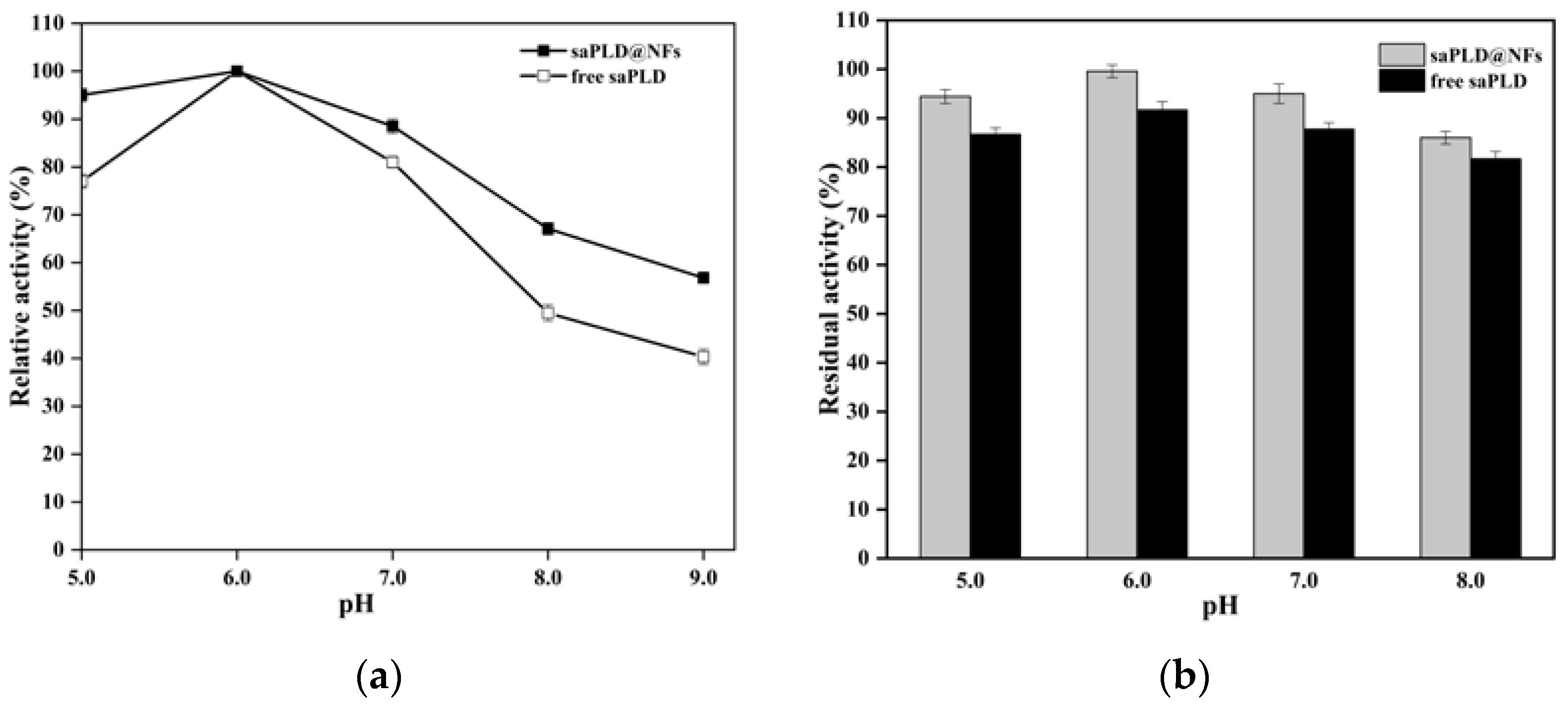
2.5. Enzymatic Properties of saPLD@NFs
2.6. Preparation of PS via Enzymatic Method
2.7. Storage Stability of saPLD@NFs
2.8. Operational Stability of saPLD@NFs
3. Results
3.1. Characterization of saPLD@NFs
3.2. Effects of Preparation Conditions on the saPLD@NFs Activity
3.3. Enzymatic Properties of saPLD@NFs
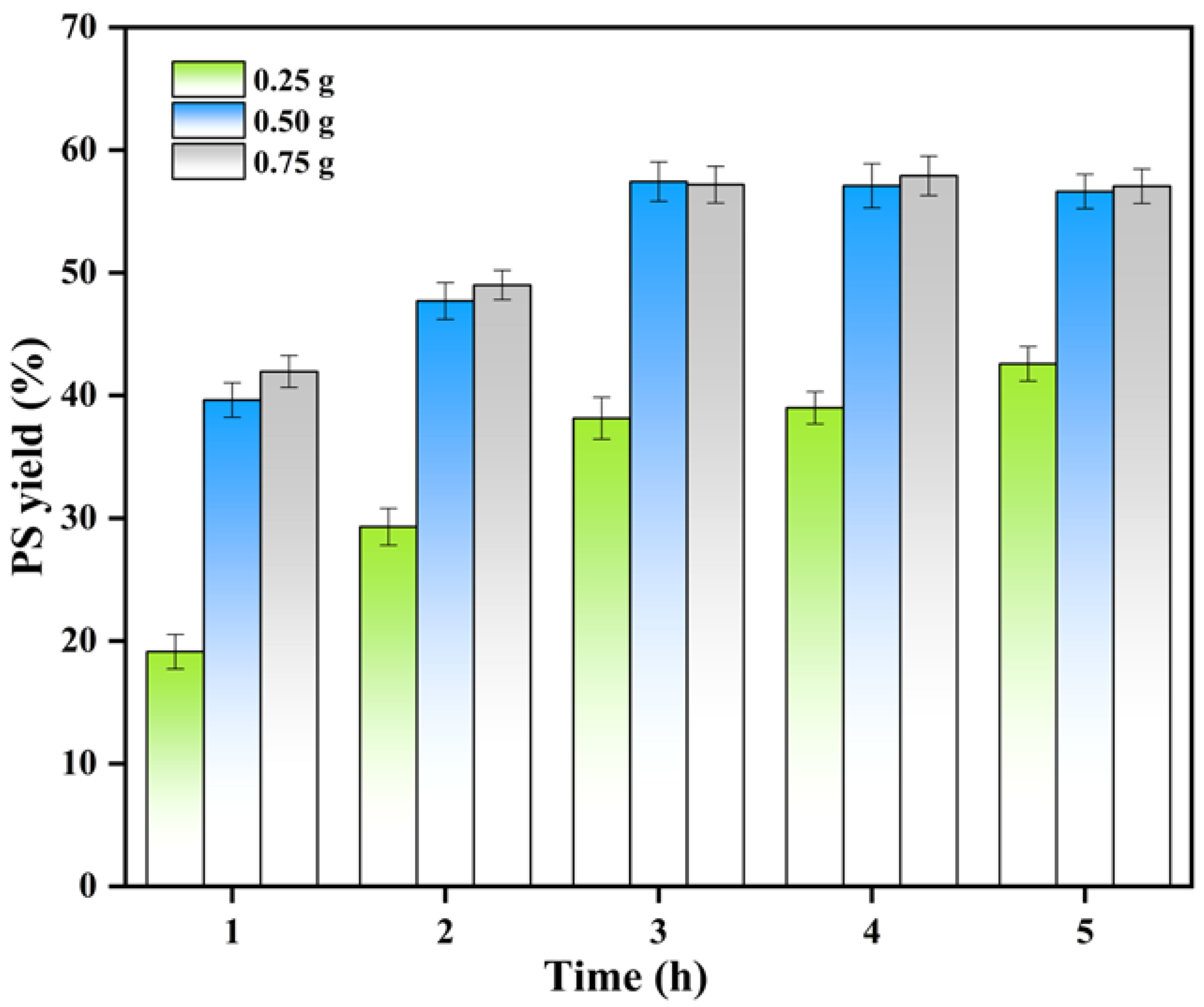
3.4. Synthesis of PS by saPLD@NFs
3.5. Storage Stability of saPLD@NFs
3.6. Operational Stability of saPLD@NFs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Liu, Y.; Huang, L.; Fu, Y.; Zheng, D.; Ma, J.; Li, Y.; Xu, Z.; Lu, F. A novel process for phosphatidylserine production using a Pichia pastoris whole-cell biocatalyst with overexpression of phospholipase D from Streptomyces halstedii in a purely aqueous system. Food Chem. 2019, 274, 535–542. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Zhang, S.; Liu, D.; Huang, Z.; Ge, Y.; Hou, J.; Lu, F.; Liu, Y. Immobilization of Phospholipase D for Production of Phosphatidylserine by a Pickering Emulsion Strategy. Catalysts 2023, 13, 1318. [Google Scholar] [CrossRef]
- Yin, C.; Zhang, H.; Mao, X. Cellulose nanofibril-stabilized Pickering emulsion as a high-performance interfacial biocatalysis system for the synthesis of phosphatidylserine. Food Chem. 2023, 399, 133865. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Y.; Yang, L.Q.; Guo, L.M. Effect of phosphatidylserine on memory in patients and rats with Alzheimer’s disease. Genet. Mol. Res. 2015, 14, 9325–9333. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.Y.; Huang, B.X.; Spector, A.A. Phosphatidylserine in the brain: Metabolism and function. Prog. Lipid Res. 2014, 56, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Glade, M.J.; Smith, K. Phosphatidylserine and the human brain. Nutrition 2015, 31, 781–786. [Google Scholar] [CrossRef] [PubMed]
- Starks, M.A.; Starks, S.L.; Kingsley, M.; Purpura, M.; Jager, R. The effects of phosphatidylserine on endocrine response to moderate intensity exercise. J. Int. Soc. Sports Nutr. 2008, 5, 11. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, T.; Qiao, J.; Liu, X.; Bo, J.; Wang, J.; Lu, F. High-yield phosphatidylserine production via yeast surface display of phospholipase D from Streptomyces chromofuscus on Pichia pastoris. J. Agric. Food Chem. 2014, 62, 5354–5360. [Google Scholar] [CrossRef]
- Li, M.; Zhou, Y.; Duan, X.; Zhou, L.; Zhang, T. Characterization of a phospholipase D from Streptomyces cinnamoneum SK43.003 suitable for phosphatidylserine synthesis. Biotechnol. Appl. Biochem. 2022, 69, 1917–1928. [Google Scholar] [CrossRef]
- Hou, H.J.; Gong, J.S.; Dong, Y.X.; Qin, J.; Li, H.; Li, H.; Lu, Z.M.; Zhang, X.M.; Xu, Z.H.; Shi, J.S. Phospholipase D engineering for improving the biocatalytic synthesis of phosphatidylserine. Bioprocess. Biosyst. Eng. 2019, 42, 1185–1194. [Google Scholar] [CrossRef]
- Morris, A.J.; Engebrecht, J.; Frohman, M.A. Structure and regulation of phospholipase D. Trends Pharmacol. Sci. 1996, 17, 182–185. [Google Scholar] [CrossRef] [PubMed]
- Imamura, S.; Horiuti, Y. Purification of Streptomyces chromofuscus phospholipase D by hydrophobic affinity chromatography on palmitoyl cellulose. J. Biochem. 1979, 85, 79–95. [Google Scholar] [CrossRef] [PubMed]
- Yu, C.H.; Liu, S.Y.; Panagia, V. The transphosphatidylation activity of phospholipase D. Mol. Cell Biochem. 1996, 157, 101–105. [Google Scholar] [CrossRef] [PubMed]
- Damnjanovic, J.; Iwasaki, Y. Phospholipase D as a catalyst: Application in phospholipid synthesis, molecular structure and protein engineering. J. Biosci. Bioeng. 2013, 116, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.F.; Freer, S.; Benson, A.A. Transphosphatidylation by Phospholipase D. J. Biol. Chem. 1967, 242, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Stanacev, N.Z.; Stuhne-Sekalec, L. On the mechanism of enzymatic phosphatidylation. Biosynthesis of cardiolipin catalyzed by phospholipase D. Biochim. Biophys. Acta 1970, 210, 350–352. [Google Scholar] [CrossRef] [PubMed]
- Veronese, F.M. Peptide and protein PEGylation: A review of problems and solutions. Biomaterials 2001, 22, 405–417. [Google Scholar] [CrossRef]
- Zhang, Y.-T.; Zhi, T.-T.; Zhang, L.; Huang, H.; Chen, H.-L. Immobilization of carbonic anhydrase by embedding and covalent coupling into nanocomposite hydrogel containing hydrotalcite. Polymer 2009, 50, 5693–5700. [Google Scholar] [CrossRef]
- Romero-Arcos, M.; Pérez-Robles, J.F.; Guadalupe Garnica-Romo, M.; Luna-Martinez, M.S.; Gonzalez-Reyna, M.A. Synthesis and functionalization of carbon nanotubes and nanospheres as a support for the immobilization of an enzyme extract from the mushroom Trametes versicolor. J. Mater. Sci. 2019, 54, 11671–11681. [Google Scholar] [CrossRef]
- Wang, M.; Qi, W.; Su, R.; He, Z. Advances in carrier-bound and carrier-free immobilized nanobiocatalysts. Chem. Eng. Sci. 2015, 135, 21–32. [Google Scholar] [CrossRef]
- Zhou, Z.; Hartmann, M. Progress in enzyme immobilization in ordered mesoporous materials and related applications. Chem. Soc. Rev. 2013, 42, 3894–3912. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Ren, S.; Lin, T.; Feng, Y.; Jia, S. Shielding effects of Fe3+-tannic acid nanocoatings for immobilized enzyme on magnetic Fe3O4@silica core shell nanosphere. Chem. Eng. J. 2018, 343, 629–637. [Google Scholar] [CrossRef]
- Sun, J.; Yendluri, R.; Liu, K.; Guo, Y.; Lvov, Y.; Yan, X. Enzyme-immobilized clay nanotube-chitosan membranes with sustainable biocatalytic activities. Phys. Chem. Chem. Phys. 2016, 19, 562–567. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Qi, X.; Wang, X.; Wang, X.; Wang, Q.; Yang, H.; Fu, Y.; Yao, S. Highly efficient enzyme immobilization by nanocomposites of metal organic coordination polymers and carbon nanotubes for electrochemical biosensing. Electrochem. Commun. 2017, 79, 18–22. [Google Scholar] [CrossRef]
- Cui, J.; Lin, T.; Feng, Y.; Tan, Z.; Jia, S. Preparation of spherical cross-linked lipase aggregates with improved activity, stability and reusability characteristic in water-in-ionic liquid microemulsion. J. Chem. Technol. Biotechnol. 2017, 92, 1785–1793. [Google Scholar] [CrossRef]
- Liu, X.; Fang, Y.; Yang, X.; Li, Y.; Wang, C. Electrospun epoxy-based nanofibrous membrane containing biocompatible feather polypeptide for highly stable and active covalent immobilization of lipase. Colloids Surf. B Biointerfaces 2018, 166, 277–285. [Google Scholar] [CrossRef]
- Ding, S.; Cargill, A.A.; Medintz, I.L.; Claussen, J.C. Increasing the activity of immobilized enzymes with nanoparticle conjugation. Curr. Opin. Biotechnol. 2015, 34, 242–250. [Google Scholar] [CrossRef]
- Ge, J.; Lu, D.; Liu, Z.; Liu, Z. Recent advances in nanostructured biocatalysts. Biochem. Eng. J. 2009, 44, 53–59. [Google Scholar] [CrossRef]
- Soni, S.; Dwivedee, B.P.; Banerjee, U.C. An Ultrafast Sonochemical Strategy to Synthesize Lipase-Manganese Phosphate Hybrid Nanoflowers with Promoted Biocatalytic Performance in the Kinetic Resolution of β-Aryloxyalcohols. ChemNanoMat 2018, 4, 1007–1020. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, Y.; Yang, C.; Ma, C.; Tang, J. Enzyme-inorganic hybrid nanoflowers: Classification, synthesis, functionalization and potential applications. Chem. Eng. J. 2021, 415, 129075. [Google Scholar] [CrossRef]
- Ge, J.; Lei, J.; Zare, R.N. Protein-inorganic hybrid nanoflowers. Nat. Nanotechnol. 2012, 7, 428–432. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Zhang, S.; Wang, X.; Yang, C.; Jiang, Z. Preparation and enzymatic application of flower-like hybrid microcapsules through a biomimetic mineralization approach. J. Mater. Chem. B 2014, 2, 4289–4296. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, F.; Wang, S.; Wu, J.; Zhang, W.; Zhang, Y.; Liu, W. Time-temperature indicators based on Lipase@Cu3(PO4)2 hybrid nanoflowers. LWT 2022, 168, 113857. [Google Scholar] [CrossRef]
- Lin, Z.; Xiao, Y.; Wang, L.; Yin, Y.; Zheng, J.; Yang, H.; Chen, G. Facile synthesis of enzyme–inorganic hybrid nanoflowers and their application as an immobilized trypsin reactor for highly efficient protein digestion. RSC Adv. 2014, 4, 13888–13891. [Google Scholar] [CrossRef]
- Hao, Y.; Li, H.; Cao, Y.; Chen, Y.; Lei, M.; Zhang, T.; Xiao, Y.; Chu, B.; Qian, Z. Uricase and Horseradish Peroxidase Hybrid CaHPO4 Nanoflower Integrated with Transcutaneous Patches for Treatment of Hyperuricemia. J. Biomed. Nanotechnol. 2019, 15, 951–965. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.; Cheon, H.J.; Adhikari, M.D.; Tran, T.D.; Yeon, K.M.; Kim, M.I.; Kim, J. Glucose oxidase-copper hybrid nanoflowers embedded with magnetic nanoparticles as an effective antibacterial agent. Int. J. Biol. Macromol. 2020, 155, 1520–1531. [Google Scholar] [CrossRef] [PubMed]
- Somturk, B.; Yilmaz, I.; Altinkaynak, C.; Karatepe, A.; Ozdemir, N.; Ocsoy, I. Synthesis of urease hybrid nanoflowers and their enhanced catalytic properties. Enzym. Microb. Technol. 2016, 86, 134–142. [Google Scholar] [CrossRef] [PubMed]
- Fu, M.; Xing, J.; Ge, Z. Preparation of laccase-loaded magnetic nanoflowers and their recycling for efficient degradation of bisphenol A. Sci. Total Environ. 2019, 651, 2857–2865. [Google Scholar] [CrossRef]
- Yu, Y.; Fei, X.; Tian, J.; Xu, L.; Wang, X.; Wang, Y. Self-assembled enzyme-inorganic hybrid nanoflowers and their application to enzyme purification. Colloids Surf. B Biointerfaces 2015, 130, 299–304. [Google Scholar] [CrossRef]
- Zhang, B.; Li, P.; Zhang, H.; Fan, L.; Wang, H.; Li, X.; Tian, L.; Ali, N.; Ali, Z.; Zhang, Q. Papain/Zn3(PO4)2 hybrid nanoflower: Preparation, characterization and its enhanced catalytic activity as an immobilized enzyme. RSC Adv. 2016, 6, 46702–46710. [Google Scholar] [CrossRef]
- Mao, S.; Zhang, Z.; Ma, X.; Tian, H.; Lu, F.; Liu, Y. Efficient secretion expression of phospholipase D in Bacillus subtilis and its application in synthesis of phosphatidylserine by enzyme immobilization. Int. J. Biol. Macromol. 2021, 169, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Lou, L.; Li, Z.; Li, Z. Rational Design to Enhance Enzyme Activity for the Establishment of an Enzyme-Inorganic Hybrid Nanoflower Co-Immobilization System for Efficient Nucleotide Production. J. Agric. Food Chem. 2022, 70, 2312–2319. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhao, J.; Zhang, Z.; Jiang, Y.; Bilal, M.; Jiang, Y.; Jia, S.; Cui, J. Self-assembly of activated lipase hybrid nanoflowers with superior activity and enhanced stability. Biochem. Eng. J. 2020, 158, 107582. [Google Scholar] [CrossRef]
- Yin, Y.; Xiao, Y.; Lin, G.; Xiao, Q.; Lin, Z.; Cai, Z. An enzyme-inorganic hybrid nanoflower based immobilized enzyme reactor with enhanced enzymatic activity. J. Mater. Chem. B 2015, 3, 2295–2300. [Google Scholar] [CrossRef]
- Damnjanović, J.; Iwasaki, Y. Enzymatic Modification of Phospholipids by Phospholipase D. In Lipid Modification by Enzymes and Engineered Microbes; AOCS Press: Champaign, IL, USA, 2018; pp. 69–88. [Google Scholar] [CrossRef]
- Nogueira, I.B.R.; Faria, R.P.V.; Rodrigues, A.E.; Loureiro, J.M.; Ribeiro, A.M. Chromatographic studies of n-Propyl Propionate, Part II: Synthesis in a fixed bed adsorptive reactor, modelling and uncertainties determination. Comput. Chem. Eng. 2019, 128, 164–173. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Sun, H.; Huang, Z.; Han, Z.; Hou, J.; Lu, F.; Liu, Y. Immobilization of Phospholipase D for Production of Phosphatidylserine via Enzyme-Inorganic Hybrid Nanoflower Strategy. Fermentation 2023, 9, 1016. https://doi.org/10.3390/fermentation9121016
Zhang S, Sun H, Huang Z, Han Z, Hou J, Lu F, Liu Y. Immobilization of Phospholipase D for Production of Phosphatidylserine via Enzyme-Inorganic Hybrid Nanoflower Strategy. Fermentation. 2023; 9(12):1016. https://doi.org/10.3390/fermentation9121016
Chicago/Turabian StyleZhang, Shujing, Hui Sun, Zhiqi Huang, Zhuoxuan Han, Jiayi Hou, Fuping Lu, and Yihan Liu. 2023. "Immobilization of Phospholipase D for Production of Phosphatidylserine via Enzyme-Inorganic Hybrid Nanoflower Strategy" Fermentation 9, no. 12: 1016. https://doi.org/10.3390/fermentation9121016
APA StyleZhang, S., Sun, H., Huang, Z., Han, Z., Hou, J., Lu, F., & Liu, Y. (2023). Immobilization of Phospholipase D for Production of Phosphatidylserine via Enzyme-Inorganic Hybrid Nanoflower Strategy. Fermentation, 9(12), 1016. https://doi.org/10.3390/fermentation9121016






