Candida albicans Adhesion Measured by Optical Nanomotion Detection
Abstract
1. Introduction
2. Materials and Methods
2.1. Yeast Cell Culture
2.2. Adhesion Assay
2.3. Optical Nanomotion Adhesion Method
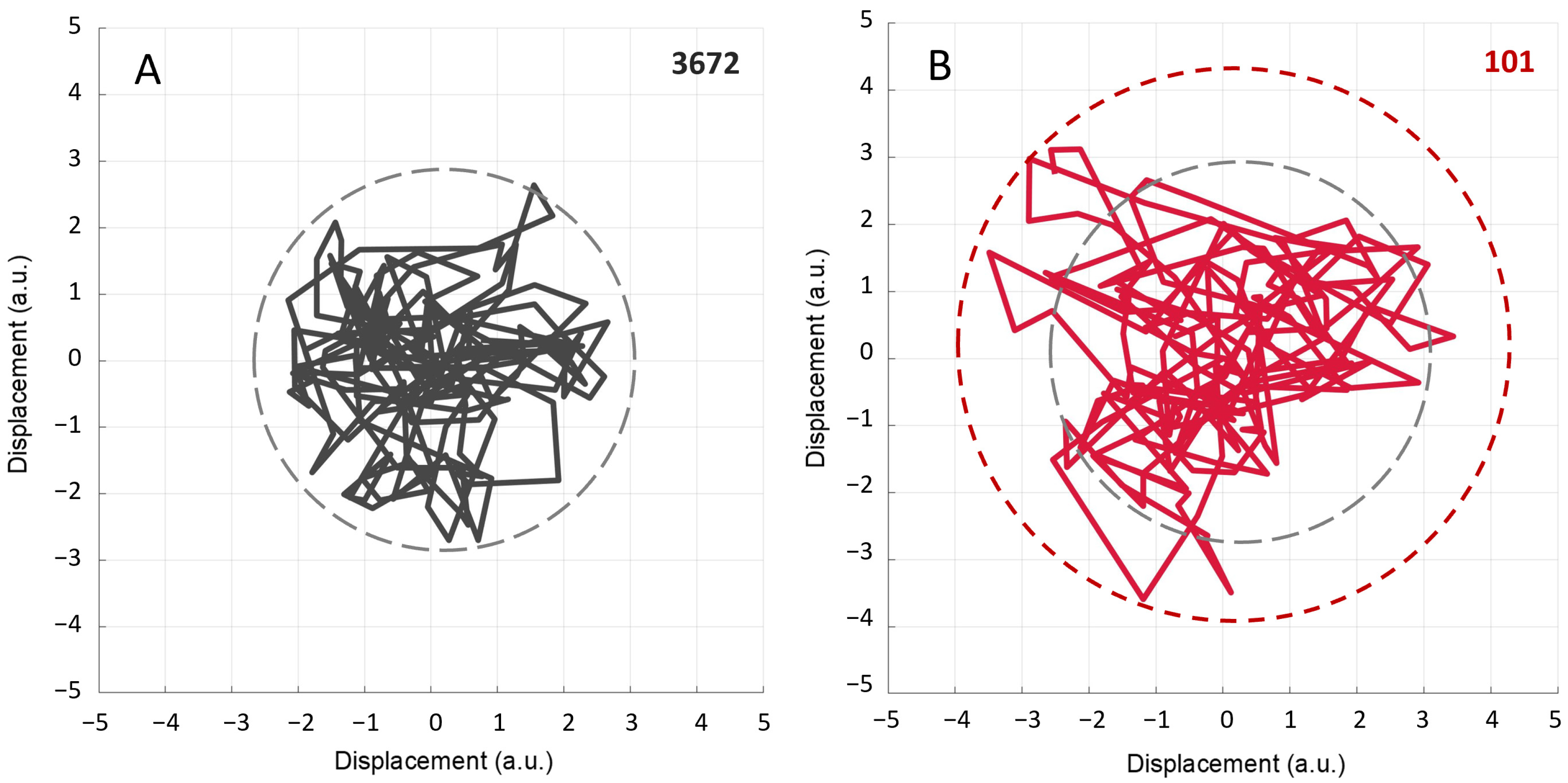
2.4. Constrained Random Walk Simulation
2.5. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Busscher, H.J.; van der Mei, H.C. Microbial Adhesion in Flow Displacement Systems. Clin. Microbiol. Rev. 2006, 19, 127–141. [Google Scholar] [CrossRef] [PubMed]
- Achinas, S.; Charalampogiannis, N.; Euverink, G.J.W. A Brief Recap of Microbial Adhesion and Biofilms. Appl. Sci. 2019, 9, 2801. [Google Scholar] [CrossRef]
- Di Martino, P. Bacterial Adherence: Much More than a Bond. AIMS Microbiol. 2018, 4, 563–566. [Google Scholar] [CrossRef] [PubMed]
- Janiszewska, M.; Primi, M.C.; Izard, T. Cell Adhesion in Cancer: Beyond the Migration of Single Cells. J. Biol. Chem. 2020, 295, 2495–2505. [Google Scholar] [CrossRef]
- Mackay, C.R.; Imhof, B.A. Cell Adhesion in the Immune System. Immunol. Today 1993, 14, 99–102. [Google Scholar] [CrossRef]
- Gunaratnam, G.; Dudek, J.; Jung, P.; Becker, S.L.; Jacobs, K.; Bischoff, M.; Hannig, M. Quantification of the Adhesion Strength of Candida Albicans to Tooth Enamel. Microorg. 2021, 9, 2213. [Google Scholar] [CrossRef]
- Hu, L.; Zhang, X.; Miller, P.; Ozkan, M.; Ozkan, C.; Wang, J. Cell Adhesion Measurement by Laser-Induced Stress Waves. J. Appl. Phys. 2006, 100, 084701. [Google Scholar] [CrossRef]
- Ahmad Khalili, A.; Ahmad, M.R. A Review of Cell Adhesion Studies for Biomedical and Biological Applications. Int. J. Mol. Sci. 2015, 16, 18149–18184. [Google Scholar] [CrossRef]
- Zhou, D.W.; García, A.J. Measurement Systems for Cell Adhesive Forces. J. Biomech. Eng. 2015, 137, 0209081–0209088. [Google Scholar] [CrossRef]
- Ungai-Salanki, R.; Peter, B.; Gerecsei, T.; Orgovan, N.; Horvath, R.; Szabo, B. A Practical Review on the Measurement Tools for Cellular Adhesion Force. Adv. Colloid Interface Sci. 2019, 269, 309–333. [Google Scholar] [CrossRef]
- Truskey, G.A.; Pirone, J.S. The Effect of Fluid Shear Stress upon Cell Adhesion to Fibronectin-Treated Surfaces. J. Biomed. Mater. Res. 1990, 24, 1333–1353. [Google Scholar] [CrossRef]
- Kucik, D.F. Measurement of Adhesion Under Flow Conditions. Curr. Protoc. Cell Biol. 2009, 43, 9.6.1–9.6.10. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, A.S.; DiMilla, P.A. Effect of Adsorbed Fibronectin Concentration on Cell Adhesion and Deformation under Shear on Hydrophobic Surfaces. J. Biomed. Mater. Res. 2002, 59, 665–675. [Google Scholar] [CrossRef] [PubMed]
- McCall, A.D.; Pathirana, R.U.; Prabhakar, A.; Cullen, P.J.; Edgerton, M. Candida Albicans Biofilm Development Is Governed by Cooperative Attachment and Adhesion Maintenance Proteins. Npj Biofilms Microbiomes 2019, 5, 21. [Google Scholar] [CrossRef] [PubMed]
- Christ, K.V.; Turner, K.T. Methods to Measure the Strength of Cell Adhesion to Substrates. J. Adhes. Sci. Technol. 2010, 24, 2027–2058. [Google Scholar] [CrossRef]
- Giacomello, E.; Neumayer, J.; Colombatti, A.; Perris, R. Centrifugal Assay for Fluorescence-Based Cell Adhesion Adapted to the Analysis of Ex Vivo Cells and Capable of Determining Relative Binding Strengths. Biotech. 1999, 26, 758–762, 764–766. [Google Scholar] [CrossRef]
- Chu, Y.-S.; Thomas, W.A.; Eder, O.; Pincet, F.; Perez, E.; Thiery, J.P.; Dufour, S. Force Measurements in E-Cadherin–Mediated Cell Doublets Reveal Rapid Adhesion Strengthened by Actin Cytoskeleton Remodeling through Rac and Cdc42. J. Cell Biol. 2004, 167, 1183–1194. [Google Scholar] [CrossRef]
- Daoudi, M.; Lavergne, E.; Garin, A.; Tarantino, N.; Debré, P.; Pincet, F.; Combadière, C.; Deterre, P. Enhanced Adhesive Capacities of the Naturally Occurring Ile249–Met280 Variant of the Chemokine Receptor CX3CR1. J. Biol. Chem. 2004, 279, 19649–19657. [Google Scholar] [CrossRef]
- Thoumine, O.; Kocian, P.; Kottelat, A.; Meister, J.-J. Short-Term Binding of Fibroblasts to Fibronectin: Optical Tweezers Experiments and Probabilistic Analysis. Eur. Biophys. J. 2000, 29, 398–408. [Google Scholar] [CrossRef]
- Castelain, M.; Rouxhet, P.G.; Pignon, F.; Magnin, A.; Piau, J.-M. Single-Cell Adhesion Probed in-Situ Using Optical Tweezers: A Case Study with Saccharomyces Cerevisiae. J. Appl. Phys. 2012, 111, 114701. [Google Scholar] [CrossRef]
- Benoit, M.; Gabriel, D.; Gerisch, G.; Gaub, H.E. Discrete Interactions in Cell Adhesion Measured by Single-Molecule Force Spectroscopy. Nat. Cell Biol. 2000, 2, 313–317. [Google Scholar] [CrossRef]
- Friedrichs, J.; Legate, K.R.; Schubert, R.; Bharadwaj, M.; Werner, C.; Müller, D.J.; Benoit, M. A Practical Guide to Quantify Cell Adhesion Using Single-Cell Force Spectroscopy. Methods 2013, 60, 169–178. [Google Scholar] [CrossRef]
- Borghi, N.; Sorokina, M.; Shcherbakova, O.G.; Weis, W.I.; Pruitt, B.L.; Nelson, W.J.; Dunn, A.R. E-Cadherin Is under Constitutive Actomyosin-Generated Tension That Is Increased at Cell–Cell Contacts upon Externally Applied Stretch. Proc. Natl. Acad. Sci. USA 2012, 109, 12568–12573. [Google Scholar] [CrossRef]
- Ramsden, J.J.; Horvath, R. Optical Biosensors for Cell Adhesion. J. Recept. Signal Transduct. 2009, 29, 211–223. [Google Scholar] [CrossRef]
- Kasas, S.; Ruggeri, F.S.; Benadiba, C.; Maillard, C.; Stupar, P.; Tournu, H.; Dietler, G.; Longo, G. Detecting Nanoscale Vibrations as Signature of Life. Proc. Natl. Acad. Sci. USA 2015, 112, 378–381. [Google Scholar] [CrossRef]
- Pleskova, S.N.; Lazarenko, E.V.; Bezrukov, N.A.; Bobyk, S.Z.; Boryakov, A.V.; Kriukov, R.N. Differences in Bacteria Nanomotion Profiles and Neutrophil Nanomotion during Phagocytosis. Front. Microbiol. 2023, 14, 1113353. [Google Scholar] [CrossRef]
- Wu, S.; Zhang, Z.; Zhou, X.; Liu, H.; Xue, C.; Zhao, G.; Cao, Y.; Zhang, Q.; Wu, X. Nanomechanical Sensors for Direct and Rapid Characterization of Sperm Motility Based on Nanoscale Vibrations. Nanoscale 2017, 9, 18258–18267. [Google Scholar] [CrossRef]
- Wu, S.; Liu, X.; Zhou, X.; Liang, X.M.; Gao, D.; Liu, H.; Zhao, G.; Zhang, Q.; Wu, X. Quantification of Cell Viability and Rapid Screening Anti-Cancer Drug Utilizing Nanomechanical Fluctuation. Biosens. Bioelectron. 2016, 77, 164–173. [Google Scholar] [CrossRef]
- Girasole, M.; Dinarelli, S.; Longo, G. Correlating Nanoscale Motion and ATP Production in Healthy and Favism Erythrocytes: A Real-Time Nanomotion Sensor Study. Front. Microbiol. 2023, 14. [Google Scholar] [CrossRef]
- Longo, G.; Alonso-Sarduy, L.; Rio, L.M.; Bizzini, A.; Trampuz, A.; Notz, J.; Dietler, G.; Kasas, S. Rapid Detection of Bacterial Resistance to Antibiotics Using AFM Cantilevers as Nanomechanical Sensors. Nat. Nanotech 2013, 8, 522–526. [Google Scholar] [CrossRef]
- Al-Madani, H.; Du, H.; Yao, J.; Peng, H.; Yao, C.; Jiang, B.; Wu, A.; Yang, F. Living Sample Viability Measurement Methods from Traditional Assays to Nanomotion. Biosens. 2022, 12, 453. [Google Scholar] [CrossRef]
- Vocat, A.; Sturm, A.; Jóźwiak, G.; Cathomen, G.; Świątkowski, M.; Buga, R.; Wielgoszewski, G.; Cichocka, D.; Greub, G.; Opota, O. Nanomotion Technology in Combination with Machine Learning: A New Approach for a Rapid Antibiotic Susceptibility Test for Mycobacterium Tuberculosis. Microbes Infect. 2023, 25, 105151. [Google Scholar] [CrossRef]
- Rosłoń, I.E.; Japaridze, A.; Steeneken, P.G.; Dekker, C.; Alijani, F. Probing Nanomotion of Single Bacteria with Graphene Drums. Nat. Nanotechnol. 2022, 17, 637–642. [Google Scholar] [CrossRef]
- Willaert, R.G.; Vanden Boer, P.; Malovichko, A.; Alioscha-Perez, M.; Radotić, K.; Bartolić, D.; Kalauzi, A.; Villalba, M.I.; Sanglard, D.; Dietler, G.; et al. Single Yeast Cell Nanomotions Correlate with Cellular Activity. Sci. Adv. 2020, 6, eaba3139. [Google Scholar] [CrossRef]
- Venturelli, L.; Kohler, A.-C.; Stupar, P.; Villalba, M.; Kalauzi, A.; Radotic, K.; Bertacchi, M.; Dinarelli, S.; Girasole, M.; Pesic, M.; et al. A Perspective View on the Nanomotion Detection of Living Organisms and Its Features. J. Mol. Recognit. 2020, 33, e2849. [Google Scholar] [CrossRef]
- Hostetter, M.K. Adhesins and Ligands Involved in the Interaction of Candida Spp. with Epithelial and Endothelial Surfaces. Clin. Microbiol. Rev. 1994, 7, 29–42. [Google Scholar] [CrossRef]
- Calderone, R.A.; Scheld, W.M. Role of Fibronectin in the Pathogenesis of Candidal Infections. Rev. Infect. Dis. 1987, 9 (Suppl. 4), S400–S403. [Google Scholar] [CrossRef]
- Klotz, S.A. Plasma and Extracellular Matrix Proteins Mediate in the Fate of Candida Albicans in the Human Host. Med. Hypotheses 1994, 42, 328–334. [Google Scholar] [CrossRef]
- Pendrak, M.L.; Klotz, S.A. Adherence of Candida Albicans to Host Cells. FEMS Microbiol. Lett. 1995, 129, 103–113. [Google Scholar] [CrossRef][Green Version]
- Kozik, A.; Karkowska-Kuleta, J.; Zajac, D.; Bochenska, O.; Kedracka-Krok, S.; Jankowska, U.; Rapala-Kozik, M. Fibronectin-, Vitronectin- and Laminin-Binding Proteins at the Cell Walls of Candida Parapsilosis and Candida Tropicalis Pathogenic Yeasts. BMC Microbiol. 2015, 15, 197. [Google Scholar] [CrossRef]
- Nett, J.E.; Cabezas-Olcoz, J.; Marchillo, K.; Mosher, D.F.; Andes, D.R. Targeting Fibronectin To Disrupt In Vivo Candida Albicans Biofilms. Antimicrob. Agents Chemother. 2016, 60, 3152–3155. [Google Scholar] [CrossRef]
- Bougnoux, M.-E.; Kac, G.; Aegerter, P.; d’Enfert, C.; Fagon, J.-Y.; Group, C.S. Candidemia and Candiduria in Critically Ill Patients Admitted to Intensive Care Units in France: Incidence, Molecular Diversity, Management and Outcome. Intensive Care Med. 2008, 34, 292–299. [Google Scholar] [CrossRef]
- Sdoudi, K.; Bougnoux, M.-E.; El Hamoumi, R.; Diogo, D.; El Mdaghri, N.; Christophe, d. Aziza Razki Phylogeny and Diversity of Candida Albicans Vaginal Isolates from Three Continents. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 471–480. [Google Scholar]
- Ropars, J.; Maufrais, C.; Diogo, D.; Marcet-Houben, M.; Perin, A.; Sertour, N.; Mosca, K.; Permal, E.; Laval, G.; Bouchier, C.; et al. Gene Flow Contributes to Diversification of the Major Fungal Pathogen Candida Albicans. Nat. Commun. 2018, 9, 2253. [Google Scholar] [CrossRef]
- Gillum, A.M.; Tsay, E.Y.; Kirsch, D.R. Isolation of the Candida Albicans Gene for Orotidine-5’-Phosphate Decarboxylase by Complementation of S. Cerevisiae Ura3 and E. Coli pyrF Mutations. Mol. Gen. Genet. 1984, 198, 179–182. [Google Scholar] [CrossRef]
- Schönherr, F.A.; Sparber, F.; Kirchner, F.R.; Guiducci, E.; Trautwein-Weidner, K.; Gladiator, A.; Sertour, N.; Hetzel, U.; Le, G.T.T.; Pavelka, N.; et al. The Intraspecies Diversity of C. Albicans Triggers Qualitatively and Temporally Distinct Host Responses That Determine the Balance between Commensalism and Pathogenicity. Mucosal Immunol. 2017, 10, 1335–1350. [Google Scholar] [CrossRef]
- Murciano, C.; Moyes, D.L.; Runglall, M.; Tobouti, P.; Islam, A.; Hoyer, L.L.; Naglik, J.R. Evaluation of the Role of Candida Albicans Agglutinin-like Sequence (Als) Proteins in Human Oral Epithelial Cell Interactions. PLoS ONE 2012, 7, e33362. [Google Scholar] [CrossRef]
- Kohler, A.-C.; Venturelli, L.; Kannan, A.; Sanglard, D.; Dietler, G.; Willaert, R.; Kasas, S. Yeast Nanometric Scale Oscillations Highlights Fibronectin Induced Changes in C. Albicans. Ferment. 2020, 6, 28. [Google Scholar] [CrossRef]
- Codling, E.A.; Plank, M.J.; Benhamou, S. Random Walk Models in Biology. J. R. Soc. Interface 2008, 5, 813–834. [Google Scholar] [CrossRef]
- Jung, P.; Mischo, C.E.; Gunaratnam, G.; Spengler, C.; Becker, S.L.; Hube, B.; Jacobs, K.; Bischoff, M. Candida Albicans Adhesion to Central Venous Catheters: Impact of Blood Plasma-Driven Germ Tube Formation and Pathogen-Derived Adhesins. Virulence 2020, 11, 1453–1465. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villalba, M.I.; LeibundGut-Landmann, S.; Bougnoux, M.-E.; d’Enfert, C.; Willaert, R.G.; Kasas, S. Candida albicans Adhesion Measured by Optical Nanomotion Detection. Fermentation 2023, 9, 991. https://doi.org/10.3390/fermentation9110991
Villalba MI, LeibundGut-Landmann S, Bougnoux M-E, d’Enfert C, Willaert RG, Kasas S. Candida albicans Adhesion Measured by Optical Nanomotion Detection. Fermentation. 2023; 9(11):991. https://doi.org/10.3390/fermentation9110991
Chicago/Turabian StyleVillalba, Maria I., Salomé LeibundGut-Landmann, Marie-Elisabeth Bougnoux, Christophe d’Enfert, Ronnie G. Willaert, and Sandor Kasas. 2023. "Candida albicans Adhesion Measured by Optical Nanomotion Detection" Fermentation 9, no. 11: 991. https://doi.org/10.3390/fermentation9110991
APA StyleVillalba, M. I., LeibundGut-Landmann, S., Bougnoux, M.-E., d’Enfert, C., Willaert, R. G., & Kasas, S. (2023). Candida albicans Adhesion Measured by Optical Nanomotion Detection. Fermentation, 9(11), 991. https://doi.org/10.3390/fermentation9110991







