Comparative Analysis of Rumen Bacterial Profiles and Functions during Adaption to Different Phenology (Regreen vs. Grassy) in Alpine Merino Sheep with Two Growing Stages on an Alpine Meadow
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Site, Design, and Sampling Date
2.2. Rumen Fluid Collection
2.3. DNA Extraction, PCR, and Sequencing
2.4. Statistical Analysis
3. Results
3.1. Rumen Bacterial Profiles
3.2. Composition of Rumen Bacterial Structure
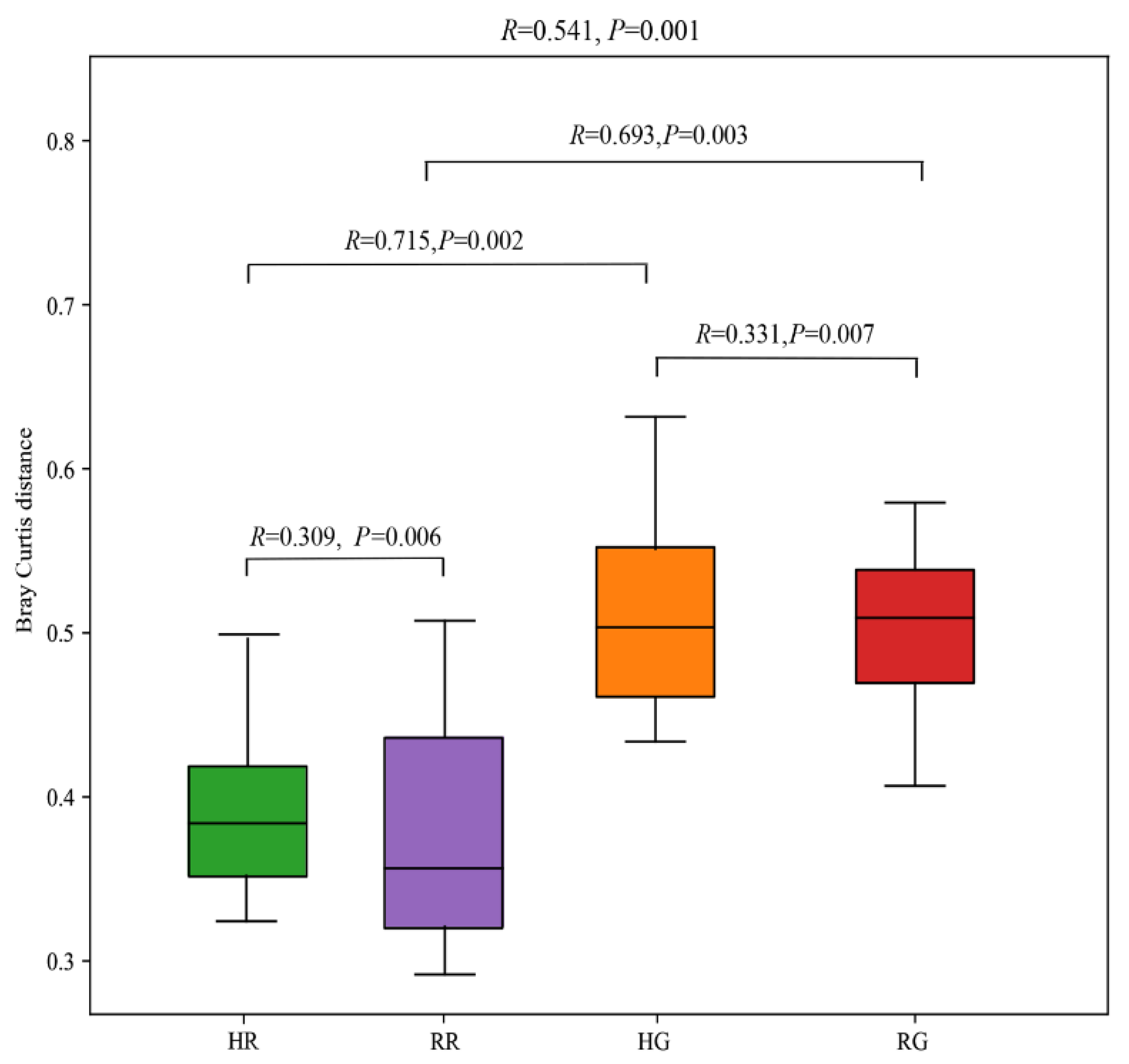
3.3. Forage Nutrition Determined Rumen Bacterial Diversity
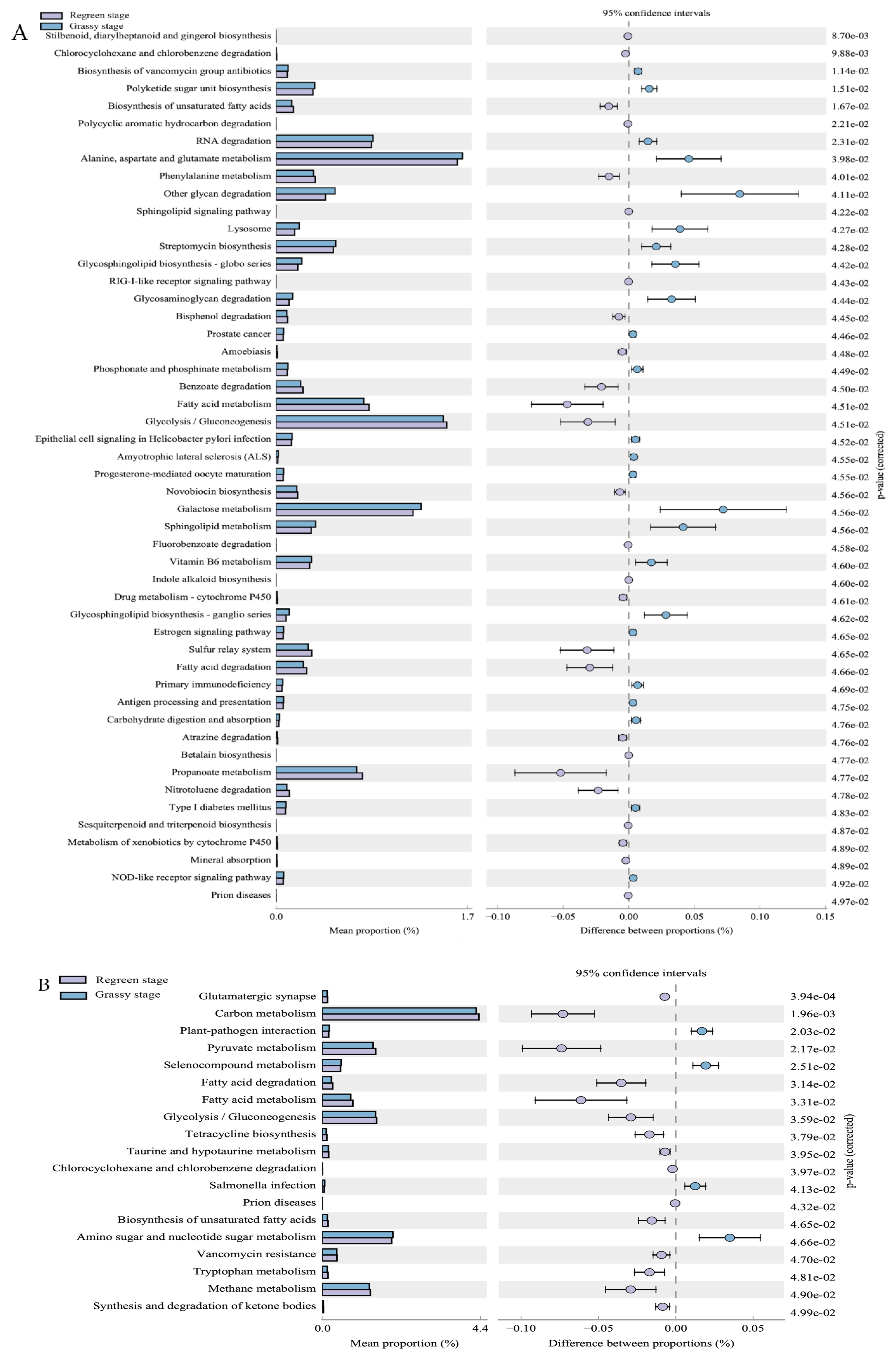
3.4. Function of Rumen Bacteria
4. Discussion
4.1. Effects of the Plant Phenology on Rumen Bacterial Profiles and Functions in Hoggets and Rams Alpine Merino Sheep
4.2. Rumen Bacterial Profiles and Functions in Hoggets and Rams Alpine Merino Sheep
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dewhurst, R.J.; Wadhwa, D.; Borgida, L.P.; Fisher, W.J. Rumen acid production from dairy feeds. 1. Effects on feed intake and milk production of dairy cows offered grass or corn silages. J. Dairy Sci. 2001, 84, 2721–2729. [Google Scholar] [CrossRef] [PubMed]
- Hua, C.; Tian, J.; Tian, P.; Cong, R.; Luo, Y.; Geng, Y.; Tao, S.; Ni, Y.; Zhao, R. Feeding a High Concentration Diet Induces Unhealthy Alterations in the Composition and Metabolism of Ruminal Microbiota and Host Response in a Goat Model. Front. Microbiol. 2017, 8, 138. [Google Scholar] [CrossRef] [PubMed]
- Schären, M.; Kiri, K.; Riede, S.; Gardener, M.; Meyer, U.; Hummel, J.; Urich, T.; Breves, G.; Dänicke, S. Alterations in the rumen liquid-, particle- and epithelium-associated microbiota of dairy cows during the transition from a silage- and concentrate-based ration to pasture in spring. Front. Microbiol. 2017, 8, 744. [Google Scholar] [CrossRef]
- Friedman, N.; Jami, E.; Mizrahi, I. Compositional and functional dynamics of the bovine rumen methanogenic community across different developmental stages. Environ. Microbiol. 2017, 19, 3365–3373. [Google Scholar] [CrossRef] [PubMed]
- Prakash, B.; Saha, S.K.; Khate, K.; Agarwal, N.; Katole, S.; Haque, N.; Rajkhowa, C. Rumen microbial variation and nutrient utilisation in mithun (Bos frontalis) under different feeding regimes. J. Anim. Physiol. An. N. 2013, 97, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Belanche, A.; Kingston-Smith, A.H.; Griffith, G.W.; Newbold, C.J. A multi-kingdom study reveals the plasticity of the rumen microbiota in response to a shift from non-grazing to grazing diets in sheep. Front. Microbiol. 2019, 10, 122. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Sanabria, E.; Goonewardene, L.A.; Wang, Z.; Zhou, M.; Moore, S.S.; Guan, L.L. Influence of sire breed on the interplay among rumen microbial populations inhabiting the rumen liquid of the progeny in beef cattle. PLoS ONE 2013, 8, e58461. [Google Scholar] [CrossRef]
- Amato, K.R.; Leigh, S.R.; Kent, A.; Mackie, R.I.; Yeoman, C.J.; Stumpf, R.M.; Wilson, B.A.; Nelson, K.E.; White, B.A.; Garber, P.A. The gut microbiota appears to compensate for seasonal diet variation in the wild black howler monkey (Alouatta pigra). Microb. Ecol. 2015, 69, 434–443. [Google Scholar] [CrossRef]
- Islam, M.; Kim, S.H.; Son, A.R.; Ramos, S.C.; Jeong, C.D.; Yu, Z.; Kang, S.H.; Cho, Y.I.; Lee, S.S.; Cho, K.K.; et al. Seasonal influence on rumen microbiota, rumen fermentation, and enteric methane emissions of holstein and jersey steers under the same total mixed ration. Animals 2021, 11, 1184. [Google Scholar] [CrossRef]
- Qiao, G.; Zhang, H.; Zhu, S.; Yuan, C.; Zhao, H.; Han, M.; Yue, Y.; Yang, B. The complete mitochondrial genome sequence and phylogenetic analysis of Alpine Merino sheep (Ovis aries). Mitochondrial. DNA B 2020, 5, 990–991. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhou, P.; Shen, X. Effects of se-enriched malt on the immune and antioxidant function in the se-deprived reclamation Merino sheep in southern Xinjiang. Biol. Trace. Elem. Res. 2022, 200, 3621–3629. [Google Scholar] [CrossRef] [PubMed]
- Wolkovich, E.M.; Cook, B.I.; Davies, T.J. Progress towards an interdisciplinary science of plant phenology: Building predictions across space, time and species diversity. New Phytol. 2014, 201, 1156–1162. [Google Scholar] [CrossRef] [PubMed]
- Long, R.J.; Apori, S.O.; Castro, F.B.; Orskov, E.R. Feed value of native forages of the Tibetan Plateau of China. Anim. Feed Sci. Tech. 1999, 80, 101–113. [Google Scholar] [CrossRef]
- Liu, H.; Hu, L.; Han, X.; Zhao, N.; Xu, T.; Ma, L.; Wang, X.; Zhang, X.; Kang, S.; Zhao, X.; et al. Tibetan sheep adapt to plant phenology in alpine meadows by changing rumen microbial community structure and function. Front. Microbiol. 2020, 11, 587558. [Google Scholar] [CrossRef] [PubMed]
- Fan, Q.; Cui, X.; Wang, Z.; Chang, S.; Wanapat, M.; Yan, T.; Hou, F. Rumen microbiota of Tibetan sheep (Ovis aries) adaptation to extremely cold season on the Qinghai-Tibetan Plateau. Front. Vet. Sci. 2021, 8, 673822. [Google Scholar] [CrossRef] [PubMed]
- Koenig, J.E.; Spor, A.; Scalfone, N.; Fricker, A.D.; Stombaugh, J.; Knight, R.; Angenent, L.T.; Ley, R.E. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. USA 2011, 108, 4578–4585. [Google Scholar] [CrossRef] [PubMed]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222. [Google Scholar] [CrossRef]
- Han, X.; Yang, Y.; Yan, H.; Wang, X.; Qu, L.; Chen, Y. Rumen bacterial diversity of 80 to 110-day-old goats using 16S rRNA sequencing. PLoS ONE 2015, 10, e0117811. [Google Scholar] [CrossRef] [PubMed]
- Jiao, J.; Huang, J.; Zhou, C.; Tan, Z. Taxonomic identification of ruminal epithelial bacterial diversity during rumen development in goats. Appl. Environ. Microb. 2015, 81, 3502–3509. [Google Scholar] [CrossRef]
- Li, H.; Yu, Q.; Li, T.; Shao, L.; Su, M.; Zhou, H.; Qu, J. Rumen microbiome and metabolome of Tibetan sheep (Ovis aries) reflect animal age and nutritional requirement. Front. Vet. Sci. 2020, 7, 609. [Google Scholar] [CrossRef]
- Kumar, S.; Indugu, N.; Vecchiarelli, B.; Pitta, D.W. Associative patterns among anaerobic fungi, methanogenic archaea, and bacterial communities in response to changes in diet and age in the rumen of dairy cows. Front. Microbiol. 2015, 6, 781. [Google Scholar] [CrossRef] [PubMed]
- Lavker, R.; Chalupa, W.; Opliger, P. Histochemical observations on rumen mucosa structure and composition. J. Dairy Sci. 1969, 52, 266–269. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Zhang, H.; Wang, H.; Sun, X.; Li, F.; Yang, B. Evaluating grazing nutrition of alpine merino growing rams in the regreening and grassy stages of pasture herbage. Pratacultural Sci. 2022, 39, 171–179. [Google Scholar] [CrossRef]
- Zhang, S.; Zhao, Z.; Sun, X.; Wang, H.; Li, F.; Yang, B. Nutritional surplus or deficiency monitoring of grazing Alpine Merino Breeding rams in regreening stage and growing stage pasture. Chin. J. Anim. Nutr. 2021, 33, 409–419. [Google Scholar] [CrossRef]
- Bainbridge, M.L.; Saldinger, L.K.; Barlow, J.W.; Alvez, J.P.; Roman, J.; Kraft, J. Alteration of rumen bacteria and protozoa through grazing regime as a tool to enhance the bioactive fatty acid content of bovine milk. Front. Microbiol. 2018, 9, 904. [Google Scholar] [CrossRef]
- Zhang, Z.; Xu, D.; Wang, L.; Hao, J.; Wang, J.; Zhou, X.; Wang, W.; Qiu, Q.; Huang, X.; Zhou, J.; et al. Convergent evolution of rumen microbiomes in high-altitude mammals. Curr. Biol. 2016, 26, 1873–1879. [Google Scholar] [CrossRef]
- Sun, B.; Wang, X.; Bernstein, S.; Huffman, M.A.; Xia, D.P.; Gu, Z.; Chen, R.; Sheeran, L.K.; Wagner, R.S.; Li, J. Marked variation between winter and spring gut microbiota in free-ranging Tibetan Macaques (Macaca thibetana). Sci. Rep. 2016, 6, 26035. [Google Scholar] [CrossRef]
- Xiao, X.; Zhang, T.; Angerer, J.P.; Hou, F. Grazing seasons and stocking rates affects the relationship between herbage traits of alpine meadow and grazing behaviors of Tibetan sheep in the Qinghai-Tibetan Plateau. Animals 2020, 10, 488. [Google Scholar] [CrossRef]
- Henderson, G.; Cox, F.; Ganesh, S.; Jonker, A.; Young, W.; Janssen, P.H. Rumen microbial community composition varies with diet and host, but a core microbiome is found across a wide geographical range. Sci. Rep. 2015, 5, 14567. [Google Scholar] [CrossRef]
- Fernando, S.C.; Purvis, H.T., 2nd; Najar, F.Z.; Sukharnikov, L.O.; Krehbiel, C.R.; Nagaraja, T.G.; Roe, B.A.; Desilva, U. Rumen microbial population dynamics during adaptation to a high-grain diet. Appl. Environ. Microb. 2010, 76, 7482–7490. [Google Scholar] [CrossRef]
- Jami, E.; White, B.A.; Mizrahi, I. Potential role of the bovine rumen microbiome in modulating milk composition and feed efficiency. PLoS ONE 2014, 9, e85423. [Google Scholar] [CrossRef] [PubMed]
- Nuriel-Ohayon, M.; Neuman, H.; Koren, O. Microbial changes during pregnancy, birth, and infancy. Front. Microbiol. 2016, 7, 1031. [Google Scholar] [CrossRef] [PubMed]
- Kaakoush, N.O. Insights into the role of Erysipelotrichaceae in the human host. Front. Cell. Infect. Mi. 2015, 5, 84. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Microbial ecology: Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef]
- Murphy, E.F.; Cotter, P.D.; Healy, S.; Marques, T.M.; O’Sullivan, O.; Fouhy, F.; Clarke, S.F.; O’Toole, P.W.; Quigley, E.M.; Stanton, C.; et al. Composition and energy harvesting capacity of the gut microbiota: Relationship to diet, obesity and time in mouse models. Gut 2010, 59, 1635–1642. [Google Scholar] [CrossRef]
- Rabee, A.E.; Alahl, A.A.S.; Lamara, M.; Ishaq, S.L. Fibrolytic rumen bacteria of camel and sheep and their applications in the bioconversion of barley straw to soluble sugars for biofuel production. PLoS ONE 2022, 17, e0262304. [Google Scholar] [CrossRef]
- Gholizadeh, M.; Fayazi, J.; Asgari, Y.; Zali, H.; Kaderali, L. Reconstruction and analysis of cattle metabolic networks in normal and acidosis rumen tissue. Animals 2020, 10, 469. [Google Scholar] [CrossRef]
- Zhu, W.; Su, Z.; Xu, W.; Sun, H.X.; Gao, J.F.; Tu, D.F.; Ren, C.H.; Zhang, Z.J.; Cao, H.G. Garlic skin induces shifts in the rumen microbiome and metabolome of fattening lambs. Animal 2021, 15, 100216. [Google Scholar] [CrossRef]
- Wang, B.; Ma, M.P.; Diao, Q.Y.; Tu, Y. Saponin-induced shifts in the rumen microbiome and metabolome of young cattle. Front. Microbiol. 2019, 10, 356. [Google Scholar] [CrossRef]
- Dervishi, E.; Serrano, C.; Joy, M.; Serrano, M.; Rodellar, C.; Calvo, J.H. The effect of feeding system in the expression of genes related with fat metabolism in semitendinous muscle in sheep. Meat Sci. 2011, 89, 91–97. [Google Scholar] [CrossRef]
- Jami, E.; Israel, A.; Kotser, A.; Mizrahi, I. Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. 2013, 7, 1069–1079. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Shah, A.M.; Wang, L.; Jin, L.; Wang, Z.; Xue, B.; Peng, Q. Relationship between the true digestibility of dietary calcium and gastrointestinal microorganisms in goats. Animals 2020, 10, 875. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.; Song, P.; Lin, G.; Huang, Y.; Wang, L.; Zhou, X.; Li, S.; Zhang, T. Gut microbiota plasticity influences the adaptability of wild and domestic animals in co-inhabited areas. Front. Microbiol. 2020, 11, 125. [Google Scholar] [CrossRef] [PubMed]
| Regreen Stage | Grassy Stage | p-Value 2 | ||||||
|---|---|---|---|---|---|---|---|---|
| Item | H | R | H | R | SEM 1 | P | G | P × G |
| OTUs no. 3 | 1057.2 a | 1034.8 a | 828.3 b | 965.8 a | 16.100 | <0.001 | 0.089 | 0.022 |
| ACE 4 | 1093.9 a | 1070.5 a | 873.2 b | 1016.4 a | 15.364 | <0.001 | 0.066 | 0.013 |
| Chao 4 | 1105.4 a | 1081.2 a | 883.7 b | 1024.8 a | 15.512 | <0.001 | 0.074 | 0.015 |
| Simpson 5 | 0.008 ab | 0.007 b | 0.012 a | 0.011 ab | 0.001 | 0.010 | 0.466 | 0.960 |
| Shannon 5 | 5.84 a | 5.86 a | 5.41 b | 5.57 b | 0.039 | <0.001 | 0.305 | 0.375 |
| Regreen Stage | Grassy Stage | p-Value 2 | ||||||
|---|---|---|---|---|---|---|---|---|
| Bacterial Taxa, % | H | R | H | R | SEM 1 | P | G | P × G |
| Bacteroidetes | 44.55 b | 49.72 ab | 55.38 a | 52.61 a | 1.030 | 0.003 | 0.566 | 0.068 |
| Prevotella_1 | 16.04 | 19.45 | 21.56 | 19.32 | 0.956 | 0.174 | 0.764 | 0.155 |
| Rikenellaceae_RC9_gut_group | 8.02 | 10.46 | 9.44 | 9.93 | 0.625 | 0.725 | 0.256 | 0.444 |
| Prevotellaceae_UCG-001 | 1.67 | 1.59 | 1.59 | 1.16 | 0.148 | 0.405 | 0.405 | 0.553 |
| Prevotellaceae_UCG-003 | 0.86 | 1.36 | 1.54 | 1.40 | 0.112 | 0.126 | 0.426 | 0.173 |
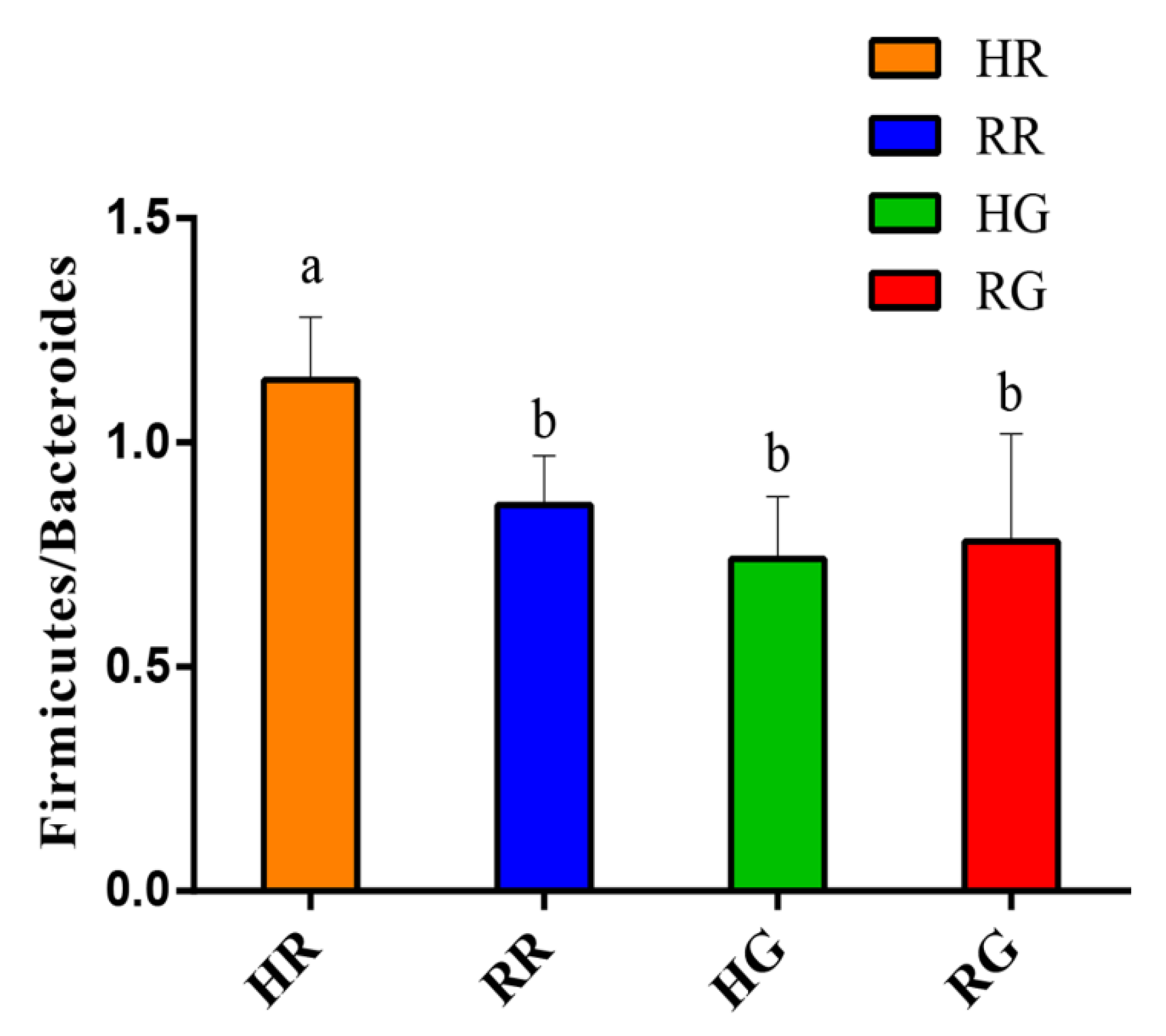
| Firmicutes | 49.62 a | 42.43 b | 40.21 b | 39.53 b | 1.185 | 0.017 | 0.112 | 0.185 |
| Christensenellaceae_R-7_group | 5.96 a | 4.37 ab | 3.14 b | 2.99 b | 0.317 | 0.004 | 0.185 | 0.271 |
| Succiniclasticum | 5.73 a | 4.89 ab | 3.92 ab | 3.05 b | 0.358 | 0.019 | 0.244 | 0.987 |
| Ruminococcaceae_NK4A214_group | 4.33 | 3.13 | 3.01 | 3.53 | 0.331 | 0.497 | 0.613 | 0.210 |
| Ruminococcus_1 | 2.45 a | 2.47 a | 1.26 b | 0.94 b | 0.122 | <0.001 | 0.546 | 0.507 |
| Butyrivibrio_2 | 2.30 | 1.69 | 2.35 | 2.44 | 0.156 | 0.212 | 0.423 | 0.274 |
| Saccharofermentans | 2.10 a | 1.98 a | 0.71 b | 0.77 b | 0.089 | <0.001 | 0.850 | 0.629 |
| Ruminococcaceae_UCG-014 | 1.76 ab | 2.45 a | 0.92 b | 1.33 b | 0.151 | 0.004 | 0.086 | 0.650 |
| Pseudobutyrivibrio | 1.72 | 1.35 | 1.67 | 2.22 | 0.141 | 0.161 | 0.748 | 0.123 |
| Selenomonas_1 | 1.72 b | 0.60 b | 3.51 a | 3.22 a | 0.237 | <0.001 | 0.151 | 0.388 |
| Lachnospiraceae_AC2044_group | 1.71 a | 1.65 a | 0.83 b | 1.15 ab | 0.100 | 0.002 | 0.522 | 0.343 |
| Ruminococcaceae_UCG-010 | 1.58 a | 1.86 a | 1.15 b | 1.11 b | 0.056 | <0.001 | 0.285 | 0.166 |
| [Eubacterium]_coprostanoligenes_group | 1.31 b | 1.73 a | 1.03 bc | 0.88 c | 0.066 | <0.001 | 0.302 | 0.041 |
| Roseburia | 0.91 | 0.57 | 1.51 | 1.32 | 0.139 | 0.025 | 0.364 | 0.785 |
| Proteobacteria | 0.71 | 0.80 | 1.05 | 1.55 | 0.220 | 0.227 | 0.516 | 0.646 |
| Candidatus_Saccharimonas | 1.16 b | 1.92 a | 0.29 c | 1.08 b | 0.070 | <0.001 | <0.001 | 0.913 |
| SR1 | 1.26 | 2.07 | 0.95 | 1.17 | 0.235 | 0.214 | 0.287 | 0.533 |
| Saccharibacteria | 1.16 b | 1.92 a | 0.29 c | 1.08 b | 0.070 | <0.001 | <0.001 | 0.913 |
| Hogget | Ram | |||
|---|---|---|---|---|
| Forage Nutrition 1 | r2 | p | r2 | p |
| Ash | 0.619 | 0.003 | 0.552 | 0.002 |
| CP | 0.566 | 0.002 | 0.520 | 0.004 |
| NDF | 0.147 | 0.145 | 0.202 | 0.080 |
| ADF | 0.163 | 0.145 | 0.071 | 0.314 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, X.; Wang, H. Comparative Analysis of Rumen Bacterial Profiles and Functions during Adaption to Different Phenology (Regreen vs. Grassy) in Alpine Merino Sheep with Two Growing Stages on an Alpine Meadow. Fermentation 2023, 9, 16. https://doi.org/10.3390/fermentation9010016
Gao X, Wang H. Comparative Analysis of Rumen Bacterial Profiles and Functions during Adaption to Different Phenology (Regreen vs. Grassy) in Alpine Merino Sheep with Two Growing Stages on an Alpine Meadow. Fermentation. 2023; 9(1):16. https://doi.org/10.3390/fermentation9010016
Chicago/Turabian StyleGao, Xiang, and Hucheng Wang. 2023. "Comparative Analysis of Rumen Bacterial Profiles and Functions during Adaption to Different Phenology (Regreen vs. Grassy) in Alpine Merino Sheep with Two Growing Stages on an Alpine Meadow" Fermentation 9, no. 1: 16. https://doi.org/10.3390/fermentation9010016
APA StyleGao, X., & Wang, H. (2023). Comparative Analysis of Rumen Bacterial Profiles and Functions during Adaption to Different Phenology (Regreen vs. Grassy) in Alpine Merino Sheep with Two Growing Stages on an Alpine Meadow. Fermentation, 9(1), 16. https://doi.org/10.3390/fermentation9010016





