Abstract
Winemaking is a well-known process that includes several steps in the production of grape must and wine. Grape marc, or bagasse, is a byproduct of wine production that can be vermicomposted and used as organic fertilizer. Grape marc vermicompost has microbial communities that are richer and more stable than grape marc alone, and its addition to a vineyard’s terroir can improve grape yields and wine quality. Here we compare the must and wine microbiota of Mencía from grapevines treated with and without (standard fertilization) vermicompost derived from Mencía grape marc. Mencía is a high-quality red wine broadly grown in Galicia, Spain, and is appreciated for its fresh acidity and fruity flavors. When Mencía grapevines are treated with vermicompost derived from its grape marc, Mencía vines increase their grape production, and the final wine improves its organoleptic properties. Metataxonomic analyses of the bacterial 16S rRNA and fungal ITS gene regions showed that Mencía must and wine have the distinct taxonomic composition (phyla, genera and ASVs—amplicon sequence variants) of bacterial and fungal groups. Must and wine bacteriotas and mycobiotas show no significant variation in alpha-diversity, while wine bacteriotas and mycobiotas show significant differences in microbial structure (beta-diversity) between treated and control grapevines. Likewise, the functional diversity and predicted metabolic pathways (biosynthesis, degradation/utilization/assimilation, generation of precursor metabolites and energy, macromolecule modification and superpathways) of the must and wine microbiota also show significant changes. Our study proposes that changes in the abundance of microbial taxa and the metabolic processes they undergo during winemaking may improve Mencía’s organoleptic properties and productivity.
1. Introduction
Winemaking is a well-developed industry with a thousand years of history, based on the growth of vines with different traits. These traits may vary by grape type, soil composition and climate []. Winemaking involves both the management of the vineyard and the production of wine in the winery. The vinification process begins with the pruning, harvesting, crushing and destemming of the grapes to produce the must, followed by the fermentation process, sediment decantation, maturation and stabilization to produce the finished wine. During wine production, grape marc (a byproduct that contains the skin, stalks and seeds that remain after pressing) is produced and accounts for up to 25% of the weight of processed grapes []. Despite being unsafe to the environment, wineries waste the great majority of the grape marc by discarding it in local environments []. Vermicomposting of grape marc has been demonstrated to be an extremely valuable methodology that yields an organic fertilizer and grape seeds as a source of bioactive compounds [,,]. Additionally, vermicomposting of grape marc can result in soil additives that are rich in nutrients, have a high capacity for retaining water and have a low C:N ratio []. Several studies have shown the positive effects of using bagasse vermicompost as a soil additive, including the potential to stimulate plant growth, seed germination and development, as well as the ability to reduce or suppress plant diseases [,,]. These positive effects have been associated with plant quality improvements, such as increased shoot and leaf area, root development, plant flowering and fruit production []. Besides, bagasse vermicompost is a microbiologically active organic amendment that includes richer and more stable microbial networks with significant functional aids for fertilized plants [,].
The microbial terroir of a specific vineyard displays central roles in stress mitigation and pathogen inhibition in the plant as well as in the availability of organic matter and essential nutrients in the soil []. Microbial terroir is also unique, with various bacterial and fungal microbiomes of the must being related to specific biogeographic regions []. Furthermore, the microbial terroir impacts the variety of microorganisms that, concomitantly, also influence the fermentation processes and wine maturation []. In this regard, it serves as the reservoir of the grapevine microbiome, which displays complex interactions and is believed to be a vital component in shaping grape attributes and wine quality []. For instance, it has been proposed that the vine microbiome impacts certain traits, for example, productivity and grape flavor, affecting the wine’s organoleptic characteristics [].
Mencía grapevines are grown mainly in the Ribeira Sacra, a Spanish Denominación de Origen Protegida (DOP) (Protected Designation of Origin) for wines located in the south of the province of Lugo and in the north of the province of Ourense, Galicia, Spain. It extends over the territories of 20 different municipalities that conform to a zone and entity called Ribeira Sacra (Sacred Shore). The vineyards are planted on the steep slopes of the valleys and canyons of the rivers Miño and Sil. It is an area of wine production with an extension of 2500 hectares of vineyards, which represents 5.2% of the land dedicated to vineyards in Galicia. The bioclimate of Ribeira Sacra is characterized by the Atlantic influence on the marked Mediterranean character of this area, which moderates the summer aridity, favoring mild temperatures and relative humidity during the ripening of the grapes. Mencía is a red wine made from the Mencía grape, a millenary variety native to northwestern Spain. Mencía grapes create wines with an intense raspberry color, penetrating fruit and floral aromas, with a characteristic velvety palate, high alcohol levels, natural acidity and aging possibilities. Ribeira Sacra has currently c.a. 100 wineries, yielding 6.5 million kg of grapes per year, resulting in 4.3 million liters of wine in 2020 [].
Given the impact of the microbial terroir on the wine’s organoleptic properties and the effect of vermicomposting on the microbiome of the soil and plant, we investigated a novel approach to vine fertilization. We collected and vermicomposted the grape marc of Mencía vines and used it to supplement the soil of the same grapevines the following year. Fertilization with and without vermicompost was then compared. Fertilization with vermicompost increased Mencía grape yield by ~14% in 2018 and 2019, as well as wine quality (unpublished data). Wine blind tastings performed by the wineries further showed noticeable contrasts in organoleptic properties (higher color intensity, overall increased aromatic and fruity expression complexity, freshness and balance with the better visual intensity and taste persistency) that gave the treated wine better ratings and reviews (unpublished data).
Given the results above, our goal here was to study the microbial communities involved in Mencía winemaking. We used amplicon sequence data and metataxonomics to characterize the bacteriota and mycobiota of the must and finished wine from Mencía grapevines treated with vermicompost derived from grape marc and controls (standard fertilization) during two consecutive years. We tested for significant differences in the composition, structure and predicted metabolic functions of those microbiotas. These new insights will improve our understanding of the microbe-grapevine interactions and the beneficial role of vermicompost supplementation in winemaking.
2. Materials and Methods
2.1. Experimental Design, Sampling and Processing
The vineyard experiment was conducted in the commercial Mencía vineyard Adegas Moure located in A Cova, O Saviñao, Lugo, Spain (Latitude: 42.56612, Longitude: −7.67693) in 2018 and 2019. The vines, Vitis vinifera cultivar Mencía, were 30 years old, with 1.2 m × 1.75 m intra- and inter-row spacing and trained to a vertical trellis on a single cordon system. Mencía grape marc was vermicomposted and used as soil supplement in the same vines for three consecutive years (from 2017 to 2019). Vermicomposting of grape bagasse from the red variety Mencía (Vitis vinifera sp.) was carried out in a rectangular metal pilot-scale vermireactor (4 m long × 1.5 m wide × 1 m high) housed in a greenhouse with no temperature control at the facilities of the soil ecology group at the University of Vigo. The vermireactor set-up has been described in previous studies (e.g., [,,,]). We used a complete block design, including 144 blocks of 5 plants in the same parcels. Thus, a total of 720 grapevines were divided into the following two groups: (a) control, subject to the standard management practices of the commercial winery (n = 72 blocks × 5 samples), and (b) treatment, same as controls with the addition of vermicompost derived from Mencía grape marc to the surrounding soil of the vines (n = 72 blocks × 5 samples).
In 2018 and 2019, harvesting was performed manually. The grapes were loaded onto tractors that brought them to the winery. Due to the short distance from the plot to the winery, the grapes did not heat up or oxidize. Once in the winery, all the grapes were selected on sorting tables. This process was carried out carefully to avoid damaging the grapes until they entered the tank. First the grapes from the vines treated with vermicompost were processed, selected, destemmed and dropped by gravity, filling the tank with approximately 1500 kg of grapes. Must samples were collected after pressing the grapes (t = 0), and densities were 1090 g/mL and 1091 g/mL (2018) and 1102 g/mL and 1098 g/mL (2019) for control and treatment musts, respectively. The tank was kept at a controlled temperature throughout the fermentation process and never exceeded 25 °C to preserve all the varietal volatile compounds. Once filled, the tank was left slightly open, and fermentation began the same day and lasted three weeks. Subsequently, the same process was carried out for the control wine, with grapes from the same plot as the grapes fertilized with vermicompost but using another tank. Fermentation took place at the same time for both wines. Selected yeasts were not seeded (i.e., spontaneous fermentation), so wild yeasts carried out the process in both the control and experimental wines. No fermentation supplements of any kind were added until the malolactic fermentation was completed. During fermentation, the cap was plunged (i.e., punch-down method) twice a day and every 12 h in both tanks at the same time. Wine samples were collected after malolactic fermentation (t = 29 days) and densities were 0983 g/mL and 0991 g/mL (2018) and 1013 g/mL and 1021 g/mL (2019) for control and treatment wines, respectively.
Adegas Moure made an experimental Mencía wine (1000 L) elaborated with grapes from vines fertilized with vermicompost derived from Mencía grape marc and a control wine (1000 L) elaborated with grapes from vines from the same plot but without vermicompost. In sum, wine samples were taken from two tanks of 1000 L each, corresponding to the two fertilization treatments. Both tanks had the same structure and jacket surface. Otherwise, both wines were made following the same standard procedures of the winery. In the control and treatment wines, the following parameters were determined in the winery: density, reducing sugars, total acidity, pH, % vol, malic acid, volatile acidity (Foss) and free and total sulfur dioxide by the Ripper method. All analyses were carried out in triplicate and in parallel for both the control and treatment wines.
Three replicates of each treatment (control and treated grapevines) were taken in two consecutive harvests in 2018 and 2019, resulting in twelve samples (50 mL) of must and twelve samples of wine. Thus, a total of 24 samples were processed using the VINEO™ Extract DNA Kit, following manufacturer instructions. Each sample was amplified and sequenced for the 16S rRNA gene (~250 bp) to characterize the bacteriota and for the ITS rRNA gene (~250 bp) to characterize the mycobiota. We followed the Earth Microbiome Project protocols for amplification and amplicon sequencing (https://www.protocols.io/workspaces/earth-microbiome-project, accessed on 11 October 2021). Amplicon libraries were created using primers for the V4 hypervariable region of the 16S rRNA gene (forward GTGYCAGCMGCCGCGGTAA and reverse GGACTACNVGGGTWTCTAAT) and a fragment of the ITS rRNA gene (ITS1f forward primer CTTGGTCATTTAGAGGAAGTAA and ITS2 reverse primer GCTGCGTTCTTCATCGATGC). All amplicon libraries were pooled and sequenced in a single run of the Illumina MiSeq sequencing platform at the Argonne National Laboratory, Illinois. The raw sequences are available in the NCBI Sequence Read Archive (SRA) database within the BioProject ID PRJNA819376.
2.2. Data Processing and Statistical Analysis
DADA2 (version 1.16) was used to estimate the amplicon sequence variants (ASVs) present in each sample []. Bioinformatics processing followed the DADA2 pipeline tutorial (https://benjjneb.github.io/dada2/tutorial.html, accessed on 25 October 2021). The 16S forward and reverse read pairs were truncated at 145 nt, no ambiguous bases allowed and each read required to have fewer than two expected errors based on their quality scores. A midpoint rooted tree of ASVs was estimated in FastTree2 []. ASVs were independently inferred from the forward and reverse of each sample using the run-specific error rates, and then read pairs were merged. Chimeras were identified in each sample and removed if identified in a sufficient fraction of the samples in which they were present. We processed ITS reads similarly, but we did not trim them. Taxonomic assignment was performed against the Silva v138 and UNITE v8.2 databases for 16S and ITS, respectively, using the implementation of the RDP naive Bayesian classifier available in the DADA2 R package [,]. We normalized ASV abundances using the negative binomial distribution [], which accounts for library size differences and biological variability. Microbial taxa showing a mean relative proportion ≥ 2% were considered part of the most abundant taxa in the bacteriota and mycobiota of each group. Differential abundances of ASVs between selected sample pairs were estimated using the Wald test with Cook’s distance correction for outliers (DESeq2 package), rare ASVs (n ≤ 10) removed and p-values adjusted by a false discovery rate of <0.01.
Microbial alpha-diversity was calculated at the ASV level using Shannon, Faith’s phylogenetic (PD) and Chao1 diversity indices as implemented in the R phyloseq package []. Microbiome structure (beta-diversity) was also estimated at the ASV level using phylogenetic UniFrac weighted and Bray–Curtis distances. Variation in microbial alpha-diversity between groups was assessed using linear models (lm) as implemented in the R stats package []. Our goal was to assess whether microbial diversity varied between fertilization methods (predictor) in must and wine across harvest years (random factor). However, since harvest years only have two levels (2018 and 2019), we fitted it as a fixed effect to control for its effect on fertilization; hence the final lm formula was expressed as follows: microbial diversity ~ fertilization + harvest years. Variation in microbial beta-diversity was assessed using permutational multivariate analysis of variance (PERMANOVA) through the Adonis function of the R vegan package []. As before, we used both cultivation and harvest years as fixed factors and ran 1000 permutations. Dissimilarity between samples was explored through principal coordinates analysis (PCoA). All analyses were performed in R-studio v4.0.2.
Bacterial metabolic functions were predicted using the metagenomic Phylogenetic Investigation of Communities by Reconstruction of Unobserved States software (PICRUSt2) [] and applying a weighted nearest sequenced taxon index (NSTI) cutoff of 0.03. Predicted metagenomes were collapsed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway metadata []. The linear discriminant analysis (LDA) implemented in LEfSe was used to identify differentially abundant metabolic pathways in the bacteriota of must and wine using fertilization method as a class, a p-value cut-off of 0.05 and an LDA effect size cutoff of 2 [].
3. Results
Data collected by the winery showed that Mencía grapevines fertilized with vermicompost derived from grape marc increased their grape yield by ~14% in 2018 and 2019 compared to standard grapevines. Blind wine tastings carried out at the winery revealed notable differences in organoleptic properties (e.g., overall increased complexity, expression, freshness and balance, with better visual intensity and taste persistency) that led to the wine made using grapes from the treated vines received better ratings and reviews. Interestingly, treated grapevines showed slightly different values for all the measured key chemical characteristics of the must and wine-to-standard (controls) grapevines (Table S1). In short, must from the treated vines was richer in sugar (glucose:fructose), lower total acidity, slightly higher pH, lower malic acid and slightly higher readily assimilable nitrogen. Wine from the treated vines was slightly poorer in reducing sugars, slightly higher in total acidity, slightly lower pH, slightly lower alcohol, slightly lower malic acid and slightly higher volatile acidity (2019), but slightly higher malic acid and slightly lower volatile acidity (2018), and slightly higher free and total sulfur dioxide (Table S2). Given these results, we characterized and compared the bacteriota and mycobiota of standard and vermicompost Mencía must and wine and present the results in the following sections.
3.1. Composition of the Bacteriota and Mycobiota of Standard and Vermicompost Mencía Must and Wine Differ
There were a total of 14 bacterial phyla and 121 bacterial genera across all samples. When looking at the mycobiota, 3 phyla and 69 genera were recovered from all samples. In the bacteriota, proteobacteria (35 ± 22%) were the most abundant bacterial phyla in all groups, while cyanobacteria (66 ± 5%) were highly abundant in must and firmicutes (62 ± 31%) were highly abundant in wine (Figure 1A). Additionally, the phylum Actinobacteriota (2%) was dominant in the wine control group in 2018 (Figure 1A). In the mycobiota, the phylum ascomycota (96 ± 6%) was highly abundant in all groups, while basidiomycota (9 ± 8%) dominated both the wine 2018 groups as well as the wine treatment group in 2019 (Figure 1B).
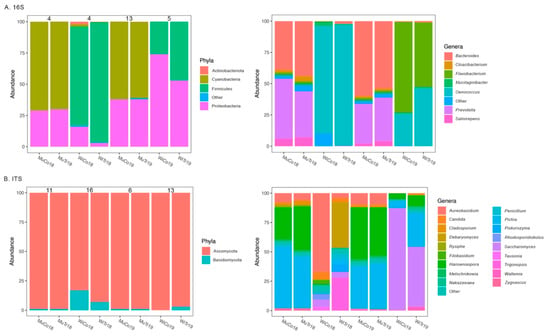
Figure 1.
Most abundant (>2%) phyla and genera of the bacteriota (A) and mycobiota (B) of the Mencía must and wine belonging to control and treatment groups harvested in 2018 and 2019. Distinct bars represent the relative abundance of each taxa. Numbers above bars indicate the number of shared ASVs between control and treatment groups for each grape state and harvest year, according to Wald test results. Each group of samples (n = 3) is labeled according to the state of the grape (Mu—must; Wi—wine), cultivation method (Co—control; Tr—treatment) and harvest year (18—2018; 19—2019).
Regarding the most abundant genera, there was a high variation between groups in both the bacteriota and mycobiota (Figure 1). Nevertheless, it was possible to see a distinction in bacterial genera dominance when comparing cultivation (control vs. treatment), grape state (must vs. wine) and harvest year (2018 vs. 2019) (Figure 1). For instance, Bacteroides and Prevotella were only dominant in the bacteriota of the must, while Oenococcus was only dominant in the bacteriota of the wine (Figure 1A). In the mycobiota, Saccharomyces was highly abundant in the wine groups (Figure 1B).
Results from the Wald test revealed that between 4 and 13 bacterial ASVs and between 6 and 16 fungal ASVs varied significantly between control and treatment groups, depending on the grape state and harvest year (Figure 1). When comparing fertilization methods in the must, there were 68 (48%) and 110 (60%) unique ASVs in control and treatment groups for the bacteriota, respectively; 36 (31%) and 46 (37%) unique ASVs in control and treatment groups for the mycobiota, respectively (Figure 2). In the bacteriota of the must, the top unique ASVs belonged to the genera Komagataeibacter, Neorhizobium, Nguyenibacter, Sphingomonas and Stenotrophomonas in the treatment group and to the genera Hymenobacter and Methylobacterium-Methylorubrum in the control group. In the mycobiota of the must, the top unique ASVs belonged to the genera Hanseniaspora and Metschnikowia in the control and treatment groups, respectively. Within the wine microbiome, there were 129 (81%) and 65 (68%) unique ASVs in the bacterial control and treatment groups, respectively; 55 (58%) and 55 (58%) unique ASVs in the mycobiota control and treatment groups, respectively (Figure 2). In the bacteriota of the wine, the top unique ASVs belonged to the genus Komagataeibacter in the treatment group and to the genera Acetobacter, Corynebacterium, Pseudomonas and Sphingomonas in the control group. In the mycobiota, the top unique ASVs belonged to the genus Candida in the control group; to the genera Candida, Pichia, Rhodotorula and Saccharomyces in the treatment group.
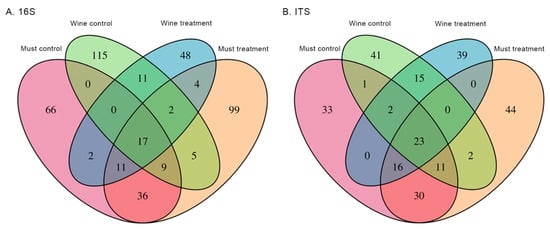
Figure 2.
Venn diagrams showing the number of shared bacterial (A) and fungal (B) ASVs between control and treatment groups of the must and wine Mencía microbiome (n = 6).
3.2. Diversity of the Bacteriota and Mycobiota of Standard and Vermicompost Mencía Must and Wine Differ
In general, must bacteriota and mycobiota showed higher mean values for the alpha-diversity indices than the wine (Figure 3). Overall, alpha-diversity estimates showed no significant differences between control and treatment groups of the bacteriota and mycobiota of the Mencía grapes, with only the following three exceptions (Figure 3; Table S3): the Chao1 diversity of the must bacteriota (p = 0.03, Figure 3A; Table S3), the Faith’s PD index of the wine bacteriota (p = 0.03, Figure 3A; Table S3) and the Shannon diversity of the wine mycobiota (p = 0.003, Figure 3B; Table S3).
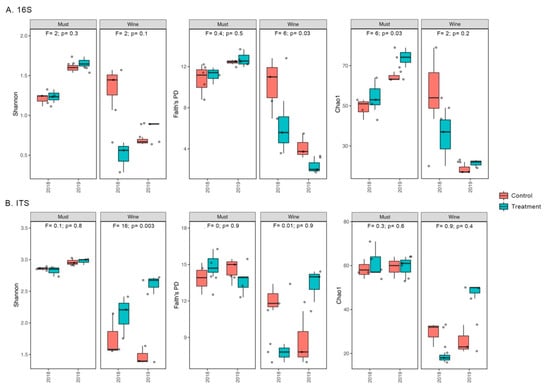
Figure 3.
Mean values and standard deviations of Shannon, Faith’s PD and Chao1 bacterial (A) and fungal (B) alpha-diversity estimates plotted for the control and treatment groups of each grape state of Mencía in each harvest year (n = 3). Results of alpha-diversity comparisons for cultivation method are presented for each grape state, showing the F value and significance (p value).
Beta-diversity estimates showed significant differences between control and treatment groups of the wine bacteriota (p = 0.01, Figure 4A; Table S3) and mycobiota (p = 0.002, Figure 4B; Table S3). Additionally, for the Bray–Curtis distance, must bacteriota showed significant differences between cultivation methods (p = 0.02, Figure 4A; Table S3).
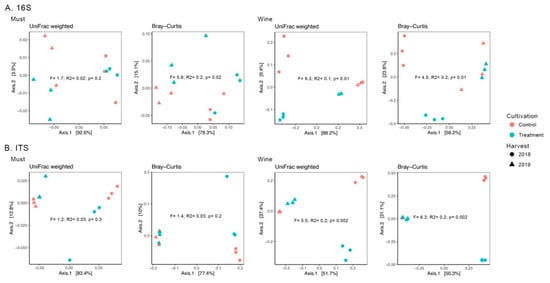
Figure 4.
PCoA plots computed using UniFrac weighted and Bray–Curtis distances. Each dot represents a bacterial (A) and fungal (B) microbiome sample of the Mencía must and wine and is colored by cultivation method. Different shapes represent different harvest years. Results of the PERMANOVA tests for beta-diversity comparisons of the cultivation method are presented, showing the F and R2 values and significance (p value).
3.3. Functional Profiles of the Bacteriota of Standard and Vermicompost Mencía Must and Wine Differ
There were 359 and 377 KEGG predicted pathways inferred in the Mencía must and wine grapes, respectively. Linear discriminant analysis of the metagenomic predictions performed in LEfSe showed that different predicted pathways were significantly enriched for each cultivation method in both must and wine (Figure 5). There were more enriched predicted pathways in the treatment group than in the control group of the must, while in the wine, only the treatment group showed enriched predicted pathways (Figure 5).
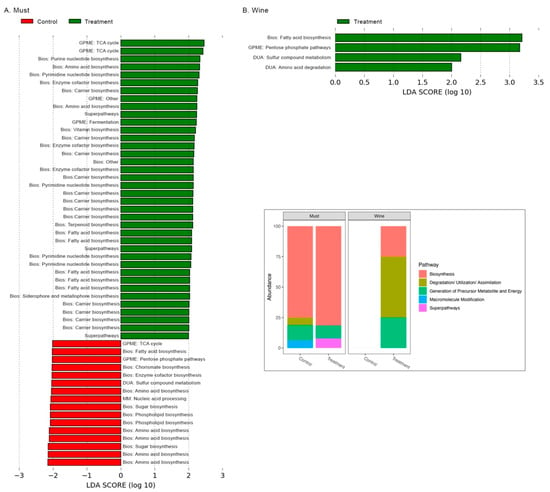
Figure 5.
LDA score and relative frequency (inlet) of differentially abundant enriched pathways in the control and treatment groups of the Mencía must (A) and wine (B). Bios: biosynthesis; DUA: degradation/utilization/assimilation; GPME: generation of precursor metabolites and energy; MM: macromolecule modification.
Predicted pathways related to biosynthesis and generation of precursor metabolites and energy were enriched in both control and treatment groups of the Mencía must (Figure 5A). Additionally, the treatment group of the Mencía must show enriched predicted superpathways, while the control group presents enriched predicted pathways related to degradation/utilization/assimilation and macromolecule modification (Figure 5A). In the treatment group of the Mencía wine, enriched predicted pathways were related to biosynthesis, degradation/utilization/assimilation and the generation of precursor metabolites and energy (Figure 5B).
4. Discussion
In this study, we compared taxonomic and functional microbial profiles in must and wine between Mencía grapevines supplemented with vermicompost derived from grape marc and controls (standard fertilization). Towards this aim, we used 16S and ITS metataxonomics to characterize the bacteriota and mycobiota, respectively, of Mencía must and finished wine harvested for two consecutive years.
4.1. Vermicompost Derived from Grape Marc Changes the Composition, Structure and Diversity of the Bacterial and Fungal Communities in Mencía Must and Wine
Vermicomposting is a highly effective technology capable of stabilizing grape marc [], originating final vermicompost that can be used as organic fertilizer after, promoting plant growth and suppressing plant diseases []. This has been shown to be a result of the microbial properties present in the vermicompost that were more diverse and associated with increases in processes related to cellulose metabolism and the synthesis of antibiotics and hormones (e.g., [,,]). Grape traits such as productivity and flavor and organoleptic characteristics of the wine are thought to be influenced by the specific microbial terroir of a vineyard [,]. In this regard, the microbial qualities of grapevines supplemented with vermicompost should be reflected in the microbiome of must and finished wine. The present study has demonstrated that the bacterial and fungal communities of the Mencía wine and must were significantly altered in the grapevines treated with vermicompost derived from grape marc. Specifically, the composition, diversity and predicted function of the Mencía must and wine microbiome changed when vermicompost was used as part of the fertilization treatment. Additionally, the majority of the ASVs retrieved from the treated Mencía must and wine microbiome were unique (56 ± 13%), indicating that the vermicompost led to a turnover of the microbial composition during the wine-making process.
Since the microbial profiles of the Mencía must and wine were so prominently different between control and treated grapevines, we suggest that the increase in grape production and the subtle improvement in quality noticed in the finished wine were due to changes in the number and abundance of specific microbial taxa. Grape juice fermentation is a process in which wine yeasts transform grape sugars into secondary products []. In this sense, microbial supplementation is a standard procedure in the wine-making industry, where the inclusion of non-Saccharomyces wine yeasts is a tool to take advantage of fermentation to improve wine properties []. For example, inoculation with a Debaryomyces strain was seen to influence the volatiles of a muscat wine, increasing its aromatic potential and flavor release []. Our metataxonomic analysis has detected an increase in the abundance of the fungal genus Debaryomyces in the treated Mencía wine, which could help to enhance the organoleptic properties of the wine. Malolactic fermentation is a bacterially driven process essential in winemaking and related to flavor modification and microbial stability of the wine []. Strains of Oenococcus are the main lactic acid bacteria for conducting malolactic fermentation and decreasing wine sourness (e.g., []). Hence, the upgrowth of Oenococcus observed in the bacteriome of the treated Mencía wine may be one of the taxa associated with the taste improvement observed. These are just two examples of taxa that are known to positively impact winemaking, but other species of the differentially abundant genera observed here in wine and must (see Figure 1 and Section 3) may also exert a positive effect on the final wine and need to be studied.
Although harvest year is not a variable of interest in this study, our analyses also indicate a strong effect of harvest year on the bacterial and fungal communities (Figure 1, Figure 3 and Figure 4). The climate in A Cova during 2018 and 2019 was slightly different. Precipitation in the vineyard from March (sprouting) to September (harvest) was 514 L·m−2 in 2018 and 302 L·m−2 in 2019. The mean temperature (13.19 °C in 2018 and 13.22 °C in 2019) and relative humidity (82.3% in 2018 and 82.05% in 2019) were quite similar in both years. Different environmental factors such as temperature, humidity and precipitation are well known to influence the microbial composition and diversity of must and wine []. In this sense, climatic differences seen in the studied or previous years can explain changes in the microbial composition, diversity and structure observed between harvest years. Ongoing longitudinal studies by our group will help to further assess the impact of climate variation on microbiota and winemaking.
4.2. Vermicompost Derived from Grape Marc Changes the Metabolic Function of the Bacterial Communities in Mencía Must and Wine
Changes observed in microbial composition were accompanied by changes in the predicted metabolic pathway diversity in both must and wine. Specifically, there were a few pathways overexpressed in the Mencía wine treated with bagasse vermicompost, suggesting higher microbial activity in this group. On the other hand, functions related to biosynthesis processes were not only higher but also more diverse on treated Mencía must, suggesting higher activity in the generation reactions of active compounds by microbes in those samples. For example, biosynthesis of terpenoids and vitamins was overexpressed in the treated Mencía must. Terpenoids are thought to play roles in pathogen defense and pollinator attraction in grape berries, as well as create complex flavor and aroma in finished wines [,]. Additionally, vitamin biosynthesis has been suggested to indicate increased nutrient competition and depletion during fermentation []. The observed increase in biosynthesis of these compounds suggests that they might be associated with grape marc vermicomposting during wine cultivation. In fact, predicted functions related to fermentation were also seen to be overexpressed in treated Mencía must. Amino acids are precursors of wine volatile compounds [] and are thought to be associated with wine aroma (e.g., [,]). On the other hand, sulfur compound metabolism can also influence wine aroma []. In the present study, biosynthesis of amino acids, such as histidine, isoleucine and valine, and pathways related to sulfur compound metabolism were overexpressed in the control Mencía must, and these alterations might have modified the wine aroma. Additionally, increased sugar concentrations during fermentation processes were seen to have adverse effects on the wine’s flavor and aroma []. Pathways related to sugar biosynthesis were enriched in the control but not in the treated Mencía must, which might also explain the organoleptic differences observed between the two groups.
5. Conclusions
This study identified significant differences in the bacteriomes and mycobiomes of Mencía must and wine from grapevines supplemented with Mencía grape marc vermicompost compared to their control counterparts—grapevines fertilized only using standard procedures. We hypothesize that observed changes in microbial composition, diversity and predicted metabolic function in the treated Mencía grapevines have contributed to increasing grape production and the quality of the finished wine. We identify some key bacterial and fungal taxa and metabolic pathways that may act as biomarkers of wine quality or benefit wine making if, for example, added as biological additives or via microbial selection. This has important implications for the wine industry on the path to a circular economy and sustainable and regenerative viticulture. Thus, our results encourage the development of an integrated cycle that allows for the conversion of the bagasse produced in the wine industry into high-quality vermicompost with biostimulant and vine defense properties. Sustainability is based on commercial strategies that allow the recovery of valuable products (vermicompost) with minimum disposal of waste streams. Application of the vermicompost to the vineyard ultimately results in the production of more “natural” wines with a distinctive personality.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fermentation8080357/s1, Table S1: Chemical characteristics of musts; Table S2: Chemical characteristics of wines; Table S3: Microbial diversity in the bacteriota (16S) and mycobiota (ITS) of the Albariño must and wine between control and treatment groups.
Author Contributions
Conceptualization, J.D. and M.P.-L.; methodology, I.R.-T., K.A.C., M.L. and D.R.; writing—original draft preparation, D.R.; writing—review and editing, D.R., I.R.-T., K.A.C., M.P.-L. and J.D. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
In The datasets generated and/or analyzed during the current study are available in the NCBI Sequence Read Archive (SRA) database within the BioProject ID PRJNA819376.
Acknowledgments
This study was supported by the Spanish Ministerio de Economía y Competitividad [AGL2017-86813-R] and the Xunta de Galicia [grant number ED431B 2019/038]. We thank Hugo Martínez and Alberto Da Silva for their help with vermicomposting, field work, sample collection and DNA extraction. We thank Adegas Moure for all the collaboration in this study and their continued partnership and interest in our work.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Gilbert, J.A.; Van Der Lelie, D.; Zarraonaindia, I. Microbial terroir for wine grapes. Proc. Natl. Acad. Sci. USA 2014, 111, 5–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beres, C.; Costa, G.N.S.; Cabezudo, I.; da Silva-James, N.K.; Teles, A.S.C.; Cruz, A.P.G.; Mellinger-Silva, C.; Tonon, R.V.; Cabral, L.M.C.; Freitas, S.P. Towards integral utilization of grape pomace from winemaking process: A review. Waste Manag. 2017, 68, 581–594. [Google Scholar] [CrossRef] [PubMed]
- Bordiga, M. Valorization of Wine Making By-Products; CRC Press, Taylor & Francis Group: Boca Raton, FL, USA, 2016. [Google Scholar]
- Domínguez, J.; Martínez-Cordeiro, H.; Álvarez-Casas, M.; Lores, M. Vermicomposting grape marc yields high quality organic biofertiliser and bioactive polyphenols. Waste Manag. Res. 2014, 32, 1235–1240. [Google Scholar] [CrossRef] [PubMed]
- Domínguez, J.; Sanchez-Hernandez, J.C.; Lores, M. Vermicomposting of Winemaking By-Products. In Handbook of Grape Processing By-Product; Academic Press: Cambridge, MA, USA, 2017; pp. 55–78. [Google Scholar] [CrossRef]
- Gómez-Brandón, M.; Lores, M.; Martínez-Cordeiro, H.; Domínguez, J. Effectiveness of vermicomposting for bioconversion of grape marc derived from red winemaking into a value-added product. Environ. Sci. Pollut. Res. 2020, 27, 33438–33445. [Google Scholar] [CrossRef]
- Domínguez, J.; Aira, M.; Kolbe, A.R.; Gómez-Brandón, M.; Pérez-Losada, M. Changes in the composition and function of bacterial communities during vermicomposting may explain beneficial properties of vermicompost. Sci. Rep. 2019, 9, 9657. [Google Scholar] [CrossRef] [Green Version]
- Lazcano, C.; Domínguez, J. The use of vermicompost in sustainable agriculture: Impact on plant growth and soil fertility. In Soil Nutrients; Miransari, M., Ed.; Nova Science Publishers: Hauppauge, NY, USA, 2011; pp. 230–254. ISBN 978-1-61324-785-3. [Google Scholar]
- Pathma, J.; Sakthivel, N. Microbial diversity of vermicompost bacteria that exhibit useful agricultural traits and waste management potential. SpringerPlus 2012, 1, 26. [Google Scholar] [CrossRef] [Green Version]
- Gómez-Brandón, M.; Domínguez, J. Recycling of Solid Organic Wastes through Vermicomposting: Microbial Community Changes Throughout the Process and Use of Vermicompost as a Soil Amendment. Crit. Rev. Environ. Sci. Technol. 2014, 44, 1289–1312. [Google Scholar] [CrossRef]
- Kolbe, A.R.; Aira, M.; Gómez-Brandón, M.; Pérez-Losada, M.; Domínguez, J. Bacterial succession and functional diversity during vermicomposting of the white grape marc Vitis vinifera v. Albariño. Sci. Rep. 2019, 9, 7472. [Google Scholar] [CrossRef]
- Belda, I.; Zarraonaindia, I.; Perisin, M.; Palacios, A.; Acedo, A. From vineyard soil to wine fermentation: Microbiome approximations to explain the “terroir” Concept. Front. Microbiol. 2017, 8, 821. [Google Scholar] [CrossRef] [Green Version]
- Bokulich, N.A.; Thorngate, J.H.; Richardson, P.M.; Mills, D.A. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proc. Natl. Acad. Sci. USA 2014, 111, E139–E148. [Google Scholar] [CrossRef] [Green Version]
- Barata, A.; Malfeito-Ferreira, M.; Loureiro, V. The microbial ecology of wine grape berries. Int. J. Food Microbiol. 2012, 153, 243–259. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Rousseaux, S.; Tourdot-Maréchal, R.; Sadoudi, M.; Gougeon, R.; Schmitt-Kopplin, P.; Alexandre, H. Wine microbiome: A dynamic world of microbial interactions. Crit. Rev. Food Sci. Nutr. 2017, 57, 856–873. [Google Scholar] [CrossRef] [PubMed]
- Xunta de Galicia. Available online: https://mediorural.xunta.gal/es/recursos/estatisticas/estatistica-agraria/2020 (accessed on 7 June 2022).
- Domínguez, J.; Gómez-Brandón, M.; Martínez-Cordeiro, H.; Lores, M. Bioconversion of Scotch broom into a high-quality organic fertiliser: Vermicomposting as a sustainable option. Waste Manag. Res. 2018, 36, 1092–1099. [Google Scholar] [CrossRef]
- Rosado, D.; Pérez-Losada, M.; Aira, M.; Domínguez, J. Bacterial succession during vermicomposting of silver wattle (Acacia dealbata link). Microorganisms 2022, 10, 65. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2–Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [Green Version]
- McMurdie, P.J.; Holmes, S. Waste Not, Want Not: Why Rarefying Microbiome Data Is Inadmissible. PLoS Comput. Biol. 2014, 10, e1003531. [Google Scholar] [CrossRef] [Green Version]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [Green Version]
- Team, R.C. R: A Language and Environment for Statistical Computing. 2012. Available online: https://www.scirp.org/(S(vtj3fa45qm1ean45vvffcz55))/reference/ReferencesPapers.aspx?ReferenceID=1462563 (accessed on 7 June 2022).
- Oksanen, J.; Kindt, R.; Legendre, P.; O’Hara, B.; Simpson, G.L.; Stevens, M.H.H.; Wagner, H. The Vegan Package: Community Ecology Package, Version 1.13-1. Available online: https://www.researchgate.net/publication/323265567_Vegan_Community_Ecology_Package_R_package_version_113-1 (accessed on 7 June 2022).
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2: An improved and extensible approach for metagenome inference. bioRxiv 2019, 672295. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Sato, Y.; Furumichi, M.; Morishima, K.; Tanabe, M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019, 47, D590–D595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [Green Version]
- Gómez-Brandón, M.; Aira, M.; Kolbe, A.R.; de Andrade, N.; Pérez-Losada, M.; Domínguez, J. Rapid Bacterial Community Changes during Vermicomposting of Grape Marc Derived from Red Winemaking. Microorganisms 2019, 7, 473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ciani, M.; Comitini, F.; Mannazzu, I.; Domizio, P. Controlled mixed culture fermentation: A new perspective on the use of non-Saccharomyces yeasts in winemaking. FEMS Yeast Res. 2010, 10, 123–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, A.; Carcel, C.; Dulau, L.; Samson, A.; Aguera, E.; Agosin, E.; Günata, Z. Influence of a mixed culture with Debaryomyces vanriji and Saccharomyces cerevisiae on the volatiles of a Muscat wine. J. Food Sci. 2002, 67, 1138–1143. [Google Scholar] [CrossRef]
- Henick-Kling, T. Malolactic fermentation. In Wine Microbiology and Biotechnology; Fleet, G., Ed.; Harwood Academic Publisher: Amsterdam, The Netherlands, 1993; pp. 289–326. [Google Scholar]
- Bartowsky, E.J.; Borneman, A.R. Genomic variations of Oenococcus oeni strains and the potential to impact on malolactic fermentation and aroma compounds in wine. Appl. Microbiol. Biotechnol. 2011, 92, 441–447. [Google Scholar] [CrossRef]
- Wedler, H.B.; Pemberton, R.P.; Tantillo, D.J. Carbocations and the Complex Flavor and Bouquet of Wine: Mechanistic Aspects of Terpene Biosynthesis in Wine Grapes. Molecules 2015, 20, 10781–10792. [Google Scholar] [CrossRef] [Green Version]
- Schwab, W.; Wüst, M. Understanding the Constitutive and Induced Biosynthesis of Mono- and Sesquiterpenes in Grapes (Vitis vinifera): A Key to Unlocking the Biochemical Secrets of Unique Grape Aroma Profiles. J. Agric. Food Chem. 2015, 63, 10591–10603. [Google Scholar] [CrossRef]
- Reiter, T.; Montpetit, R.; Byer, S.; Frias, I.; Leon, E.; Viano, R.; Mcloughlin, M.; Halligan, T.; Hernandez, D.; Runnebaum, R.; et al. Saccharomyces Cerevisiae Gene Expression during Fermentation of Pinot Noir Wines at an Industrially Relevant Scale. Appl. Environ. Microbiol. 2021, 87, e00036-21. [Google Scholar] [CrossRef]
- Styger, G.; Prior, B.; Bauer, F.F. Wine flavor and aroma. J. Ind. Microbiol. Biotechnol. 2011, 38, 1145. [Google Scholar] [CrossRef] [PubMed]
- Bouzas-Cid, Y.; Trigo-Córdoba, E.; Orriols, I.; Falqué, E.; Mirás-Avalos, J.M. Influence of Soil Management on the Red Grapevine (Vitis vinifera L.) Mencía Must Amino Acid Composition and Wine Volatile and Sensory Profiles in a Humid Region. Beverages 2018, 4, 76. [Google Scholar] [CrossRef] [Green Version]
- Hernández-Orte, P.N.; Cacho, J.F.; Ferreira, V. Relationship between Varietal Amino Acid Profile of Grapes and Wine Aromatic Composition. Experiments with Model Solutions and Chemometric Study. J. Agric. Food Chem. 2002, 50, 2891–2899. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).