Abstract
The ecological uniqueness of the Great Bitter Lake ecosystem makes its bacterial population interesting for investigation. Here, we present the first trial to evaluate the biosynthetic capacity of the bacterial population at the lake as a source of novel antimicrobials. We collected different samples from various locations throughout the lake including the oxic sediment, anoxic sediment, shore water, and off-shore water. We modified a molecular approach to compare and choose the samples with the highest bacterial biosynthetic capacity by quantifying the polyketide synthase gene clusters in their total community DNA. Furthermore, we screened the bacterial isolates recovered from these samples and their metabolic extracts for antimicrobial activity. We tried to tentatively investigate the identity of the active metabolites by PCR screening and LC–MS. The bacterial population in the oxic sediment had the highest biosynthetic capacity compared to other sample types. Four active Bacillus isolates were identified. The isolated Bacillus species were expected to produce numerous probable bioactive metabolites encoded by biosynthetic gene clusters related to the polyketide synthases (either individual or hybrid with non-ribosomal peptide synthetase), such as Bacillomycin D, Iturin A, Bacilosarcin B, Bacillcoumacin G and Macrolactin (N and G). These results suggest that the under-explored bacterial community of the Great Bitter Lake has a prospective biosynthetic capacity and can be a promising source for novel antibiotics.
1. Introduction
The serious health and economic impacts of antimicrobial resistance (AMR) introduce a major concern and global threat facing humans [1]. Therefore, it is more necessary than ever to find novel antibiotics [2,3].
In recent decades, natural products have played a vital role in drug discovery [4]. In addition to animals and plants, microorganisms including bacteria, fungi, and algae are a huge resource for secondary metabolites [5,6]. Over 22,000 bioactive microbial secondary metabolites were identified, and many played a role in drug discovery [7].
These metabolites are synthesized by a wide range of gene clusters called biosynthetic gene clusters (BGCs). The most popular BGCs are the ones coding for polyketide synthases (PKSs) including PKS-I, and PKS-II gene clusters [8,9]. Bacteria, in particular those from the phylum of Actinobacteria and the family Bacillaceae, were described to produce numerous polyketides that have antimicrobial activities [9,10].
With more than two-thirds of Earth’s surface covered with water, where the most primitive forms of life originated, marine microorganisms and their bioactive metabolites have attracted researchers during their active search for new drugs [11,12]. Marine microorganisms are major producers of bioactive compounds that exhibit a wide range of activities such as antimicrobial, antioxidant, antitumor, anti-inflammatory, antivirus, antihypertensive, and anti-diabetic activity [6].
Egypt has a variable collection of water bodies: the Red Sea in the east, and the Mediterranean Sea in the north, in addition to several lakes. The Great Bitter Lake, in the northeast of Egypt, is a saltwater lake. What makes this lake interesting is that it is considered as a live bridge between two seas—the Red and the Mediterranean Seas, each with a remarkably different ecosystem. The two ecosystems were connected by the Suez Canal and through the Great Bitter Lake in 1869.
Currently, the salinity level of the lake is between 41.1 and 44.6%, which is higher than the salinity level of the sea when the canal is open and much higher when it is closed. The salinity of the lake is affected by the water flow from both the Red and the Mediterranean seas and the season, as the salinity is higher in summer than in winter [13,14].
Different culture-dependent and culture-independent approaches were adopted by microbiologists to investigate marine microbial metabolites [15,16].
This study aimed to determine the ability of the marine microbial community in the Great Bitter Lake to be a promising source for novel antimicrobial metabolites by measuring the levels of the bacterial PKSs and BGCs in the samples’ total community DNA and screening the antimicrobial activity of some bacterial isolates recovered from the lake.
2. Materials and Methods
2.1. Samples Collection
Samples were collected on the 5th of April 2020 from the Great Bitter Lake, Fayed, Ismailia, Egypt, at three different locations: L1, L2, and L3 (Figure 1a). Four types of samples were collected from the three locations: shore water (SW) and muddy anoxic sediment (MS) from L1 (2 m depth), sandy oxic sediment (SS) from L2 (6 m depth), and off-shore water (OSW) from L3 (Figure 1b). Each sample was collected in triplicate and stored in sterile falcons with a volume of 50 mL. We started working on these samples within 8 h. A step-wise scheme was followed to evaluate the bacterial population biosynthetic capacity and antimicrobial activity potential.
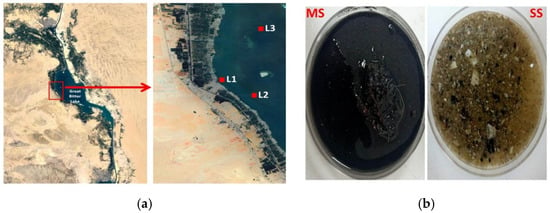
Figure 1.
(a) The map indicates the sampling locations from the Great Bitter Lake—L1: 30°20′08″ N 32°18′25″ E, L2: 30°19′31″ N 32°19′38″ E and L3: 30°21′52″ N 32°19′48″ E. (b) Difference in the physical appearance between the collected sediment types. MS: muddy sediment. SS: sandy sediment.
2.2. Quantification of PKSs Gene Clusters in Samples’ Total Community DNA
Total community DNA was extracted from samples using the DNeasy PowerSoil kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Extracted DNA was stored at −20 °C for further use.
Polyketide synthases (PKS-I and PKS-II), Firmicutes 16S rRNA, and total bacterial 16S rRNA genes were quantified by qPCR using StepOne PlusTM Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA, USA) and sets of degenerate primers. The total bacterial 16S rRNA gene was amplified using Eub338 (forward) and Eub518 (reverse) primers. The Firmicutes 16S rRNA gene was amplified using Firm352F (forward) and Firm525R (reverse) primers. PKS-I was amplified using PKS1-F (forward) and PKS1-R (reverse) primers. PKS-II was amplified using PF6 (forward) and PR6 (reverse) primers. All primers used in this study were obtained from (Invitrogen, Thermo Fisher Scientific, USA). Primers’ sequences are illustrated in (Table 1).

Table 1.
List of primers used in the quantitative PCR.
The qPCR was performed in a 20 µL reaction mixture containing 4 µL of 5× Hot FIREPol® EvaGreen® qPCR Mix Plus (ROX) (Solis BioDyne, Tartu, Estonia), 0.5 µL forward primer (10 pmol·µL−1), 0.5 µL reverse primer (10 pmol·µL−1) and 1 µL from TcDNA. For all, the PCR conditions were carried out as follows: 95 °C for 12 min; 40 cycles with a denaturation step of 95 °C for 15 s, annealing for 30 s at 58 °C; elongation at 72 °C for 30 s; and a final step of 72 °C for 8 min. The purity of the PCR products was checked with the melting curve plots. A standard curve was drawn using ten-fold serial dilutions of the bacterial 16S rRNA gene. The absolute quantity of the total bacterial 16S rRNA and the relative abundance of Firmicutes 16S rRNA and PKSs genes were calculated in the same way as described in Yang et al. [21]. In brief, a standard curve was drawn using ten-fold serial dilutions of the standard bacterial 16S rRNA gene. The absolute of total bacterial 16S rRNA was derived from the curve automatically by the thermocycler. The relative abundance was calculated using the following equation:
where x is the relative abundance. The Eff.Univ and Eff.Spec are the efficacy of the universal and specific primers, respectively. The Ct.univ and Ct.spec are the threshold cycles of universal and specific primers, respectively.
x = 〖(Eff.Univ)〗^(Ct.univ)/〖(Eff.Spec)〗^(Ct.spec) × 100
The statistical analysis of data was performed using one-way ANOVA and Tukey’s test for multiple comparisons. Significance was considered at a p-value of <0.05. Pearson’s correlation analysis was performed between PKS-I and PKS-II.
2.3. Isolation and Purification of Culturable Bacteria
For sediment samples, 1.0 g of each was vortexed with 10 mL of filtered, sterilized seawater for 2 min. For the water ones, they were vortexed for 1 min. A volume of 1 mL of each sample was ten-fold serially diluted with sterile pre-filtered seawater. A volume of 100 µL from each dilution was placed and carefully spread with a sterile glass spreader onto plates of nutrient agar (NA) (Oxoid TM, Basingstoke, UK), Reasoner’s 2A agar (R2A) (DifcoTM, Detroit, MI, USA), Actinomycete isolation agar (AIA) (Oxoid TM, UK), and starch casein agar (SCA) (Sigma-Aldrich, St. Louis, MO, USA). The plates were incubated at 30 °C for 24–72 h, and then colonies were isolated based on the difference in colony morphology. The plates were re-incubated for a longer time (2 to 3 weeks) to observe any new slow-growing strain.
All media were constituted and prepared in filtered, pre-sterilized seawater obtained from the Great Bitter Lakes area, except the R2A, and supplied with 0.001% nystatin to inhibit fungal growth. The total aerobic viable count on nutrient agar was calculated. After that, isolates were purified by repetitive sub-culturing on the corresponding relevant media and then on the R2A medium.
2.4. Standard Indicator Microorganisms
Five different standard microbial strains were used for screening the antimicrobial activity: Escherichia coli ATCC 5087 (E.C), Staphylococcus aureus ATCC 6538 (S.A), Bacillus cereus ATCC 33018 (B.C), Pseudomonas aeurginosa ATCC 27853 (P.A), and Candida albicans ATCC 60193 (C.A).
2.5. Evaluation of the Antimicrobial Activity
Isolates from samples with higher PKS levels were evaluated for their antimicrobial activity. We have followed two levels for screening the antimicrobial activity of the bacterial isolates. First, we screened the antagonistic behavior of isolates. Second, the cell-free extract of isolates, which showed positive microbial antagonism, was screened for antimicrobial activity.
2.5.1. Screening the Antagonistic Behavior of the Bacteria Isolates
The antimicrobial activity of the bacterial isolates was determined by the drop-plate technique as described in Ismail et al. [22] with some modifications. In brief, the tested isolate was inoculated into semisolid nutrient agar and incubated at 30 °C for 48–72 h, then spotted onto agar plates seeded with 500 µL of 0.5 McFarland of the standard microbial strains and incubated at 37 °C for 24 h, except for the standard C. albicans which was incubated at 30 °C for 48 h. The results of microbial antagonism were divided into a clear inhibition zone, attenuated growth, or no inhibition.
2.5.2. Screening the Antimicrobial Activity of the Cell-Free Bacterial Metabolic Extract
The metabolic extract of bacterial isolates that showed positive antagonistic activity was prepared as follows: a fresh colony was inoculated into 100 mL nutrient broth (Oxoid TM, UK) and then incubated with shaking at 160 rpm (C25KC incubator shaker, New Brunswick Scientific, Enfield, CT, USA) at 30 °C for 5–7 days.
The broth culture was centrifuged at 3000 rpm for 10 min (Biobase 800 Centrifuge) and filtered using a 0.22 µm syringe filter.
The cell-free supernatant was then extracted with an equal volume of ethyl acetate (HIMEDIA, AS051-500ML, INDIA) by incubating at room temperature while shaking at 240 rpm for 24 h.
Ethyl acetate was then separated using a separating funnel and evaporated to dryness using a rotavap device (BUCHI 011, Uster, Switzerland) at 45 °C. The residual material was dissolved into 10% dimethyl sulfoxide (DMSO) with a concentration of 20 mg·mL−1. The antimicrobial activity of the extract was assayed by the standard well-diffusion method. A well of 6 mm in a Muller–Hinton agar plate seeded with 500 µL of 0.5 McFarland of the standard microbial strains was filled with 100 µL of the 20 mg·mL−1 of the extract then incubated at 30 °C for 24 h except for C.A, which was incubated for 48 h. A negative control of 10% DMSO was used. For positive control, gentamicin (20 µg·mL−1) was used as an antibacterial and Itraconazol (20 µg·mL−1) was used as an antifungal.
2.6. Isolates Identification and Phylogenetic Analysis
2.6.1. Genomic DNA Extraction and Amplification and Sequencing of 16S rRNA Gene PCR Products
Genomic DNA was extracted from bacterial isolates using the GeneJET Genomic DNA Purification kit (Thermo Fisher Scientific, USA) according to the manufacturer’s instructions. The 16S rRNA gene was partially amplified using primers U8-27 (AGAGTTTGATC(AC)TGGCTCAG) and L1494–1514 (CTA CGG (AG)TACCT TGT TAC GAC) [23]. PCR reactions were performed in BIO-RAD T100TM Thermal cycler, U.S. The annealing temperature was 56 °C. PCR amplicons were visualized using gel electrophoresis. The PCR products of the partially amplified 16S rRNA gene were sent for Sanger sequencing (Macrogen, Inc., Seoul, Korea). All sequences were analyzed using the blastn tool at NCBI against the 16S ribosomal RNA (Bacteria and Archaea type strains).
2.6.2. Phylogenetic Analysis
For the construction of a phylogenetic tree, the most related type strains’ 16S rRNA sequences were aligned using MUSCLE [24], and then the tree was constructed using Molecular Evolutionary Genetics Analysis MEGA X [25] using the Maximum Likelihood method and the Kimura 2-parameter model [26]. A Bootstrap test (for 1000 replicates) was performed.
2.7. Screening of Bacillus Iturin Family Lipopeptides
Bacillus species are known to produce several high-molecular-weight lipopeptides (>1000 daltons) with antibacterial and antifungal properties. Most of them are encoded by non-ribosomal peptide synthases (NRPS) BGCs except for the Iturin family which is encoded by a hybrid PKS-I/NRPS system [27]. The isolated active Bacillus species were screened for the presence of two of the most common Iturin family lipopeptides (Iturin A and Bacillomycin D). Iturin A coding gene (ituD) was amplified using ItuD1F (GATGCGATCTCCTTGGATGT) and ItuD1R (ATCGTCATGTGCTGCTTGAG) primers. Bacillomycin D coding gene (bamC) was amplified using Bacc1F (GAAGGACACGGAGAGAGTC) and Bacc1R (CGCTGATGACTGTTCATGCT) primers [28]. The PCR conditions for both reactions were carried out as follows: 95 °C for 5 min; 35 cycles with a denaturation step of 94 °C for 1 min, annealing for 1 min at 58 °C; elongation at 72 °C for 1 min; and a final step of 72 °C for 10 min.
2.8. Liquid Chromatography–Mass Spectroscopy Analysis of the Metabolic Extracts
LC–MS was used for the tentative investigation of the low-molecular-weight metabolites (<1000 daltons) in the isolates’ extracts. The dried ethyl acetate metabolic extracts of the four active Bacillus isolates were dissolved in 5 mL of methanol and then left to dry at room temperature. The residual materials were dissolved in HPLC grade water and purified through ultra-filtration using Vivaspin® (SARTORIUS, Gottingen, Germany) with a molecular weight cut-off of 10,000 daltons. The Acquity® LC–MS System (Waters®, Milford, MA, USA) equipped with the ACQUITY UPLC® BEH C18 1.7 m column in positive ion mode was used for separation and mass spectrometry analysis. The injected volume was 6 µL, the average column temperature was 40 °C, and the run time was 32 min. The elution was carried out by a component solvent system in which solvent A was water, and solvent B was methanol. Both were injected with 0.1% formic acid.
The natural products atlas database (NPAtlas) [29] was searched for the probable identity of each detected compound using the exact mass in daltons (±0.02 daltons). The search parameters were limited to the bacterial metabolites of the Bacillus genus to improve the accuracy.
3. Results
3.1. Biosynthetic Genes Abundance in Total Community-DNA (TC-DNA)
Shore water (SW), off-shore water (OSW), muddy anoxic sediments (MS), and sandy oxic sediment (SS) samples have shown variable levels of the total bacterial 16S rRNA gene, the Firmicutes 16S rRNA gene, NRPSs, and PKSs.
The total bacterial 16S rRNA gene showed an average of 12.702 × 106 copies.g−1, 3.485 × 106, 35.171 × 106 and 3.948 × 106 copies.g−1 of SW, OSW, MS, and SS samples, respectively. The number of 16S rRNA gene copies per gram MS samples was significantly higher than other samples. SW samples were significantly higher than both OSW and SS samples. There was no statistically significant difference between OSW and SS samples (Figure 2a).


Figure 2.
(a) Absolute quantification for bacterial 16S rRNA gene copies/gram. (b) Relative abundance of Firmicutes 16S rRNA. (c) Relative abundance of polyketide synthase type I. (d) Relative abundance of polyketide synthase type II in each sample type—SW: shore water, OSW: off-shore water, MS: muddy anoxic sediment and SS: sandy oxic sediment. (e) Correlation between the relative abundance of PKS-I gene and PKS-II gene. The statistical analysis of data was performed using one-way ANOVA and Tukey’s test for multiple comparisons. Significance was considered at a p-value of <0.05, p < 0.05 (*), p < 0.01 (**), p < 0.001 (***), and p < 0.0001 (****); non-significant (ns).
The relative abundance of the Firmicutes 16S rRNA gene was in an average of 2.769%, 0.232%, 0.462%, and 0.778% for SW, OSW, MS, and SS samples, respectively.
SW samples significantly had the highest relative abundance in Firmicutes 16S rRNA gene copies followed by SS samples. There was no significant difference between MS and OSW samples (Figure 2b).
PKS-I was higher in all samples than PKS-II by approximately 6.2 fold on average. SS samples showed the highest relative abundance for both BGCs encoding PKS-I and PKS-II among all samples (Figure 2c,d).
There was a linear correlation between the relative abundance of PKS-I and PKS-II in our samples (Figure 2e). A similar correlation was described in a terrestrial ecosystem [17].
3.2. Isolation of Marine Bacteria
A total of 64 bacterial isolates were recovered from all the samples. A total of 52% of the isolates were recovered by R2A medium, 31% were recovered by nutrient agar (NA), and only 17% were recovered by both AIA and SCA media (Figure 3).
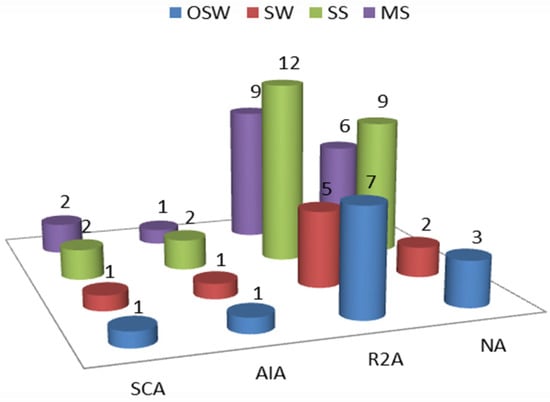
Figure 3.
The number of isolates recovered by each medium—NA: nutrient agar, R2A: Reasoner’s 2A agar. AIA; Actinomycete isolation agar, and SCA; starch casein agar. OSW: off-shore water, SW: shore water, SS: sandy sediment, and MS: muddy sediment.
The average total aerobic viable count recovered by NA medium after 72 h of incubation was 12 × 105 CFU·mL−1, 84 × 105 CFU·mL−1, 165 × 105 CFU·g−1, and 147 × 107 CFU·g−1 for OSW, SW, SS, and MS samples, respectively. Although MS was the highest in the viable count, it has shown a lower diversity in colony morphology in comparison to SS.
3.3. The Antagonistic Behavior and Antimicrobial Activity of the Bacterial Isolates and Their Metabolic Extracts
A total of ten isolates have shown antagonism against at least one of the indicator microbes using the drop-plate method (Table 2). Seven out of the ten active isolates were found to be active only against the Gram-positive bacteria.

Table 2.
Results of the positive microbial antagonism isolates.
R-4 isolate showed the highest antimicrobial activity against all tested indicator strains, including both Gram-positive and Gram-negative organisms in addition to the yeast strain. Isolates S-41 and S-30 showed only attenuated growth against one of the indicator strains, C. albicans and S. aureus, respectively. In addition to the production of antimicrobial secondary metabolites, bacterial antagonistic behavior can be mediated by other mechanisms, such as the production of lactic acid as observed in the case of Lactobacilli or the release of bacteriocins and lysozymes [30]. To only focus on the isolates with antimicrobial secondary metabolites, we further screened the antimicrobial activity of the cell-free ethyl acetate filtrates of these ten bacterial isolates.
Four cell-free ethyl acetate filtrates have shown antimicrobial activity against more than one of the tested standard strains using the well-diffusion assay (Figure 4). The cell-free ethyl acetate filtrates of isolates R-4 and R-11 showed broad activity against both tested Gram-positive strains (Staphylococcus aureus and Bacillus cereus) and Gram-negative strain (Escherichia coli). For the positive controls, the average inhibition zones were 20, 19, 20, 18, and 17 mm, for E.C, S.A, B.C, P.A, and C.A, respectively.

Figure 4.
Zones of inhibition of the six active bacterial metabolic crude extracts. E.C.: Escherichia coli, S.A: Staphylococcus aureus, B.C: Bacillus cereus, P.A: Pseudomonas aeruginosa and C.A: Candida albicans.
3.4. Phylogenetic Analysis
The analysis of the partial 16S rRNA sequences of the active bacterial isolates has indicated these isolates belong to the phylum Firmicutes. They were identified as Bacillus licheniformis, Bacillus pumilus, Bacillus subtilis, and Bacillus haynesii for isolates R-1, R-4, R-11, and R-19, respectively. The NCBI blastn search results are illustrated in Table 3. The phylogenetic tree is represented in Figure 5.

Table 3.
Results of the NCBI Blast search.
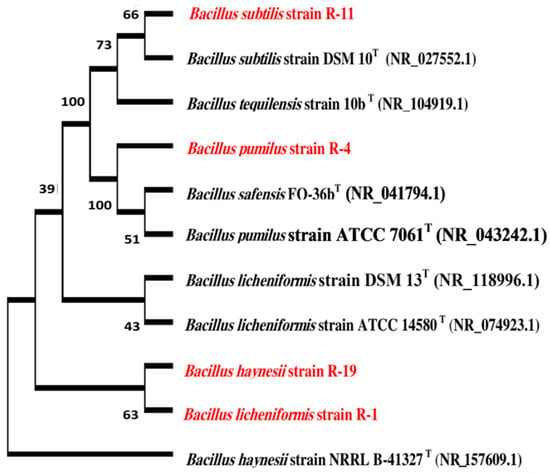
Figure 5.
Phylogenetic relationship between our identified isolates (colored in red) and the most related type strains (labeled by (T)).
3.5. Amplification of Iturin Family Lipopeptides Coding Genes
Bacillus species are known to produce a diverse collection of bioactive secondary metabolites [31]. Among these metabolites are Bacillus lipopeptides.
By conventional PCR, the isolated B. subtilis strain R-11 has shown sharp bands for ituD and bamC (Figure 6). By the presence of these two genes, B. subtilis strain R-11 is expected to produce both Iturin A and Bacillomycin D lipopeptides. Iturin A and Bacillomycin D are known for their antifungal properties [32].

Figure 6.
Bacillus subtilis strain R-11 shows sharp bands for ItuD and BamC genes in gel electrophoresis at 647 and 875, respectively. Lanes 1, 2, 3, 4 and 5 represent Bacillus licheniformis strain R-1, Bacillus pumilus strain R-4, Bacillus subtilis strain R-11, Bacillus haynesii strain R-19 and negative control, respectively; the molecular marker used is (GeneRuler 1 kb DNA Ladder, Thermo Fisher Scientific).
3.6. Liquid Chromatography–Mass Spectroscopy Analysis of the Metabolic Extracts
The numbers of detected masses were 31, 37, 39 and 42 (<1000 daltons) for the cell-free ethyl acetate extracts of isolates R-1, R-4, R-11, and R-19, respectively (see supplementary material). A total of 2, 2, 3, and 2 probable active metabolites for the metabolic extracts of isolates R-1, R-4, R-11 and R-19, respectively, were obtained from the NPAtlas database. The activity of these metabolites ranged between antimicrobial, cytotoxic, antioxidant, plant growth inhibitor and nitrogen monoxide (NO) production inhibitor (Table 4).

Table 4.
The probable metabolite for each exact mass detected in the metabolic extracts of the active isolated strains from the Natural Products Atlas.
4. Discussion
In this study, we modified the method used before by Peng et al. [42] in investigating the antibiotic-related BGCs levels in a newly explored ecosystem, to compare the biosynthetic capacity of different types of samples taken from our study area, and choose the most promising ones. This method secures a time-saving approach and increases the success rates to isolate active microbial strains.
D’hondt et al. reported that the total microbial count is usually higher in the organic-rich anoxic sediment than in the organic-poor oxic sediment [43]. This agrees with our findings that both the total 16S rRNA gene copies and the total aerobic viable count were higher in the MS than in SS samples.
The relative abundance and diversity of PKSs BGCs in total community DNAs can provide a picture of the ability of a system to produce new bioactive compounds [15,42].
We have found that the oxic sediment had the greatest levels of both PKS-I and PKS-II BGCs.
Generally, it is well observed that bacteria, especially the spore-forming ones such as the phylum of Actinobacteria and the family Bacillaceae, present in a higher relative abundance in the marine sediment than in the water column. This can be explained by the static nature of marine sediment that facilitates the accumulation of these bacteria and their spores and the higher availability of organic matters in the marine sediment than in the water column [44,45]. This explains our observation that both PKS-I and PKS-II were significantly higher in SS samples than in water samples. The higher relative abundance of PKS-I and PKS-II in SS samples than in MS samples may be explained by the aerobic nature of these bacterial families.
R2A was the most efficient of the four used media in recovering a diverse collection of isolates. This may be attributed to the low nutrient composition of the medium that controls the growth of the fast-growing bacteria, thus preventing them from suppressing the slow growers and allowing the growth of oligotrophs [46].
All the identified active strains were from the genus Bacillus. Although Actinobacteria are known to be the primary producers of bioactive compounds possessing antimicrobial activity, Firmicutes, especially the genus Bacillus, are the most involved in the research for antimicrobials from bacteria isolated from the marine environment. This may be due to the special and, most of the time, challenging culture requirements of marine actinobacteria [47].
B. licheniformis was reported to have antimicrobial activity against Gram-positive bacteria and a few Gram-negative bacteria [48,49]. In addition to the antimicrobial activity of marine B. licheniformis’ metabolites, other activities such as antiviral and immune-regulatory activities have also been reported [50]. The antimicrobial protein BLDZ1 from marine B. licheniformis inhibits the biofilm formation of microbial pathogens such as P. aeurginosa [51]. The genome mining of a marine B. pumilus strain showed at least twelve BGCs, including PKS-I, PKS-II, and NRPS which indicates a high bioactivity potential [52]. Marine B. pumilus was reported to produce an active non-ribosomal lipopeptide Pumilacidin (~3 KD) with anti-Staphylococcus aureus activity [53]. The Marine B. pumilus was also reported as a source of antitrypanosomal active metabolites [54]. Exopolysaccharide of marine B. haynesii has many applications in food industries as an antioxidant and emulsifying agent [55].
In addition, B. subtilis and B. pumilus were reported for their broad-spectrum antibacterial and antifungal properties [56,57,58,59,60]. This partly agrees with the results of our study, where only B. pumilus and B. subtilis showed broad antimicrobial activity, while B. licheniformis possessed only a narrow spectrum activity against the tested standard indicator microorganisms.
Marine bacteria are active producers of secondary metabolites. They produce these metabolites as a response to the marine extreme conditions, such as pressure, temperature, salinity, and depletion of micronutrients [61].
Originally, these lipopeptides are produced by some marine Bacillus as biosurfactants to aid in the transport of hydrophobic low water-soluble substrates and enhance their bioavailability [29]. Applications such as food preservation (control of spoilage yeast), and plant disease biocontrol (against fungal infections) are possible applications for our B. subtilis strain R-11.
Macrolactins are lactone, natural polyketides mainly derived from marine bacteria. They are biosynthesized through the PKS-I system. Many bioactivities were reported regarding macrolactins including, antibacterial, antifungal, antiviral, anticancer, anti-inflammatory, anti-angiogenic, and other activities [62]. Macrolactin N produced by B. subtilis was reported for its antimicrobial activity against S. aureus and E. coli as it inhibits the peptide deformylase enzyme [38]. The marine Bacillus antimicrobial fatty acids (Ieodomycins) were reported for antibacterial and anti-yeast properties [34]. Bacillamidins are a group of marine natural products, originally derived from marine Bacillus pumilus. They were reported for their potent antimicrobial activity against Gram-negative bacteria through the inhibition of the citrate synthase type II enzyme [37].
It is well observed that the majority of the detected active metabolites are encoded by either individual PKS-I or hybrid PKS-I/NRPS systems, which was expected based on the used approach.
Based on the findings of this study, we suggest that this approach can be modified to target microbial strains with specific biosynthetic systems. It is also observed that most of the low-molecular-weight metabolites detected by the LC–MS originated from marine Bacillus strains, which proves the uniqueness of the marine environment.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/fermentation8070309/s1, All LC–MS spectra are provided as a supplementary material.
Author Contributions
Conceptualization, A.M.S., M.M.I., T.R.E. and M.A.R.; data curation, A.M.S., M.M.I. and T.R.E.; formal analysis, A.M.S., M.M.I. and T.R.E.; investigation, A.M.S., M.M.I., T.R.E. and M.A.R.; methodology, A.M.S., M.M.I., T.R.E. and M.A.R.; project administration, M.M.I., T.R.E. and M.A.R.; resources, A.M.S., M.M.I. and T.R.E.; software, A.M.S. and T.R.E.; supervision, M.M.I., T.R.E. and M.A.R.; validation, A.M.S., T.R.E. and M.A.R.; visualization, A.M.S., M.M.I., T.R.E. and M.A.R.; writing—original draft, A.M.S., M.M.I., T.R.E. and M.A.R.; writing—review and editing, A.M.S., M.M.I., T.R.E. and M.A.R. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The 16S rRNA nucleotide sequence data are available on NCBI GenBank as follows: B. licheniformis strain R-1 (accession number: ON667781), B. pumilus strain R-4 (accession number: ON667807), B. subtilis strain R-11 (accession number: ON677749), and B. haynesii strain R-19 (accession number: ON667855).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Dadgostar, P. Antimicrobial resistance: Implications and costs. Infect. Drug Resist. 2019, 12, 3903–3910. [Google Scholar] [CrossRef] [PubMed]
- Aminov, R.I. A brief history of the antibiotic era: Lessons learned and challenges for the future. Front. Microbiol. 2010, 1, 134. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, P.S. Antibacterial discovery: 21st century challenges. Antibiotiotics 2020, 9, 213. [Google Scholar] [CrossRef]
- Atanasov, A.G.; Zotchev, S.B.; Dirsch, V.M.; Orhan, I.E.; Banach, M.; Rollinger, J.M.; Barreca, D.; Weckwerth, W.; Bauer, R.; Bayer, E.A.; et al. Natural products in drug discovery: Advances and opportunities. Nat. Rev. Drug Discov. 2021, 20, 200–216. [Google Scholar] [CrossRef]
- Singh, B.P.; Rateb, M.E.; Rodriguez-Couto, S.; De Lourdes Teixeira De Moraes Polizeli, M.; Li, W.J. Editorial: Microbial secondary metabolites: Recent developments and technological challenges. Front. Microbiol. 2019, 10, 914. [Google Scholar] [CrossRef] [PubMed]
- Barzkar, N.; Jahromi, S.T.; Poorsaheli, H.B.; Vianello, F. Metabolites from marine microorganisms, micro, and macroalgae: Immense scope for pharmacology. Mar. Drugs 2019, 17, 464. [Google Scholar] [CrossRef]
- Bérdy, J. Bioactive microbial metabolites: A personal view. J. Antibiot. 2005, 58, 1–26. [Google Scholar] [CrossRef]
- Tran, P.N.; Yen, M.R.; Chiang, C.Y.; Lin, H.C.; Chen, P.Y. Detecting and prioritizing biosynthetic gene clusters for bioactive compounds in bacteria and fungi. Appl. Microbiol. Biotechnol. 2019, 103, 3277–3287. [Google Scholar] [CrossRef]
- Ridley, C.P.; Ho, Y.L.; Khosla, C. Evolution of polyketide synthases in bacteria. Proc. Natl. Acad. Sci. USA 2008, 105, 4595–4600. [Google Scholar] [CrossRef]
- Olishevska, S.; Nickzad, A.; Déziel, E. Bacillus and Paenibacillus secreted polyketides and peptides involved in controlling human and plant pathogens. Appl. Microbiol. Biotechnol. 2019, 103, 1189–1215. [Google Scholar] [CrossRef]
- Penesyan, A.; Kjelleberg, S.; Egan, S. Development of novel drugs from marine surface associated microorganisms. Mar. Drugs 2010, 8, 438–459. [Google Scholar] [CrossRef] [PubMed]
- Wiese, J.; Imhoff, J.F. Marine bacteria and fungi as promising source for new antibiotics. Drug Dev. Res. 2019, 80, 24–27. [Google Scholar] [CrossRef] [PubMed]
- Elton, C.S. Charles Elton the Ecology of Invasions by Animals and Plants; University of Chicago Press: Chicago, IL, USA, 2000. [Google Scholar]
- El-serehy, H.A.; Abdallah, H.S.; Al-misned, F.A.; Irshad, R.; Al-farraj, S.A.; Almalki, E.S. Aquatic ecosystem health and trophic status classification of the Bitter Lakes along the main connecting link between the Red Sea and the Mediterranean. Saudi J. Biol. Sci. 2018, 25, 204–212. [Google Scholar] [CrossRef]
- Mahapatra, G.P.; Raman, S.; Nayak, S.; Gouda, S.; Das, G.; Patra, J.K. Metagenomics Approaches in Discovery and Development of New Bioactive Compounds from Marine Actinomycetes. Curr. Microbiol. 2020, 77, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Z.; Wang, J.; Hao, Y.; Wang, Y. Recent Advances in the Discovery and Development of Marine Microbial Natural Products. Mar. Drugs 2013, 11, 700–717. [Google Scholar] [CrossRef]
- Le, T.H.; Sivachidambaram, V.; Yi, X.; Li, X.; Zhou, Z. Quantification of polyketide synthase genes in tropical urban soils using real-time PCR. J. Microbiol. Methods 2014, 106, 135–142. [Google Scholar] [CrossRef]
- Metsä-Ketelä, M.; Salo, V.; Halo, L.; Hautala, A.; Hakala, J.; Mäntsälä, P.; Ylihonko, K. An efficient approach for screening minimal PKS genes from Streptomyces. FEMS Microbiol. Lett. 1999, 180, 1–6. [Google Scholar] [CrossRef]
- Fierer, N.; Jackson, J.A.; Vilgalys, R.; Jackson, R.B. Assessment of soil microbial community structure by use of taxon-specific quantitative PCR assays. Appl. Environ. Microbiol. 2005, 71, 4117–4120. [Google Scholar] [CrossRef]
- Joint, I.; Mühling, M.; Querellou, J. Culturing marine bacteria—An essential prerequisite for biodiscovery: Minireview. Microb. Biotechnol. 2010, 3, 564–575. [Google Scholar] [CrossRef]
- Yang, Y.W.; Chen, M.K.; Yang, B.Y.; Huang, X.J.; Zhang, X.R.; He, L.Q.; Zhang, J.; Hua, Z.C. Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in mouse feces. Appl. Environ. Microbiol. 2015, 81, 6749–6756. [Google Scholar] [CrossRef]
- Ismail, A.; Ktari, L.; Ahmed, M.; Bolhuis, H.; Boudabbous, A.; Stal, L.J.; Cretoiu, M.S.; El Bour, M. Antimicrobial activities of bacteria associated with the brown alga padina pavonica. Front. Microbiol. 2016, 7, 1072. [Google Scholar] [CrossRef]
- Elsayed, T.R.; Grosch, R.; Smalla, K. Potato plant spheres and to a lesser extent the soil type influence the proportion and diversity of bacterial isolates with in vitro antagonistic activity towards Ralstonia solanacearum. FEMS Microbiol. Ecol. 2021, 97, fiab038. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Raaijmakers, J.M.; de Bruijn, I.; Nybroe, O.; Ongena, M. Natural functions of lipopeptides from Bacillus and Pseudomonas: More than surfactants and antibiotics. FEMS Microbiol. Rev. 2010, 34, 1037–1062. [Google Scholar] [CrossRef]
- Gond, S.K.; Bergen, M.S.; Torres, M.S.; White, J.F. Endophytic Bacillus spp. produce antifungal lipopeptides and induce host defence gene expression in maize. Microbiol. Res. 2015, 172, 79–87. [Google Scholar] [CrossRef]
- Van Santen, J.A.; Jacob, G.; Singh, A.L.; Aniebok, V.; Balunas, M.J.; Bunsko, D.; Neto, F.C.; Castaño-Espriu, L.; Chang, C.; Clark, T.N.; et al. The Natural Products Atlas: An Open Access Knowledge Base for Microbial Natural Products Discovery. ACS Cent. Sci. 2019, 5, 1824–1833. [Google Scholar] [CrossRef]
- Feichtmayer, J.; Deng, L.; Griebler, C. Antagonistic microbial interactions: Contributions and potential applications for controlling pathogens in the aquatic systems. Front. Microbiol. 2017, 8, 2192. [Google Scholar] [CrossRef]
- Mondol, M.A.M.; Shin, H.J.; Islam, M.T. Diversity of secondary metabolites from marine bacillus species: Chemistry and biological activity. Mar. Drugs 2013, 11, 2846–2872. [Google Scholar] [CrossRef]
- Zhao, H.; Shao, D.; Jiang, C.; Shi, J.; Li, Q.; Huang, Q.; Rajoka, M.S.R.; Yang, H.; Jin, M. Biological activity of lipopeptides from Bacillus. Appl. Microbiol. Biotechnol. 2017, 101, 5951–5960. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liu, S.; Jiang, Z.; Sun, C. Catechol amide iron chelators produced by a mangrove-derived Bacillus subtilis. Tetrahedron 2017, 73, 5245–5252. [Google Scholar] [CrossRef]
- Mondol, M.A.M.; Kim, J.H.; Lee, M.A.; Tareq, F.S.; Lee, H.S.; Lee, Y.J.; Shin, H.J. Ieodomycins A-D, antimicrobial fatty acids from a marine Bacillus sp. J. Nat. Prod. 2011, 74, 1606–1612. [Google Scholar] [CrossRef] [PubMed]
- Azumi, M.; Ogawa, K.-I.; Fujita, T.; Takeshita, M.; Yoshida, R.; Furumai, T.; Igarashi, Y. Bacilosarcins A and B, novel bioactive isocoumarins with unusual heterocyclic cores from the marine-derived bacterium Bacillus subtilis. Tetrahedron 2008, 64, 6420–6425. [Google Scholar] [CrossRef]
- Asolkar, R.N.; Kamat, V.P.; Wagner-Döbler, I.; Laatsch, H. Limnazine, the first bacterial azine derivative from Bacillus sp. GW90a. J. Nat. Prod. 2002, 65, 1664–1666. [Google Scholar] [CrossRef]
- Zhou, S.Y.; Hu, Y.J.; Meng, F.C.; Qu, S.Y.; Wang, R.; Andersen, R.J.; Liao, Z.H.; Chen, M. Bacillamidins A-G from a marine-derived bacillus pumilus. Mar. Drugs 2018, 16, 326. [Google Scholar] [CrossRef]
- Yoo, J.S.; Zheng, C.J.; Lee, S.; Kwak, J.H.; Kim, W.G. Macrolactin N, a new peptide deformylase inhibitor produced by Bacillus subtilis. Bioorg. Med. Chem. Lett. 2006, 16, 4889–4892. [Google Scholar] [CrossRef]
- Bai, J.; Liu, D.; Yu, S.; Proksch, P.; Lin, W. Amicoumacins from the marine-derived bacterium Bacillus sp. with the inhibition of NO production. Tetrahedron Lett. 2014, 55, 6286–6291. [Google Scholar] [CrossRef]
- Hai, Y.; Wei, M.Y.; Wang, C.Y.; Gu, Y.C.; Shao, C.L. The intriguing chemistry and biology of sulfur-containing natural products from marine microorganisms (1987–2020). Mar. Life Sci. Technol. 2021, 3, 488–518. [Google Scholar] [CrossRef]
- Nagao, T.; Adachi, K.; Sakai, M.; Nishijima, M.; Sano, H. Novel macrolactins as antibiotic lactones from a marine bacterium. J. Antibiot. 2001, 54, 333–339. [Google Scholar] [CrossRef]
- Peng, C.; Wang, H.; Jiang, Y.; Yang, J.; Lai, H.; Wei, X. Exploring the Abundance and Diversity of Bacterial Communities and Quantifying Antibiotic-Related Genes Along an Elevational Gradient in Taibai Mountain, China. Microb. Ecol. 2018, 76, 1053–1062. [Google Scholar] [CrossRef]
- D’hondt, S.; Inagaki, F.; Zarikian, C.A.; Abrams, L.J.; Dubois, N.; Engelhardt, T.; Evans, H.; Ferdelman, T.; Gribsholt, B.; Harris, R.N.; et al. Presence of oxygen and aerobic communities from sea floor to basement in deep-sea sediments. Nat. Geosci. 2015, 8, 299–304. [Google Scholar] [CrossRef]
- Ghanem, N.B.; Sabry, S.A.; El-Sherif, Z.M.; Abu El-Ela, G.A. Isolation and enumeration of marine actinomycetes from seawater and sediments in Alexandria. J. Gen. Appl. Microbiol. 2000, 46, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Vijayabaskara Sethubathi, G.V.; Sivakumar, K.; Thangaradjou, T.; Kannan, L. Ecology and population density of the marine actinobacteria of little Andaman island, India. Indian. J. Mar. Sci. 2013, 42, 390–401. [Google Scholar]
- Sedeek, A.M.; Ismail, M.M.; Elsayed, T.R.; Ramadan, M.A. Recent methods for discovering novel bioactive metabolites, specifically antimicrobial agents, from marine-associated microorganisms. Lett. Appl. Microbiol. 2022, in press. [Google Scholar] [CrossRef]
- Stincone, P.; Brandelli, A. Marine bacteria as source of antimicrobial compounds. Crit. Rev. Biotechnol. 2020, 40, 306–319. [Google Scholar] [CrossRef]
- Devi, P.; Wahidullah, S.; Rodrigues, C.; Souza, L.D. The sponge-associated bacterium Bacillus licheniformis SAB1: A source of antimicrobial compounds. Mar. Drugs 2010, 8, 1203–1212. [Google Scholar] [CrossRef]
- Alvarez-Ordóñez, A.; Begley, M.; Clifford, T.; Deasy, T.; Considine, K.; O’Connor, P.; Paul Ross, R.; Hill, C. Investigation of the Antimicrobial Activity of Bacillus licheniformis Strains Isolated from Retail Powdered Infant Milk Formulae. Probiotics Antimicrob. Proteins 2014, 6, 32–40. [Google Scholar] [CrossRef]
- Arena, A.; Maugeri, T.L.; Pavone, B.; Iannello, D.; Gugliandolo, C.; Bisignano, G. Antiviral and immunoregulatory effect of a novel exopolysaccharide from a marine thermotolerant Bacillus licheniformis. Int. Immunopharmacol. 2006, 6, 8–13. [Google Scholar] [CrossRef]
- Hamza, F.; Kumar, A.R.; Zinjarde, S. Antibiofilm potential of a tropical marine Bacillus licheniformis isolate: Role in disruption of aquaculture associated biofilms. Aquac. Res. 2016, 47, 2661–2669. [Google Scholar] [CrossRef]
- Freitas-Silva, J.; de Oliveira, B.F.R.; Vigoder, F.D.M.; Muricy, G.; Dobson, A.D.W.; Laport, M.S. Peeling the Layers Away: The Genomic Characterization of Bacillus pumilus 64-1, an Isolate with Antimicrobial Activity From the Marine Sponge Plakina cyanorosea (Porifera, Homoscleromorpha). Front. Microbiol. 2021, 11, 592735. [Google Scholar] [CrossRef] [PubMed]
- Saggese, A.; Culurciello, R.; Casillo, A.; Corsaro, M.M.; Ricca, E.; Baccigalupi, L. A marine isolate of bacillus pumilus secretes a pumilacidin active against staphylococcus aureus. Mar. Drugs 2018, 16, 180. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Luis, S.; Gómez, J.F.; Spadafora, C.; Guzmán, H.M.; Gutiérrez, M. Antitrypanosomal alkaloids from the marine bacterium Bacillus pumilus. Molecules 2012, 17, 11146–11155. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Mohammed Breig, S.J.; Gómez, A.; Sánchez-Arévalo, I.; González-Faune, P.; Sarkar, S.; Bandopadhyay, R.; Vuree, S.; Cornejo, J.; Tapia, J.; et al. Optimization and Characterization of a Novel Exopolysaccharide from Bacillus haynesii CamB6 for Food Applications. Biomolecules 2022, 12, 834. [Google Scholar] [CrossRef]
- Peypoux, F.; Guinand, M.; Michel, G.; Delcambe, L.; Das, B.C.; Lederer, E. Structure of Iturine A, a Peptidolipid Antibiotic from Bacillus subtilis. Biochemistry 1978, 17, 3992–3996. [Google Scholar] [CrossRef]
- Ramachandran, R.; Chalasani, A.G.; Lal, R.; Roy, U. A broad-spectrum antimicrobial activity of bacillus subtilis RLID 12.1. Sci. World J. 2014, 2014, 1–10. [Google Scholar] [CrossRef]
- Ouoba, L.I.I.; Diawara, B.; Jespersen, L.; Jakobsen, M. Antimicrobial activity of Bacillus subtilis and Bacillus pumilus during the fermentation of African locust bean (Parkia biglobosa) for Soumbala production. J. Appl. Microbiol. 2007, 102, 963–970. [Google Scholar] [CrossRef]
- Sawale, A.; Kadam, T.A.; Karale, M.A.; Kadam, O.A. Antimicrobial Activity of Secondary Metabolites from Halophilic Bacillus pumilus sp. Int. J. Curr. Microbiol. App. Sci. 2014, 3, 506–512. [Google Scholar]
- Todorova, S.; Kozhuharova, L. Characteristics and antimicrobial activity of Bacillus subtilis strains isolated from soil. World J. Microbiol. Biotechnol. 2010, 26, 1207–1216. [Google Scholar] [CrossRef]
- De Carvalho, C.C.C.R.; Fernandes, P. Production of metabolites as bacterial responses to the marine environment. Mar. Drugs 2010, 8, 705–727. [Google Scholar] [CrossRef]
- Wu, T.; Xiao, F.; Li, W. Macrolactins: Biological activity and biosynthesis. Mar. Life Sci. Technol. 2021, 3, 62–68. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).