Identification and Transcriptional Profiling of SNARE Family in Monascus ruber M7 Reveal Likely Roles in Secondary Metabolism
Abstract
1. Introduction
2. Materials and Methods
2.1. Microbial Strains and Culture Conditions
2.2. Genome-Wide Identification of SNARE Genes in Monascus ruber M7
2.3. Characterization and Phylogenetic Analysis of Identified MrSNAREs
2.4. MPs and CIT Production Analysis
2.5. Gene Expression Analysis by Real-Time Quantitative PCR
2.6. Statistical Analysis
3. Results
3.1. Monascus ruber M7 Has 20 SNARE-Encoding Genes
3.2. Classification of SNARE Genes in Monascus ruber M7
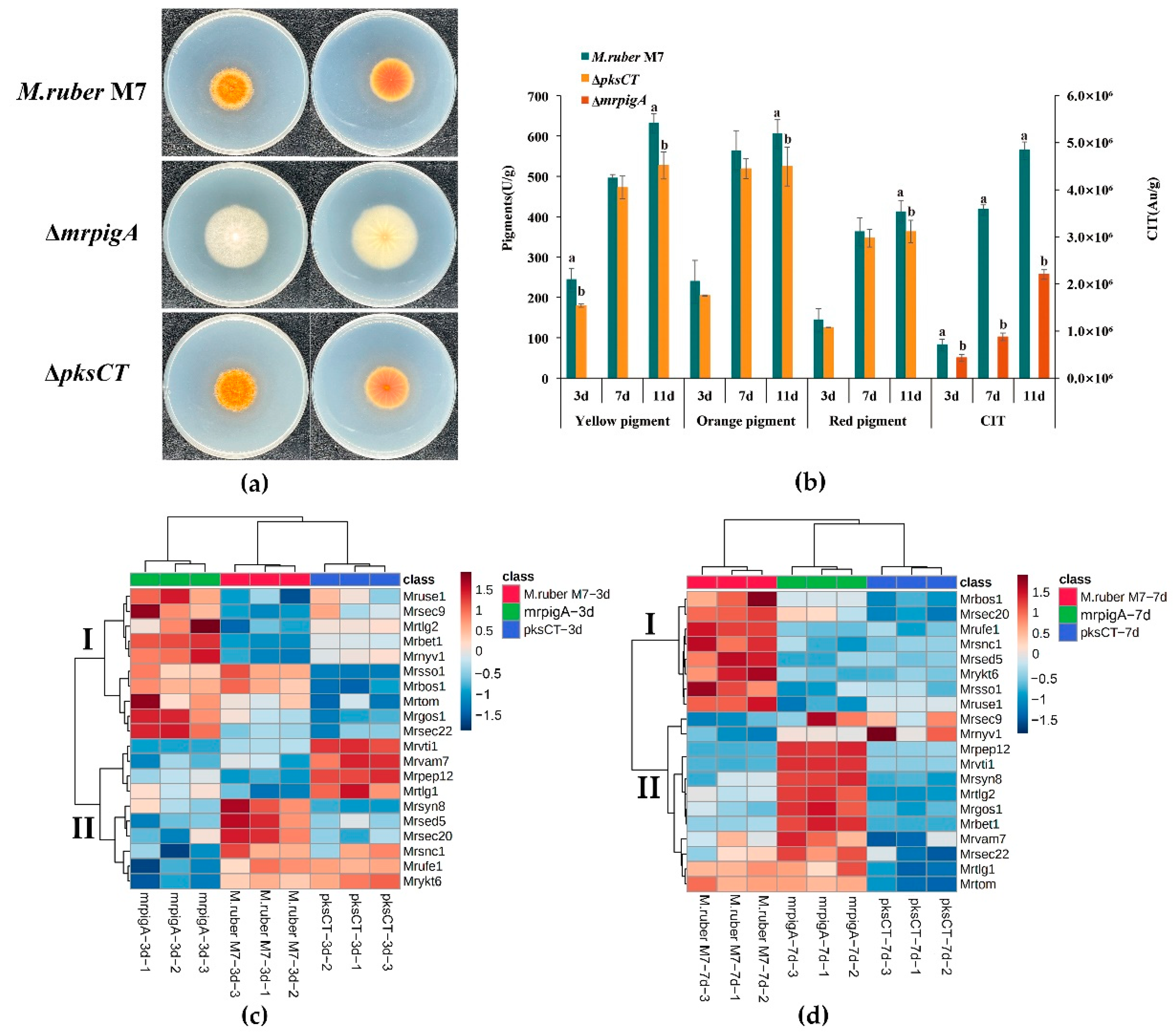
3.3. Expression Profiles of 20 MrSNAREs in Three Monascus Strains
3.4. Analysis of Potential MrSNAREs Related to MPs and CIT Synthesis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Behnia, R.; Munro, S. Organelle identity and the signposts for membrane traffic. Nature 2005, 438, 597–604. [Google Scholar] [CrossRef] [PubMed]
- Spang, A. The life cycle of a transport vesicle. Cell Mol. Life Sci. 2008, 65, 2781–2789. [Google Scholar] [CrossRef] [PubMed]
- Higuchi, Y. Membrane traffic related to endosome dynamics and protein secretion in filamentous fungi. Biosci. Biotechnol. Biochem. 2021, 85, 1038–1045. [Google Scholar] [CrossRef] [PubMed]
- Gillingham, A.K.; Munro, S. Transport carrier tethering—How vesicles are captured by organelles. Curr. Opin. Cell Biol. 2019, 59, 140–146. [Google Scholar] [CrossRef]
- Margiotta, A. Membrane fusion and snares: Interaction with ras proteins. Int. J. Mol. Sci. 2022, 23, 8067. [Google Scholar] [CrossRef]
- Koike, S.; Jahn, R. SNARE proteins: Zip codes in vesicle targeting? Biochem. J. 2022, 479, 273–288. [Google Scholar] [CrossRef]
- Heo, P.; Coleman, J.; Fleury, J.B.; Rothman, J.E.; Pincet, F. Nascent fusion pore opening monitored at single-SNAREpin resolution. Proc. Natl. Acad. Sci. USA 2021, 118, e2024922118. [Google Scholar] [CrossRef]
- Hong, W.; Lev, S. Tethering the assembly of SNARE complexes. Trends Cell Biol. 2014, 24, 35–43. [Google Scholar] [CrossRef]
- Van den Bogaart, G.; Lang, T.; Jahn, R. Chapter Six—Microdomains of Snare Proteins in the Plasma Membrane, in Current Topics in Membranes; Bennett, V., Ed.; Academic Press: Cambridge, MA, USA, 2013; pp. 193–230. [Google Scholar]
- Fasshauer, D.; Sutton, R.B.; Brunger, A.T.; Jahn, R. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc. Natl. Acad. Sci. USA 1998, 95, 15781–15786. [Google Scholar] [CrossRef]
- Jahn, R.; Scheller, R.H. SNAREs—Engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 2006, 7, 631–643. [Google Scholar] [CrossRef]
- Han, J.; Pluhackova, K.; Böckmann, R.A. The multifaceted role of snare proteins in membrane fusion. Front. Physiol. 2017, 8, 5. [Google Scholar] [CrossRef] [PubMed]
- Hong, W. SNAREs and traffic. BBA-Mol. Cell Res. 2005, 1744, 120–144. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.A.; Scheller, R.H. SNARE-mediated membrane fusion. Nat. Rev. Mol. Cell Biol. 2001, 2, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Söllner, T.; Whiteheart, S.W.; Brunner, M.; Erdjument-Bromage, H.; Geromanos, S.; Tempst, P.; Rothman, J.E. SNAP receptors implicated in vesicle targeting and fusion. Nature 1993, 362, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Burri, L.; Lithgow, T. A complete set of SNAREs in yeast. Traffic 2004, 5, 45–52. [Google Scholar] [CrossRef]
- Pratelli, R.; Sutter, J.U.; Blatt, M.R. A new catch in the SNARE. Trends Plant Sci. 2004, 9, 187–195. [Google Scholar] [CrossRef]
- Adnan, M.; Islam, W.; Noman, A.; Hussain, A.; Anwar, M.; Khan, M.U.; Akram, W.; Ashraf, M.F.; Raza, M.F. Q-SNARE protein FgSyn8 plays important role in growth, DON production and pathogenicity of Fusarium graminearum. Microb. Pathog. 2020, 140, 103948. [Google Scholar] [CrossRef]
- Li, S.; Zhang, S.; Li, B.; Li, H. The SNARE protein cfvam7 is required for growth, endoplasmic reticulum stress response, and pathogenicity of Colletotrichum fructicola. Front. Microbiol. 2021, 12, 736066. [Google Scholar] [CrossRef]
- Li, B.; Gao, Y.; Mao, H.Y.; Borkovich, K.A.; Ouyang, S.Q. The SNARE protein FolVam7 mediates intracellular trafficking to regulate conidiogenesis and pathogenicity in Fusarium oxysporum f. sp. lycopersici. Environ. Microbiol. 2019, 21, 2696–2706. [Google Scholar] [CrossRef]
- Su, Z.Z.; Dai, M.D.; Zhu, J.N.; Liu, X.H.; Li, L.; Zhu, X.M.; Wang, J.Y.; Yuan, Z.L.; Lin, F.C. Dark septate endophyte Falciphora oryzae-assisted alleviation of cadmium in rice. J. Hazard. Mater. 2021, 419, 126435. [Google Scholar] [CrossRef]
- Adnan, M.; Islam, W.; Zhang, J.; Zheng, W.; Lu, G.D. Diverse role of SNARE protein Sec22 in vesicle trafficking, membrane fusion, and autophagy. Cells 2019, 8, 337. [Google Scholar] [CrossRef] [PubMed]
- O’Mara, S.P.; Broz, K.; Boenisch, M.; Zhong, Z.; Dong, Y.; Kistler, H.C. The Fusarium graminearum t-SNARE Sso2 is involved in growth, defense, and don accumulation and virulence. Mol. Plant Microbe Interact. 2020, 33, 888–901. [Google Scholar] [CrossRef] [PubMed]
- Qi, Z.; Liu, M.; Dong, Y.; Zhu, Q.; Li, L.; Li, B.; Yang, J.; Li, Y.; Ru, Y.; Zhang, H.; et al. The syntaxin protein (MoSyn8) mediates intracellular trafficking to regulate conidiogenesis and pathogenicity of rice blast fungus. New Phytol. 2016, 209, 1655–1667. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; He, Y.; Zhou, Y.; Shao, Y.; Feng, Y.; Li, M.; Chen, F. Edible filamentous fungi from the species Monascus: Early traditional fermentations, modern molecular biology, and future genomics. Compr. Rev. Food Sci. Food Saf. 2015, 14, 555–567. [Google Scholar] [CrossRef]
- Chen, W.; Feng, Y.; Molnár, I.; Chen, F. Nature and nurture: Confluence of pathway determinism with metabolic and chemical serendipity diversifies Monascus azaphilone pigments. Nat. Prod. Rep. 2019, 36, 561–572. [Google Scholar] [CrossRef]
- Zhang, C.; Chen, M.; Zang, Y.; Wang, H.; Wei, X.; Zhu, Q.; Yang, X.; Sun, B.; Wang, C. Effect of arginine supplementation on Monacolin K yield of Monascus purpureus. J. Food Compos. Anal. 2022, 106, 104252. [Google Scholar] [CrossRef]
- Hou, Y.; Liu, J.; Shao, Y.; Peng, X.; Zhang, D.; Hu, L.; Chen, F.; Zhou, Y. Evaluation of the underestimation of citrinin content in Hongqu using hydrolysis treatments and UPLC-FLD. Food Control 2021, 130, 108245. [Google Scholar] [CrossRef]
- Righetti, L.; Dall’Asta, C.; Bruni, R. Risk assessment of ryr food supplements: Perception vs. reality. Front. Nutr. 2021, 8, 792529. [Google Scholar] [CrossRef]
- Liu, W.; An, C.; Shu, X.; Meng, X.; Yao, Y.; Zhang, J.; Chen, F.; Xiang, H.; Yang, S.; Gao, X.; et al. A dual-plasmid crispr/cas system for mycotoxin elimination in polykaryotic industrial fungi. ACS Synth. Biol. 2020, 9, 2087–2095. [Google Scholar] [CrossRef]
- Ning, Z.Q.; Cui, H.; Xu, Y.; Huang, Z.B.; Tu, Z.; Li, Y.P. Deleting the citrinin biosynthesis-related gene, ctnE, to greatly reduce citrinin production in Monascus aurantiacus Li AS3.4384. Int. J. Food Microbiol. 2017, 241, 325–330. [Google Scholar] [CrossRef]
- Xie, N. Identification and Exploration of Pigment Gene Cluster and Metabolic Pathway in Monascus ruber M-7. Ph.D. Thesis, Huazhong Agricultural University, Wuhan, China, 2013; pp. 52–57. [Google Scholar]
- Chen, W.; Chen, R.; Liu, Q.; He, Y.; He, K.; Ding, X.; Kang, L.; Guo, X.; Xie, N.; Zhou, Y.; et al. Orange, red, yellow: Biosynthesis of azaphilone pigments in Monascus fungi. Chem. Sci. 2017, 8, 4917–4925. [Google Scholar] [CrossRef] [PubMed]
- He, Y. Construction of a High-Efficiency Gene Knockout System of Monascus ruber M7 and Analysis of the Biosynthetic Pathway of Citrinin. Ph.D. Thesis, Huazhong Agricultural University, Wuhan, China, 2015; pp. 113–118. [Google Scholar]
- He, Y.; Cox, R.J. The molecular steps of citrinin biosynthesis in fungi. Chem. Sci. 2016, 7, 2119–2127. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Li, Y.; Yu, W.; Li, A.; Wang, Y. Study on production of yellow pigment from potato fermented by Monascus. Food Biosci. 2022, 50, 102088. [Google Scholar] [CrossRef]
- Huang, Z.; Hu, T.; Liu, H.; Xie, H.; Tian, X.; Wu, Z. Biosynthesis and polyketide oxidation of Monascus red pigments in an integrated fermentation system with microparticles and surfactants. Food Chem. 2022, 394, 133545. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xiong, D.; Tian, C. Genome-wide identification, phylogeny and transcriptional profiling of SNARE genes in Cytospora chrysosperma. J. Phytopathol. 2021, 169, 471–485. [Google Scholar] [CrossRef]
- Khurana, G.K.; Vishwakarma, P.; Puri, N.; Lynn, A.M. Phylogenetic analysis of the vesicular fusion SNARE machinery revealing its functional divergence across Eukaryotes. Bioinformation 2018, 14, 361–368. [Google Scholar] [CrossRef]
- Grissom, J.H.; Segarra, V.A.; Chi, R.J. New perspectives on snare function in the yeast minimal endomembrane system. Genes 2020, 11, 899. [Google Scholar] [CrossRef]
- Tsui, M.; Banfield, D. Yeast Golgi SNARE interactions are promiscuous. J. Cell Sci. 2000, 113, 145–152. [Google Scholar] [CrossRef]
- Dilcher, M.; Veith, B.; Chidambaram, S.; Hartmann, E.; Schmitt, H.D.; von Mollard, G.F. Use1p is a yeast SNARE protein required for retrograde traffic to the ER. EMBO J. 2003, 22, 3664–3674. [Google Scholar] [CrossRef]
- Kádková, A.; Radecke, J.; Sørensen, J.B. The SNAP-25 protein family. Neuroscience 2019, 420, 50–71. [Google Scholar] [CrossRef]
- Li, L.; Xu, N.; Chen, F. Inactivation of mrpigH gene in Monascus ruber M7 results in increased Monascus pigments and decreased citrinin with mrpyrG selection marker. J. Fungi 2021, 7, 1094. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Huang, Y.; Shao, Y. From traditional application to genetic mechanism: Opinions on Monascus research in the new milestone. Front. Microbiol. 2021, 12, 659907. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Li, S. Compartmentalized biosynthesis of fungal natural products. Curr. Opin. Biotechnol. 2021, 69, 128–135. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Zhou, G.; Ewald, J.; Chang, L.; Hacariz, O.; Basu, N.; Xia, J. Using MetaboAnalyst 5.0 for LC–HRMS spectra processing, multi-omics integration and covariate adjustment of global metabolomics data. Nat. Protoc. 2022, 17, 1735–1761. [Google Scholar] [CrossRef]
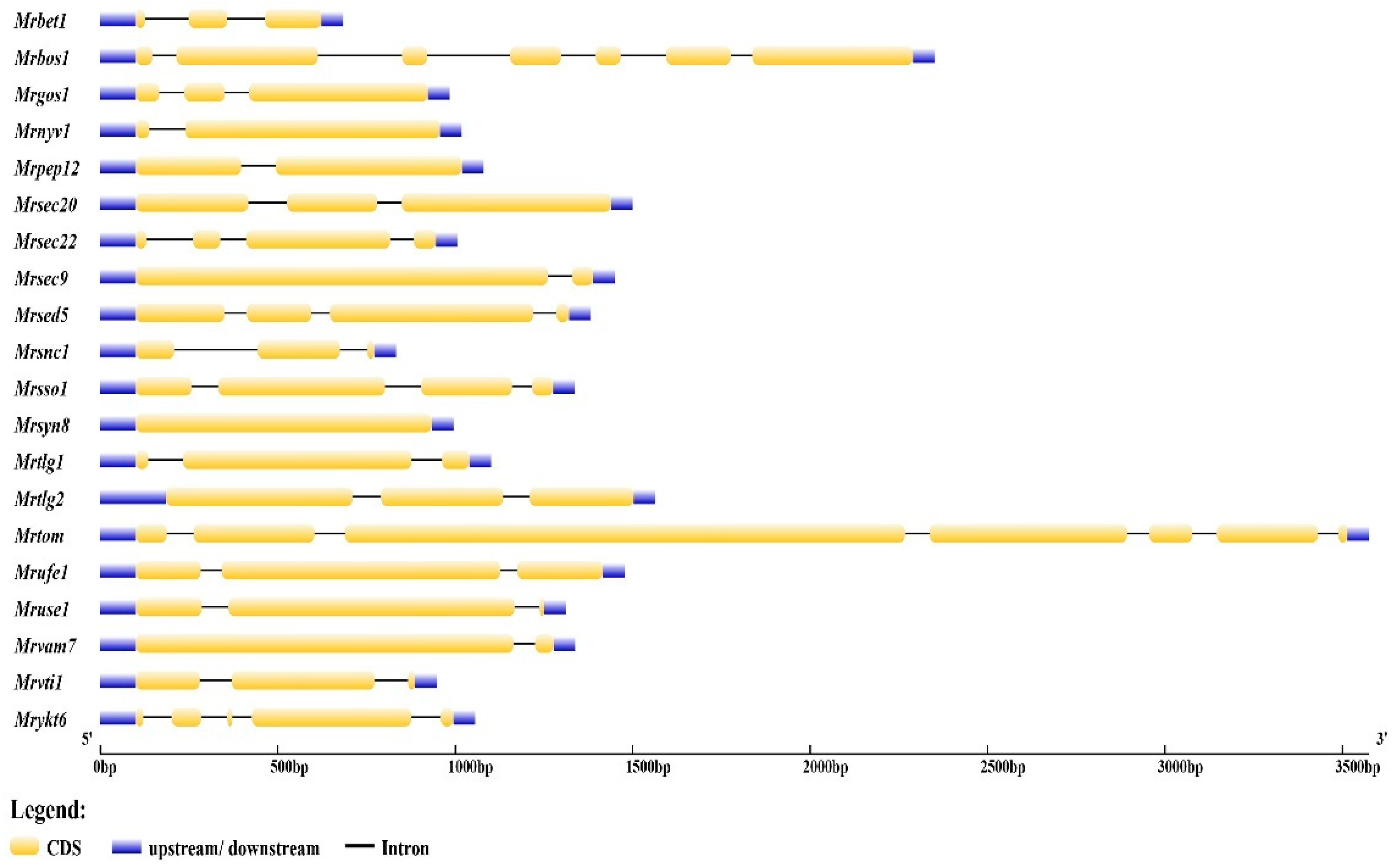
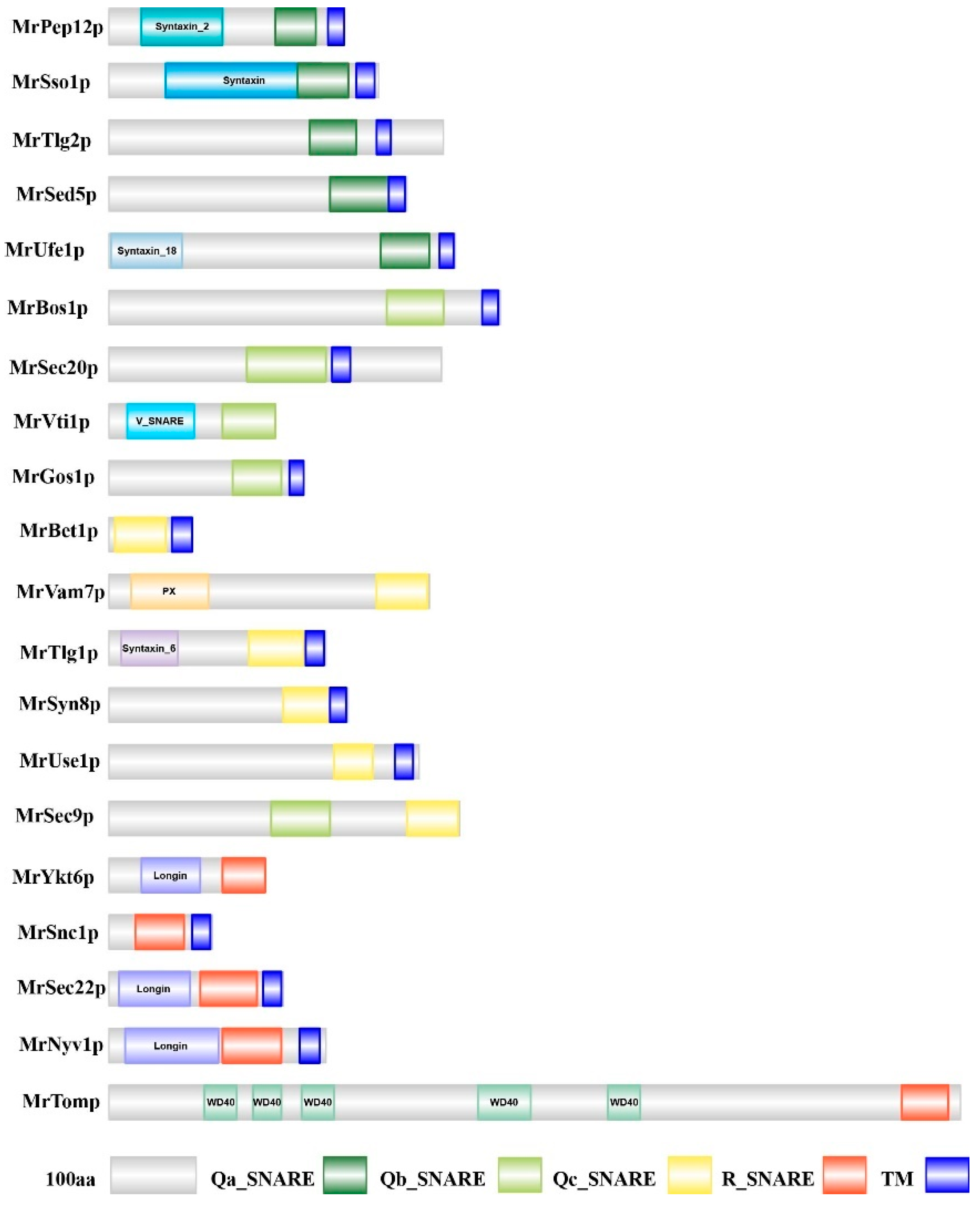

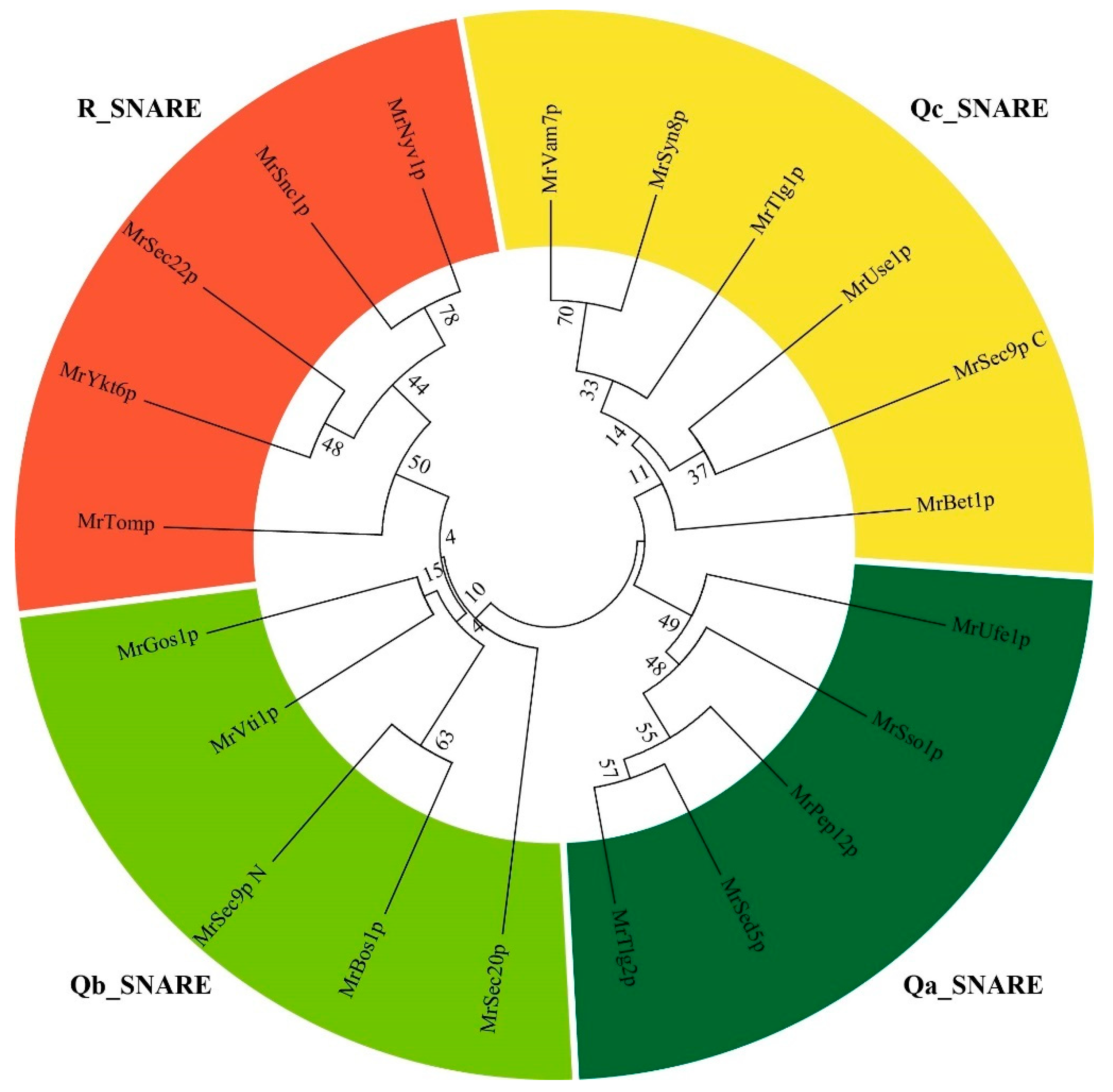

| Gene Name | GenBank | Length of CDS (bp) | Protein Length (aa) | Molecular Weight (Da) | Isoelectric Point | A. nidulans Identity | S. cerevisiae Identity |
|---|---|---|---|---|---|---|---|
| Mrpep12 | OP620680.1 | 822 | 273 | 30,719.09 | 5.02 | Pep12 (76%) | Pep12 (29%) |
| Mrsso1 | OP620682.1 | 942 | 313 | 34,792.64 | 5.18 | Sso (39%) | Sso1 (25%) |
| Mrtlg2 | OP620686.1 | 1041 | 346 | 45,169.38 | 6.75 | Tlg2 (78%) | Tlg2 (33%) |
| Mrsed5 | OP620683.1 | 1164 | 387 | 38,021.88 | 9.04 | Sed5 (88%) | Sed5 (37%) |
| Mrufe1 | OP620691.1 | 1209 | 402 | 44,159.32 | 7.21 | Ufe1 (49%) | Ufe1 (34%) |
| Mrbos1 | OP620676.1 | 1365 | 454 | 51,257.33 | 9.83 | - | Bos1 (29%) |
| Mrsec20 | OP620689.1 | 1161 | 386 | 43,724.39 | 5.52 | Sec20 (58%) | Sec20 (26%) |
| Mrvti1 | OP620690.1 | 603 | 200 | 22,885.51 | 5.63 | Vti1 (79%) | Vti1 (41%) |
| Mrgos1 | OP620693.1 | 684 | 227 | 25,819.33 | 9.75 | Gos1 (86%) | Gos1 (35%) |
| Mrbet1 | OP620679.1 | 294 | 97 | 10,397.74 | 5.16 | Bet1 (29%) | Bet1 (22%) |
| Mrvam7 | OP620681.1 | 1116 | 371 | 41,239.48 | 9.42 | Vam7 (66%) | Vam7 (26%) |
| Mrtlg1 | OP620685.1 | 756 | 251 | 27,983.68 | 4.46 | Tlg1 (70%) | Tlg1 (27%) |
| Mrsyn8 | OP620684.1 | 834 | 277 | 30,575.95 | 5.04 | Syn8 (65%) | Syn8 (24%) |
| Mruse1 | OP620687.1 | 1080 | 359 | 40,026.02 | 5.14 | Use1 (58%) | - |
| Mrsec9 | OP620692.1 | 1221 | 406 | 44,644.99 | 6.71 | Sec9 (63%) | Sec9 (40%) |
| Mrykt6 | OP620694.1 | 603 | 200 | 22,882.07 | 6.83 | Ykt6 (57%) | Ykt6 (53%) |
| Mrsnc1 | OP620677.1 | 360 | 119 | 13,012.98 | 9.39 | Snc (71%) | Snc1 (59%) |
| Mrsec22 | OP620678.1 | 576 | 171 | 23,645.99 | 8.35 | Sec22 (79%) | Sec22 (49%) |
| Mrnyv1 | OP620695.1 | 756 | 223 | 27,838.05 | 9.21 | Nyv1 (82%) | Nyv1 (39%) |
| Mrtom | OP620688.1 | 2994 | 988 | 107,978.53 | 6.98 | Tomosyn (67%) | Sro7 (30%) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meng, C.; Zhou, Y.; Liu, J.; Chen, F. Identification and Transcriptional Profiling of SNARE Family in Monascus ruber M7 Reveal Likely Roles in Secondary Metabolism. Fermentation 2022, 8, 750. https://doi.org/10.3390/fermentation8120750
Meng C, Zhou Y, Liu J, Chen F. Identification and Transcriptional Profiling of SNARE Family in Monascus ruber M7 Reveal Likely Roles in Secondary Metabolism. Fermentation. 2022; 8(12):750. https://doi.org/10.3390/fermentation8120750
Chicago/Turabian StyleMeng, Chenchen, Youxiang Zhou, Jiao Liu, and Fusheng Chen. 2022. "Identification and Transcriptional Profiling of SNARE Family in Monascus ruber M7 Reveal Likely Roles in Secondary Metabolism" Fermentation 8, no. 12: 750. https://doi.org/10.3390/fermentation8120750
APA StyleMeng, C., Zhou, Y., Liu, J., & Chen, F. (2022). Identification and Transcriptional Profiling of SNARE Family in Monascus ruber M7 Reveal Likely Roles in Secondary Metabolism. Fermentation, 8(12), 750. https://doi.org/10.3390/fermentation8120750







