Uncovering the Effects of Ammonium Sulfate on Neomycin B Biosynthesis in Streptomyces fradiae SF-2
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Plasmids, and Growth Conditions
2.2. Comparative Transcriptomic Sequencing
2.3. RT-qPCR Analysis
2.4. Optimizing the Conjugation between E. coli and S. fradiae SF-2
2.5. Detection of Neomycin and Residual Sugar
3. Results and Discussion
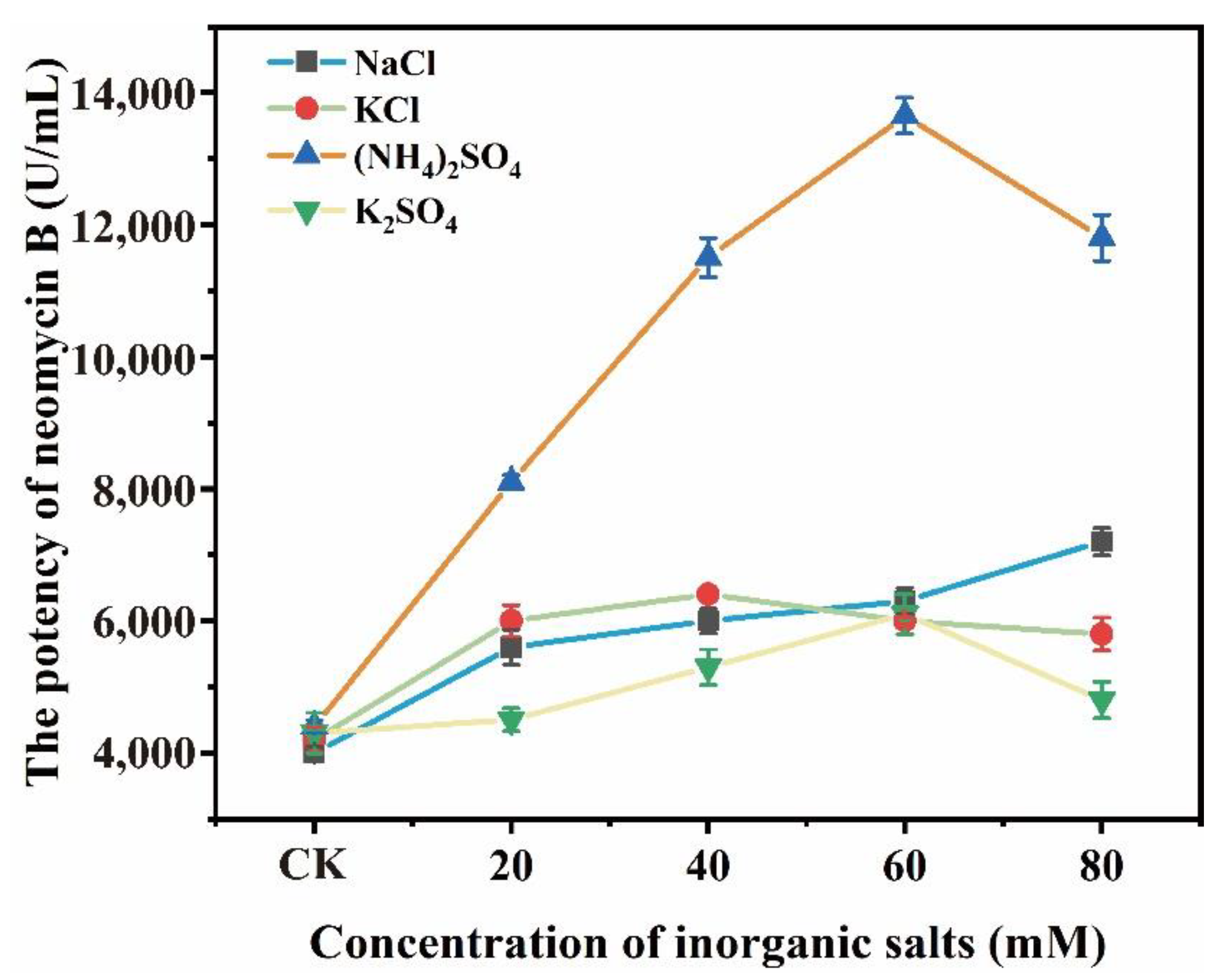
3.1. (NH4)2SO4 Promoted the Biosynthesis of Neomycin B
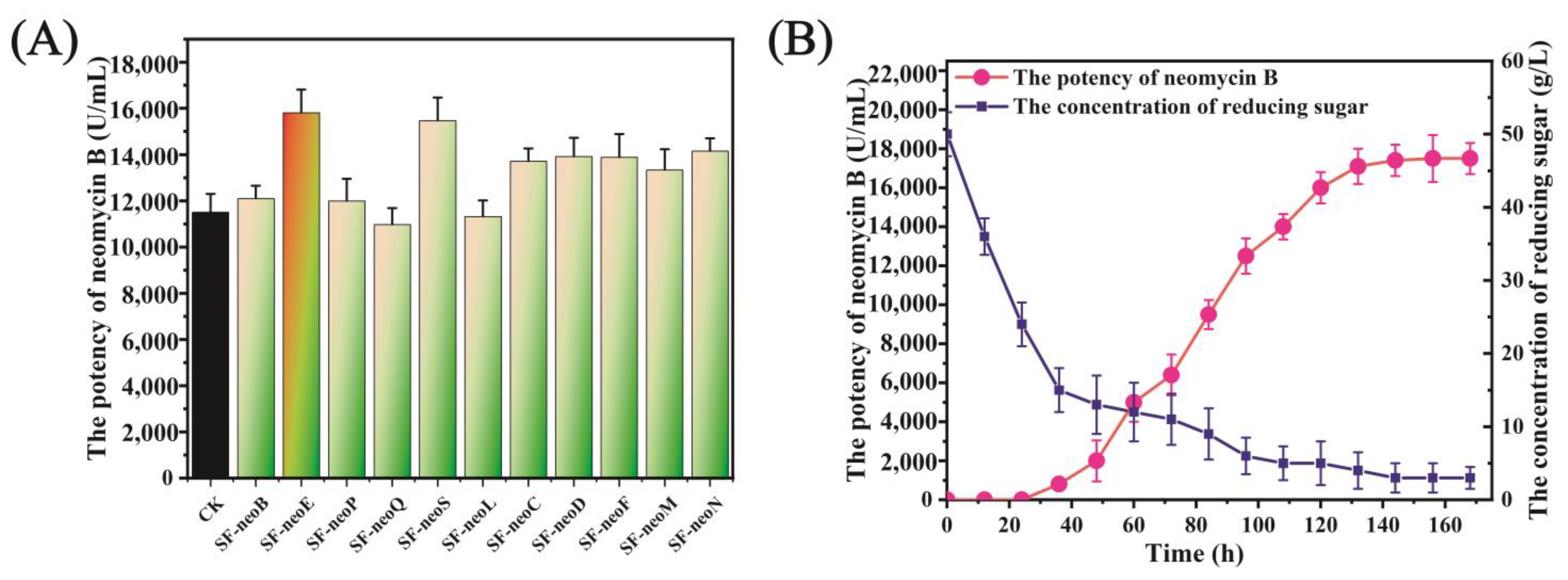
3.2. (NH4)2SO4 Enhanced Cell Growth and Utilization of Reducing Sugar
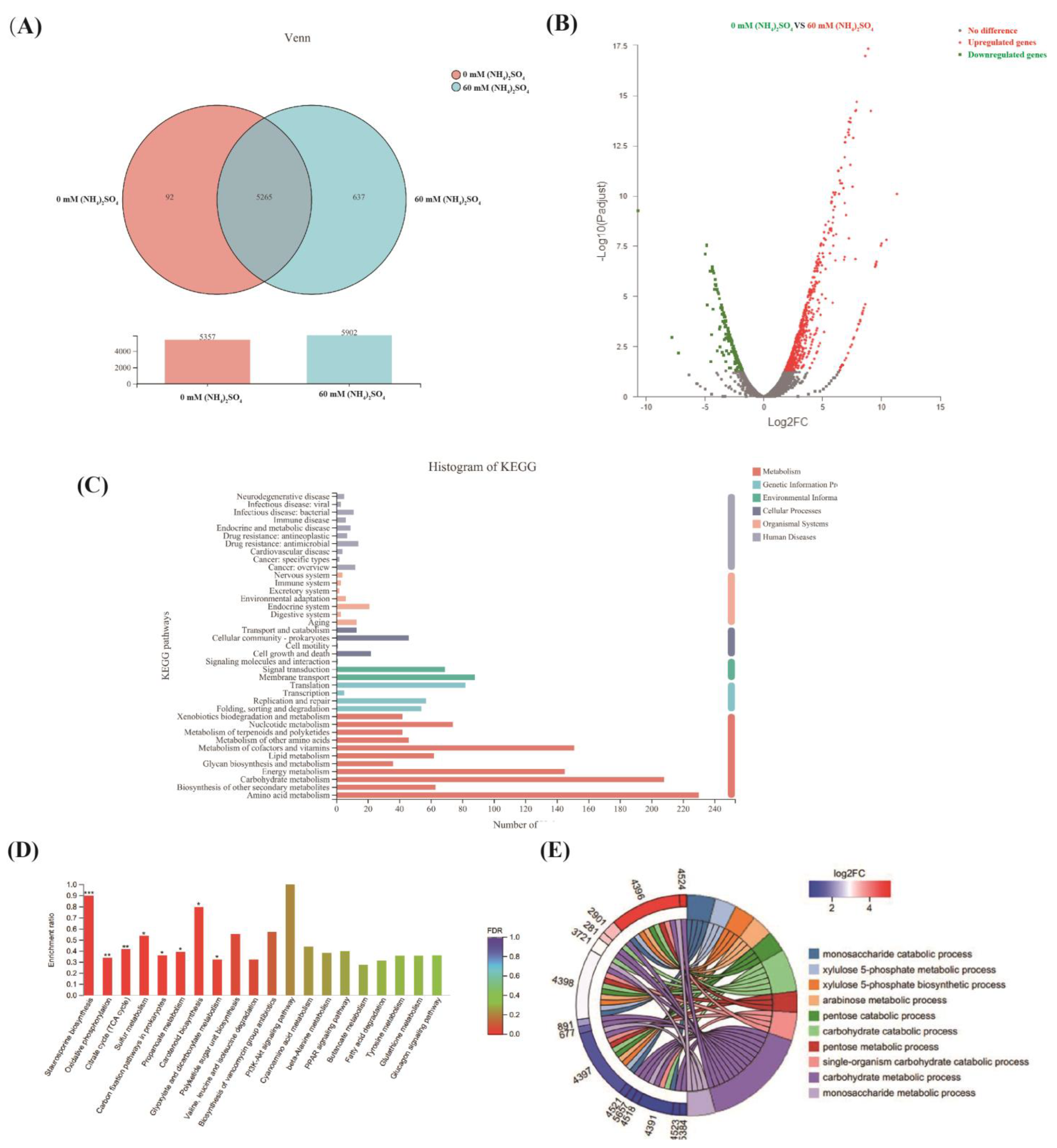
3.3. Comparative Transcriptomics Revealed the Mechanisms Underlying the Effect of (NH4)2SO4 on Neomycin B Biosynthesis
3.4. neoE Overexpression Improved the Biosynthesis of Neomycin B
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Zheng, J.; Li, Y.; Guan, H.; Li, J.; Li, D.; Zhang, J.; Tan, H. Component optimization of neomycin biosynthesis via the reconstitution of a combinatorial mini-gene-cluster in Streptomyces fradiae. ACS Synth. Biol. 2020, 9, 2493–2501. [Google Scholar] [CrossRef]
- Wright, F.; Bibb, M.J. Codon usage in the G+C-rich Streptomyces genome. Gene 1992, 113, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Bekker, O.B.; Klimina, K.M.; Vatlin, A.A.; Zakharevich, N.V.; Kasianov, A.S.; Danilenko, V.N. Draft genome sequence of Streptomyces fradiae ATCC 19609, a strain highly sensitive to antibiotics. Genome Announc. 2014, 2, e01247-14. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Ji, L.; Tang, W.; Guo, L.; Gao, C.; Chen, X.; Liu, J.; Hu, G.; Liu, L. Metabolic engineering of Streptomyces to enhance the synthesis of valuable natural products. Eng. Microbiol. 2022, 2, 100022. [Google Scholar] [CrossRef]
- Champney, W.S. Antibiotics targeting bacterial ribosomal subunit biogenesis. J. Antimicrob. Chemother. 2020, 75, 787–806. [Google Scholar] [CrossRef]
- Foster, C.; Champney, W.S. Characterization of a 30S ribosomal subunit assembly intermediate found in Escherichia coli cells growing with neomycin or paromomycin. Arch. Microbiol. 2008, 189, 441–449. [Google Scholar] [CrossRef] [PubMed]
- Mehta, R.; Champney, W.S. Neomycin and paromomycin inhibit 30S ribosomal subunit assembly in Staphylococcus aureus. Curr. Microbiol. 2003, 47, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Xiao, C.; Liu, J.; Yang, A.; Zhao, H.; He, Y.; Li, X.; Yuan, Z. Colorimetric determination of neomycin using melamine modified gold nanoparticles. Microchim. Acta 2015, 182, 1501–1507. [Google Scholar] [CrossRef]
- Swiatkowska, A.; Dutkiewicz, M.; Machtel, P.; Janecki, D.M.; Kabacinska, M.; żydowicz-Machtel, P.; Ciesiołka, J. Regulation of the p53 expression profile by hnRNP K under stress conditions. RNA Biol. 2020, 17, 1402–1415. [Google Scholar] [CrossRef]
- Jaiswal, S.K.; Oh, J.J.; Depamphilis, M.L. Cell cycle arrest and apoptosis are not dependent on p53 prior to p53-dependent embryonic stem cell differentiation. Stem Cells 2020, 9, 1091–1106. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, M.Z.; Zia, Q.; Haque, A.; Alqahtani, A.S.; Almarfadi, O.M.; Banawas, S.; Alqahtani, M.S.; Ameta, K.L.; Haque, S. Aminoglycosides as potential inhibitors of SARS-CoV-2 main protease: An in silico drug repurposing study on FDA-approved antiviral and anti-infection agents. J. Infect. Public Health 2021, 14, 611–619. [Google Scholar] [CrossRef] [PubMed]
- Vastrad, B.M.; Neelagund, S.E. Optimization of medium composition for the production of neomycin by Streptomyces fradiae NCIM 2418 in solid state fermentation. Biotechnol. Res. Int. 2014, 2014, 674286. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Rho, Y.T. Improvement of tylosin fermentation by mutation and medium optimization. Lett. Appl. Microbiol. 1998, 28, 142–144. [Google Scholar] [CrossRef]
- Yang, X.; Yang, Y.; Huang, J.; Man, D.; Guo, M. Comparisons of urea or ammonium on growth and fermentative metabolism of Saccharomyces cerevisiae in ethanol fermentation. World J. Microbiol. Biotechnol. 2021, 37, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Shah, N.N.; Taylor, R.T.; Droege, M.W. Batch cultivation of Methylosinus trichosporium OB3b: II. Production of particulate methane monooxygenase. Biotechnol. Bioeng. 1992, 40, 151–157. [Google Scholar] [CrossRef] [PubMed]
- Commichau, F.M.; Forchhammer, K.; Stulke, J. Regulatory links between carbon and nitrogen metabolism. Curr. Opin. Microbiol. 2006, 9, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Majumdar, M.K.; Majumdar, S.K. Utilization of carbon and nitrogen-containing compounds for neomycin production by Streptomyces fradiae. Appl. Microbiol. 1967, 15, 744–749. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Wang, W.; Xie, Z.; Li, P.; Li, Y.; Guo, Z.; Lu, Y.; Yang, J.; Guan, K.; Lu, Z.; et al. Neomycin biosynthesis is regulated positively by AfsA-g and NeoR in Streptomyces fradiae CGMCC 4.7387. Sci. China Life Sci. 2017, 60, 980–991. [Google Scholar] [CrossRef]
- Kudo, F.; Eguchi, T. Aminoglycoside antibiotics: New insights into the biosynthetic machinery of old drugs. Chem. Rec. 2016, 16, 4–18. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Chen, D.; Wu, J.; Chu, X.; Yang, Y.; Fang, L.; Zhang, W. Comparative transcriptome analysis demonstrates the positive effect of the cyclic AMP receptor protein Crp on daptomycin biosynthesis in Streptomyces roseosporus. Front. Bioeng. Biotechnol. 2021, 9, 401. [Google Scholar] [CrossRef] [PubMed]
- Pinilla, L.; Toro, L.F.; Laing, E.; Alzate, J.F.; Ríos-Estepa, R. Comparative transcriptome analysis of Streptomyces clavuligerus in response to favorable and restrictive nutritional conditions. Antibiotics 2019, 8, 96. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Huang, Z.; Gui, X.; Xiang, W.; Jin, Y.; Chen, J.; Zhao, J. Multi-omics comparative analysis of Streptomyces mutants obtained by iterative atmosphere and room-temperature plasma mutagenesis. Front. Microbiol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Bao, T.; Osire, T.; Qiao, Z.; Liu, J.; Zhang, X.; Xu, M.; Yang, T.; Rao, Z. MarR-type transcription factor RosR regulates glutamate metabolism network and promotes accumulation of L-glutamate in Corynebacterium glutamicum g01. Bioresour. Technol. 2021, 342, 125945. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.; Zhang, M.; Sun, J.; Wang, F.; Li, X.; Liu, Y.; Wang, Z.; Zhao, X.; Li, J.; Chen, J.; et al. Improved neomycin sulfate potency in streptomyces fradiae using atmospheric and room temperature plasma (ARTP) mutagenesis and fermentation medium optimization. Microorganisms 2022, 10, 94. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Li, Y.; Yu, J.; Spencer, J.B. Biosynthesis of aminoglycoside antibiotics: Cloning, expression and characterisation of an aminotransferase involved in the pathway to 2-deoxystreptamine. Chem. Commun. 2002, 23, 2860–2861. [Google Scholar] [CrossRef] [PubMed]
- Dam, B.; Dam, S.; Kim, Y.; Liesack, W. Ammonium induces differential expression of methane and nitrogen metabolism-related genes in Methylocystis sp. Strain SC2. Environ. Microbiol. 2014, 16, 3115–3127. [Google Scholar] [CrossRef] [PubMed]
- Cueto-Rojas, H.F.; Maleki, S.R.; Ten, P.A.; van Helmond, W.; Pieterse, M.M.; Heijnen, J.J.; Wahl, S.A. In vivo analysis of NH4+ transport and central nitrogen metabolism in Saccharomyces cerevisiae during aerobic nitrogen-limited growth. Appl. Environ. Microbiol. 2016, 82, 6831–6845. [Google Scholar] [CrossRef] [PubMed]
- Stitt, M. Nitrate regulation of metabolism and growth. Curr. Opin. Plant Biol. 1999, 2, 178–186. [Google Scholar] [CrossRef] [PubMed]
- Technikova-Dobrova, Z.; Damiano, F.; Tredici, S.M.; Vigliotta, G.; Di Summa, R.; Palese, L.; Abbrescia, A.; Labonia, N.; Gnoni, G.V.; Alifano, P. Design of mineral medium for growth of Actinomadura sp. ATCC 39727, producer of the glycopeptide A40926: Effects of calcium ions and nitrogen sources. Appl. Microbiol. Biot. 2004, 6, 671–677. [Google Scholar] [CrossRef]
- Gonzalez, R.; Islas, L.; Obregon, A.M.; Escalante, L.; Sanchez, S. Gentamicin formation in Micromonospora purpurea: Stimulatory effect of ammonium. J. Antibiot. 1995, 48, 479–483. [Google Scholar] [CrossRef]
- Zhu, C.H.; Lu, F.P.; He, Y.N.; Han, Z.L.; Du, L.X. Regulation of avilamycin biosynthesis in Streptomyces viridochromogenes: Effects of glucose, ammonium ion, and inorganic phosphate. Appl. Microbiol. Biotechnol. 2007, 73, 1031–1038. [Google Scholar] [CrossRef] [PubMed]
- Hong, M.; Mou, H.; Liu, X.; Huang, M.; Chu, J. 13C-assisted metabolomics analysis reveals the positive correlation between specific erythromycin production rate and intracellular propionyl-CoA pool size in Saccharopolyspora erythraea. Bioprocess Biosyst. Eng. 2017, 40, 1337–1348. [Google Scholar] [CrossRef] [PubMed]


| Strains and Plasmids | Description | Source |
|---|---|---|
| Strains | ||
| E. coli DH5α | General cloning host | The lab |
| E. coli ET12567 | Demethylated strain containing pUZ8002 plasmid for conjugative transfer with actinomycetes | The lab |
| S. fradiae SF-2 | Streptomyces fradiae, neomycin B-producing strains generated by ARTP | [24] |
| ET12567/PPR-NeoB | E. coli ET12567 derivative harboring PPR-NeoB | This study |
| ET12567/PPR-NeoE | E. coli ET12567 derivative harboring PPR-NeoE | This study |
| ET12567/PPR-NeoP | E. coli ET12567 derivative harboring PPR-NeoP | This study |
| ET12567/PPR-NeoQ | E. coli ET12567 derivative harboring PPR-NeoQ | This study |
| ET12567/PPR-NeoS | E. coli ET12567 derivative harboring PPR-NeoS | This study |
| ET12567/PPR-NeoL | E. coli ET12567 derivative harboring PPR-NeoL | This study |
| ET12567/PPR-NeoC | E. coli ET12567 derivative harboring PPR-NeoC | This study |
| ET12567/PPR-NeoD | E. coli ET12567 derivative harboring PPR-NeoD | This study |
| ET12567/PPR-NeoF | E. coli ET12567 derivative harboring PPR-NeoF | This study |
| ET12567/PPR-NeoM | E. coli ET12567 derivative harboring PPR-NeoM | This study |
| ET12567/PPR-NeoN | E. coli ET12567 derivative harboring PPR-NeoN | This study |
| SF-NeoB | S. fradiae SF-2 derivative the expression of NeoB | This study |
| SF-NeoE | S. fradiae SF-2 derivative the expression of NeoE | This study |
| SF-NeoP | S. fradiae SF-2 derivative the expression of NeoP | This study |
| SF-NeoQ | S. fradiae SF-2 derivative the expression of NeoQ | This study |
| SF-NeoS | S. Fradiae SF-2 derivative the expression of NeoS | This study |
| SF-NeoL | S. fradiae SF-2 derivative the expression of NeoL | This study |
| SF-NeoC | S. fradiae SF-2 derivative the expression of NeoC | This study |
| SF-NeoD | S. fradiae SF-2 derivative the expression of NeoD | This study |
| SF-NeoF | S. fradiae SF-2 derivative the expression of NeoF | This study |
| SF-NeoM | S. fradiae SF-2 derivative the expression of NeoM | This study |
| SF-NeoN | S. fradiae SF-2 derivative the expression of NeoN | This study |
| Plasmids | ||
| PPR (pSET152-PermE*) | E. coli-S. fradiae shuttle vector for the expression of target protein | The lab |
| PPR-NeoB | Derived from PPR, for the expression of NeoB | This study |
| PPR-NeoE | Derived from PPR, for the expression of NeoE | This study |
| PPR-NeoP | Derived from PPR, for the expression of NeoP | This study |
| PPR-NeoQ | Derived from PPR, for the expression of NeoQ | This study |
| PPR-NeoS | Derived from PPR, for the expression of NeoS | This study |
| PPR-NeoL | Derived from PPR, for the expression of NeoL | This study |
| PPR-NeoC | Derived from PPR, for the expression of NeoC | This study |
| PPR-NeoD | Derived from PPR, for the expression of NeoD | This study |
| PPR-NeoF | Derived from PPR, for the expression of NeoF | This study |
| PPR-NeoM | Derived from PPR, for the expression of NeoM | This study |
| PPR-NeoN | Derived from PPR, for the expression of NeoN | This study |
| Primer Name | Sequences | Digest Sites |
|---|---|---|
| NeoB F | ATAGCGGCCGCGATGACGAAAAACTCTTCCCTGC | NotI |
| NeoB R | CGAGATATCTCAGTCGTCCAGCAGCCG | EcoRV |
| NeoE F | ATAGCGGCCGCGATGAAGGCTCTGGTGTTCGAGG | NotI |
| NeoE R | CGAGATATCTCAGGCCCGGAGGTTGAAGTA | EcoRV |
| NeoP F | ATAGCGGCCGCGATGACGGCCGCCCAGC | NotI |
| NeoP R | CGAGATATCTCATGCCGTCCTGGCCAG | EcoRV |
| NeoQ F | ATAGCGGCCGCGATGAAGCGCCTTCGAGGCAC | NotI |
| NeoQ R | CGAGATATCTCAGACGTGCGCGGTGTGC | EcoRV |
| NeoS F | ATAGCGGCCGCGATGGTCTCCCCGTTGGCA | NotI |
| NeoS R | CGAGATATCTCAAGTGGCCAGGTCGGC | EcoRV |
| NeoL F | ATAGCGGCCGCGGTGGTGACGACCGGCGTGGC | NotI |
| NeoL R | CGAGATATCTCAGGCCAGTGCGGCGAC | EcoRV |
| NeoC F | ATAGCGGCCGCGATGCAGACCACCCGCAT | NotI |
| NeoC R | CGAGATATCTTACGGCACGGGTCCGGC | EcoRV |
| NeoD F | ATAGCGGCCGCGGTGGGTGAGCCGACGTGG | NotI |
| NeoD R | CGAGATATCTCACCGGGCACCCGCCG | EcoRV |
| NeoF F | ATAGCGGCCGCGGTGGCTGAGGCGCCTGC | NotI |
| NeoF R | CGAGATATCTCACCCACCGTGCTCCTCC | EcoRV |
| NeoM F | ATAGCGGCCGCGGTGCTGCGGCTCACCC | NotI |
| NeoM R | CGAGATATCTCACGGCGCCCACCCG | EcoRV |
| NeoN F | ATAGCGGCCGCGATGACCACCGAC | NotI |
| NeoN R | CGAGATATCTCATACGAGCG | EcoRV |
| RT-NeoE F | TGACGGCCACCTTCTCGC | |
| RT-NeoE R | ACCTGCTCCTGCGGCACCT | |
| RT-NeoS F | TAGGTGTAGTACGTACGGG | |
| RT-NeoS R | ATGGGCAGCAACCGCTGCCT | |
| RT-NeoC F | GTTCTTGACGAGCGCGGT | |
| RT-NeoC R | GGACACGCCATCGAGCACG | |
| RT-NeoD F | CTCGGTCTCGTCGTCGTA | |
| RT-NeoD R | ACGCGACGCTCCTGACGGTGT | |
| RT-NeoM F | TGGACGTGCACCAGGT | |
| RT-NeoM R | AGCAGCTCGTCATGACCGT | |
| RT-NeoN F | TGGTAGTTGTAGCCGTTGGT | |
| RT-NeoN R | ACTGCTCCACTTCATGCCGC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Yu, F.; Liu, K.; Zhang, M.; Cheng, Y.; Wang, F.; Wang, S.; Han, R.; Xue, Z. Uncovering the Effects of Ammonium Sulfate on Neomycin B Biosynthesis in Streptomyces fradiae SF-2. Fermentation 2022, 8, 678. https://doi.org/10.3390/fermentation8120678
Li X, Yu F, Liu K, Zhang M, Cheng Y, Wang F, Wang S, Han R, Xue Z. Uncovering the Effects of Ammonium Sulfate on Neomycin B Biosynthesis in Streptomyces fradiae SF-2. Fermentation. 2022; 8(12):678. https://doi.org/10.3390/fermentation8120678
Chicago/Turabian StyleLi, Xiangfei, Fei Yu, Kun Liu, Min Zhang, Yihan Cheng, Fang Wang, Shan Wang, Rumeng Han, and Zhenglian Xue. 2022. "Uncovering the Effects of Ammonium Sulfate on Neomycin B Biosynthesis in Streptomyces fradiae SF-2" Fermentation 8, no. 12: 678. https://doi.org/10.3390/fermentation8120678
APA StyleLi, X., Yu, F., Liu, K., Zhang, M., Cheng, Y., Wang, F., Wang, S., Han, R., & Xue, Z. (2022). Uncovering the Effects of Ammonium Sulfate on Neomycin B Biosynthesis in Streptomyces fradiae SF-2. Fermentation, 8(12), 678. https://doi.org/10.3390/fermentation8120678






